Abstract
Functional RNA structures play an important role both in the context of noncoding RNA transcripts as well as regulatory elements in mRNAs. Here we present a computational study to detect functional RNA structures within the ENCODE regions of the human genome. Since structural RNAs in general lack characteristic signals in primary sequence, comparative approaches evaluating evolutionary conservation of structures are most promising. We have used three recently introduced programs based on either phylogenetic–stochastic context-free grammar (EvoFold) or energy directed folding (RNAz and AlifoldZ), yielding several thousand candidate structures (corresponding to ∼2.7% of the ENCODE regions). EvoFold has its highest sensitivity in highly conserved and relatively AU-rich regions, while RNAz favors slightly GC-rich regions, resulting in a relatively small overlap between methods. Comparison with the GENCODE annotation points to functional RNAs in all genomic contexts, with a slightly increased density in 3′-UTRs. While we estimate a significant false discovery rate of ∼50%–70% many of the predictions can be further substantiated by additional criteria: 248 loci are predicted by both RNAz and EvoFold, and an additional 239 RNAz or EvoFold predictions are supported by the (more stringent) AlifoldZ algorithm. Five hundred seventy RNAz structure predictions fall into regions that show signs of selection pressure also on the sequence level (i.e., conserved elements). More than 700 predictions overlap with noncoding transcripts detected by oligonucleotide tiling arrays. One hundred seventy-five selected candidates were tested by RT-PCR in six tissues, and expression could be verified in 43 cases (24.6%).
The goal of The ENCODE Project Consortium (Encyclopedia of DNA Elements [ENCODE]) is the comprehensive analysis of functional elements in the human genome. One of its main goals is the thorough annotation of transcripts in terms of structure and function. Both genome-wide studies (Bertone et al. 2004; Carninci et al. 2005; Cheng et al. 2005) and the far more detailed studies targeted to the ENCODE regions (The ENCODE Project Consortium 2007) show a much more extensive and complex transcriptional map than previously anticipated, comprising a mosaic of overlapping transcription, antisense transcripts, abundant alternative splicing, and a plethora of novel transcribed elements. Using a series of sensitive methods, it was demonstrated that 93% of the ENCODE regions exist in primary nuclear transcripts in at least one of the tested tissues.
An as-yet not satisfactorily resolved question is whether novel transcripts lacking protein-coding capacity (noncoding transcripts) have biological function as such, or whether they rather represent “biological noise” (i.e., selectively neutral transcription). Analogous to the analysis of protein-coding genes, a combination of both experimental and computational techniques seems necessary to address this question.
On the experimental side, we can draw upon the evidence from large-scale oligonucleotide tiling array studies performed on the ENCODE regions as well as a small set of verification experiments (The ENCODE Project Consortium 2007). Unfortunately, there is at present no general way to predict noncoding transcripts in eukaryotic genomes. A few methods exploit weak statistical signals like mutational strand bias, strand-specific selection against polyadenylation signals, or exclusion of repeat elements to predict transcribed regions in the genome (Semon and Duret 2004; Glusman et al. 2006). Such approaches are limited to very large transcripts and cannot define functional elements within a transcript, for example, as do protein gene finders by predicting coding exons. A subclass of noncoding transcripts, however, appears to function in the context of ribonucleoprotein complexes that require specific RNA secondary structures. This is the case, in particular, for many of the “classical” noncoding RNAs (ncRNAs), including snoRNAs, snRNAs, or the signal recognition particle RNA. Other sources of structural constraints may derive from particular processing pathways, such as the hairpin-shaped precursors of microRNAs, specific steric requirements as in the case of tRNAs, or from structural requirements for the catalytic function of the RNA itself, as in the case of rRNAs, RNaseP RNA, and group I and II introns (Bompfünewerer et al. 2005).
RNA secondary structures are known to play an important functional role not only in noncoding transcripts, but also in the context of protein-coding mRNAs. Structural motifs serve regulatory functions in untranslated regions (Mignone et al. 2002), lead to genetic reprogramming of coding regions (Hubert et al. 1996; Namy et al. 2004), and can influence splicing of pre-mRNAs (Buratti and Baralle 2004).
The comprehensive knowledge of encoded secondary structures in the genome is important to determine at which level DNA is actually functional, and without it, an “encyclopedia” of functional elements would be incomplete.
In this study, we use different comparative approaches to predict functional RNA secondary structures and provide a detailed comparison with the results of other ENCODE subprojects; in particular, experimental data from oligonucleotide tiling array studies. The computational approach is based on predicting consensus structures and the observation that structural constraints imply specific mutational patterns visible at the sequence level. EvoFold (Pedersen et al. 2006) analyzes substitution patterns and models RNA structures directly in the framework of a phylogenetic–stochastic context-free grammar (phylo-SCFG) (Knudsen and Hein 1999, 2003), while RNAz (Washietl et al. 2005b) and AlifoldZ (Washietl and Hofacker 2004) consider structural conservation and stability of the putative structures in terms of predicted folding energies (Hofacker et al. 2002). Both EvoFold and RNAz have been used in genome-wide computational screens for structured RNAs (Washietl et al. 2005a; Pedersen et al. 2006), limited, however, on a preselected set of sequence-constrained elements (Siepel et al. 2005), and also based on a much smaller number of genomes. In the ENCODE regions, we not only have access to alignments of up to 28 species, which greatly enhances the power of such comparative approaches, but more importantly, there is also a dense set of additional data to which we can compare our data.
Results
Three approaches
Almost all RNA molecules form secondary structures. The challenge is thus to recognize those sections of the genome in which the structure is more conserved than one would expect from primary sequence conservation alone. We employ here three fairly different methods that are designed to recognize evolutionarily conserved secondary structures. All three are based on given multiple-sequence alignments and attempt to (1) predict a consensus secondary structure for aligned sequences, and then (2) apply a test of whether or not the consensus structure found is unusual.
Consensus structures can be inferred either by means of energy-directed folding or using a phylo-SCFG model. The RNAalifold algorithm computes the most stable secondary structure that is compatible with the input alignment (Hofacker et al. 2002). Pfold uses a phylo-SCFG to predict the most likely common secondary structure based on a model of secondary structure formation combined with a phylogenetic analysis of the observed substitution pattern (Knudsen and Hein 1999, 2003). Both approaches yield comparable accuracies for consensus secondary structure prediction (Gardner and Giegerich 2004). Recently, these algorithms have been used for ncRNA prediction by augmenting them with significance measures.
AlifoldZ uses a random shuffle approach to estimate the expected background distribution (Washietl and Hofacker 2004). It expresses the significance of a hit in terms of a normalized Z-score. Negative Z-scores indicate that an observed fold is more stable and conserved than expected by chance. AlifoldZ is relatively slow and nondeterministic, and fairly sensitive to alignment errors since it depends on a strictly conserved fold.
These limitations are overcome by RNAz (Washietl et al. 2005b), which uses a different approach to evaluate the RNAalifold prediction. Structure conservation is measured here directly as the ratio of the unconstrained folding energies relative to the folding energies under the constraint that all aligned sequences are forced to fold into a common structure. If no common structure can be found, this results in a low conservation score. Thermodynamic stability is measured independently for each sequence and then averaged over the alignment. Both measures are interpreted by a support vector machine (SVM) classification algorithm. Since the thermodynamic component is completely independent of the alignment, this method is relatively robust against alignment errors. In its current implementation, it is, however, limited to six sequences.
EvoFold is based on two competing phylo-SCFG models of RNA sequence evolution: a structural model, similar to the Pfold model, and a nonstructural model (Pedersen et al. 2006).15 Structure is only predicted when a segment of the alignment is better described by the structural model than the nonstructural model. The two models describe alignments with identical properties, except that the nonstructural model assumes a higher substitution rate and does not include correlated base-pair changes, as found in RNA helices. Each structure prediction is assigned a score based on the relative likelihood of the alignment under the combined structural/nonstructural model and a purely nonstructural model. For the purpose of this study, the structure predictions are ranked according to their scores.
Screening multispecies alignments of the ENCODE regions
We used TBA/MultiZ (Blanchette et al. 2004) multiple sequence alignments with up to 28 species as prepared by the ENCODE alignment group (Margulies et al. 2007). The nonrepeat regions were scanned using the three algorithms as described in detail in Methods. We predict local secondary structures, performing the analysis in overlapping windows of size 120 and slide 40.
For AlifoldZ, we used a sample of a maximum of 10 sequences from the alignments. The consensus minimum free energy (MFE) quantifying the stability of the consensus fold predicted by RNAalifold of all scanned windows is shown in Figure 1. This shows that some sort of consensus fold can be found in almost all alignments. It is not possible to discriminate on the basis of this score; therefore, the Z-score is calculated to assess its significance. We only considered Z-scores for alignments with consensus MFE <−15, since Z-scores can be unstable for low levels of consensus MFE. This filter is the most stringent one and leaves us with 660 and 348 hits, respectively, for the two significance cutoffs Z < −3.5 and Z < −4, which have been used by Washietl and Hofacker (2004).
Figure 1.
Score distribution of AlifoldZ, RNAz, and EvoFold computed for all input alignments. (A) Minimum free energies of the consensus structures as computed by RNAalifold. Note that more negative scores correspond to more stable/conserved consensus structures. (B) The significance of the consensus MFEs are estimated by AlifoldZ for all consensus structures with MFE < −15 resulting in normalized Z-scores. Also here negative values mean more stable and conserved structures. The two significance cutoffs used throughout this work are indicated. (C) RNAz classifies alignments using a support vector machine. The distribution of SVM decision variables is shown as well as the two significance cutoffs, which are expressed as “classification probabilities,” P. (D) Enlarged tail of C. (E) Raw EvoFold scores on the original input alignments. (F) EvoFold scores after extracting the predicted substructure, filtering weak structures (see Methods), and rescoring. The histogram shows all predictions of which the top-scoring 50% were chosen as the high significance prediction set.
In the case of the RNAz screen, we selected up to six sequences; if there were more than 10 sequences in the alignment, we selected three different samples of six. These were classified using the SVM. The SVM score distributions can be seen in Figure 1. For convenience, the SVM scores are converted to “RNA class probabilities,” and we used two cutoffs, 0.5 and 0.9, as introduced by Washietl et al. (2005b). This results in 7093 and 3707 predictions, respectively.
All sequences of the alignments were used for EvoFold. First the regions were screened in fixed-size windows, then the predicted substructures were rescored and filtered for spurious predictions (short predictions with <10 base pairs [bp] were discarded). Based on the EvoFold score, we defined two sets: one with all predicted structures and one with the top 50% high-scoring structures, consisting of 9953 and 4986 predictions, respectively.
From the score distributions in Figure 1 and the results in Table 1, one can see that all three methods apply a relatively stringent filter on the data: On the high-significance level, RNAz and EvoFold predict 1.4% and 1.3% of the ENCODE regions to form structural RNAs, which is in both cases <5% of the scored input alignments. Note that the input varies between RNAz and EvoFold because specific schemes were used to filter the raw alignments (for details, see Methods).
Table 1.
Statistics of predictions
aAlifoldZ: Z < −3.5; RNAz: P > 0.5; EvoFold: all predictions.
bAlifoldZ: Z < −4; RNAz: P > 0.9; EvoFold: top 50% predictions.
cZ-scores corrected to compensate for the genomic background signal.
Estimating background signal
An important issue in any genome-wide screen, be it experimental or computational, is the estimation of the false discovery rate. To this end, we repeated the analysis with randomly shuffled alignments (see Methods). This procedure is designed to remove correlations arising from secondary structures while leaving other characteristics of the aligned sequences untouched. Score distributions for the randomized data are shown in Figure 1, and the results of the randomized screens are summarized in Table 1.
An important aspect in the context of randomizing RNA secondary structures is dinucleotide content (Workman and Krogh 1999). Since energy-directed folding is based on stacking interactions of neighboring base pairs, dinucleotide content can affect stability scores considerably. RNAz uses a mononucleotide shuffling model to compute the energy Z-scores, which are used as stability measures for the single sequences in the alignment. Indeed, we observe that the randomized alignments on average lead to slightly negative Z-scores rather than being centered around zero. This signal disappears when using dinucleotide shuffling. It is interesting to ask why the natural dinucleotide content of the genome results in more stable secondary structures and whether this has a biological meaning given that a large fraction of the genome is transcribed. However, conservatively, we have to consider this effect as a bias. Randomization procedures for entire alignments that respect dinucleotide content do not seem feasible; hence we cannot correct for the dinucleotide frequency effect in the case of AlifoldZ. For RNAz, however, the energy Z-score is independent of the alignment. We can compensate for the dinucleotide bias in the random control by shifting all Z-scores by the observed background Z-score of 0.5 and reevaluating the adjusted values by the SVM. EvoFold is not directly affected by dinucleotide content since the SCFG does not explicitly model stacking base pairs.
We observe a relatively high false discovery rate for both RNAz and EvoFold (Table 2). On the highly significant set, the false discovery rate (after dinucleotide correction) is 50.0% for RNAz and 70.9% for EvoFold, respectively. Since the shuffling approach comes with uncertainties (Washietl and Hofacker 2004; Washietl et al. 2005a; Pedersen et al. 2006), the real false positive rate could conceivably be even higher.
Table 2.
False discovery rates estimated on shuffled alignments
Comparison of different predictions
Figure 2 shows the overlap between different methods. Of the AlifoldZ hits, 70.9% overlap with the RNAz predictions. Since false positives are estimated to be at least 20% in AlifoldZ and false positives for RNAz and AlifoldZ arise for different reasons, this overlap is what can be expected. The 247 overlapping hits thus can be regarded as predictions with very high confidence. On the other hand, due to the very restrictive consensus MFE and Z-score cutoff used for AlifoldZ, many true RNAz hits will not yield an AlifoldZ signal.
Figure 2.
Overlap of predictions from different methods (high significance level). The sets are drawn to scale for overlap in terms of nucleotides, and numbers indicate overlapping predictions. In addition, we give the total number of items outside the respective sets. (Left) All predictions; (right) predictions without coding exons and UTRs according to GENCODE annotation.
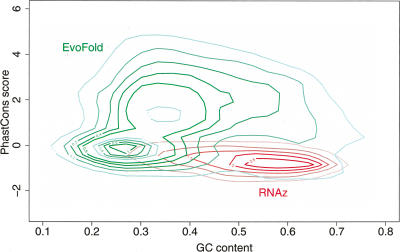
The overlap between RNAz and EvoFold is extremely low. Only 7.2% of the RNAz hits overlap with EvoFold predictions. While this constitutes a 1.6-fold enrichment over the randomly expected overlap, and although the high estimated false discovery rates limit the best possible overlap to about one-third, this small overlap was unexpected. Close inspection of the data, however, revealed the interesting fact that RNAz and EvoFold essentially detect complementary RNA structures: While RNAz is sensitive to alignments with moderate and high GC content and relatively low sequence similarity, EvoFold has its peak sensitivities for low GC content and high sequence similarity (Fig. 3). Both methods were trained on structurally diverse subsets of the Rfam database with average GC contents of ∼50%. However, the parametrization of EvoFold’s nonstructural submodel creates a bias in its structural predictions toward AT-rich regions. The human genome has an overall GC content of ∼42%. Many of the known structured RNAs, such as microRNAs and H/ACA box snoRNAs, have an average GC content close to 50%; however, some have a relatively low GC content, such as tRNAs, which have an average GC content of 34%.
Figure 3.
Densities of EvoFold and RNAz predictions (high significance level) as a function of GC content and sequence conservation measured by the phastCons program (Siepel et al. 2005). While most RNAz predictions have elevated GC content and moderate sequence conservation, EvoFold is most sensitive at low GC contents and high sequence conservation.
The second clear difference is that a large fraction of EvoFold predictions are within highly conserved alignments, while RNAz predictions essentially follow the conservation distribution found in the input regions. EvoFold, as opposed to RNAz, explicitly models the rate of substitution and was trained to detect slowly evolving RNA structures. Since many known ncRNAs are highly conserved not only in structure but also in sequence, this part of the conservation spectrum is of particular interest. However, due to the lack of sequence variation in these alignments, discriminating between true- and false-positive predictions is difficult. EvoFold is more sensitive for highly conserved alignments than RNAz, at the expense of a higher rate of false positives.
Detection of known ncRNAs
The ENCODE regions are surprisingly poor in annotated ncRNAs. Of 74 loci with sequence similarity to ncRNAs in the Rfam database (Griffiths-Jones et al. 2005), 60 are repeat masked and hence excluded from this study, and seven are annotated as “ncRNA related” (i.e., putative pseudogenes). Thus, there are only eight well-characterized ncRNAs within the ENCODE regions: three H/ACA snoRNAs; four microRNAs; and H19, an imprinted, developmentally regulated mRNA-like noncoding transcript in human and mouse that is not contained in Rfam (Gabory et al. 2006; see Table 3). There is, for example, not a single tRNA or C/D-box snoRNA in any of the ENCODE-selected regions. The eight well-characterized examples are generally detectable by all three methods with high significance (AlifoldZ, Z < −4.7; RNAz, P > 0.95; and EvoFold, top 25%). For the few examples missed, the reason is always because the ncRNA is not represented in the input alignment and simple manual editing of the alignment would have resulted in positive predictions. This shows the importance of the underlying genomic alignments.
Table 3.
Known ncRNAs in ENCODE regions
Scores for RNAs that have been missed in this screen owing to problems in the input alignments or the prescreening process are shown in parentheses.
An interesting example is H19, which shows that long spliced transcripts can have structural “domains” and that structural ncRNAs are not necessarily small RNAs with a global structure as seen for tRNAs or snoRNAs. In addition to these well-described examples, we found seven overlapping EvoFold/RNAz hits with significant sequence similarity (BLAST E < 10−6) to the set of putative ncRNAs from the mouse Fantom2 project (Okazaki et al. 2002), supporting the role of these transcripts as functional ncRNAs.
Comparison with other ENCODE data
Sites of transcription can be empirically determined using oligonucleotide tiling-array techniques resulting in maps of transcriptionally active regions (TARs) (Bertone et al. 2004) or transcribed fragments (Transfrags) (Cheng et al. 2005). We compared predicted RNA structures with a union of TARs/Transfrags generated in the course of the ENCODE project using 11 human tissues (Fig. 4; The ENCODE Project Consortium 2007). One has to keep in mind that these maps were derived from RNA fractions longer than 200 nucleotides (nt), and therefore a large fraction of small structured ncRNAs should be missed. However, many ncRNAs like miRNAs and snoRNAs are processed from longer precursor transcripts and are very well detectable by these methods (see below).
Figure 4.
Overlap of predicted structured RNAs (high significance level) with the union of TARs/Transfrags and the “moderate” set of sequence-constrained elements. Hits in coding exons and UTRs are excluded.
Of the high-significance RNAz hits, 22.3% overlap with experimentally detected sites of transcription. This includes UTR elements and the predictions in coding regions (see below). Without these regions (i.e., counting only intergenic and intronic), 15.7% of the RNAz hits overlap with TARs/Transfrags. This corresponds to a significant enrichment of approximately twofold. However, this must be interpreted with caution since TARs/Transfrags are very GC rich (unannotated Transfrags: 56%). It is unclear to what extent this bias has biological reasons or is the result of the hybridization technique, and consequently, it is difficult to interpret the significance of these enriched overlaps. GC content seems to be an important issue since we do not see any enrichment but, in fact, a small negative correlation of EvoFold hits and TARs/Transfrags (only 5.8% of the intergenic and intronic EvoFold hits overlap TARs/Transfrags). The sensitivity of tiling arrays on AU-rich sequences may be lower than for GC-rich sequences.
Another important issue in this context is that it is unclear how secondary structure affects detection performance on tiling arrays. Similar to previous studies (Clote et al. 2005), which reported that functional RNAs are more stable than other sequences, we systematically compared Z-scores of folding stability of single sequences while taking dinucleotide content into account. We compared different annotation groups (introns, intergenic, CDS, UTRs, TARs/no TARs) to see if there are any general trends. Somewhat surprisingly, we found only one single statistically significant signal, which we interpret to be a technical rather than a biological effect: Regions detected by TARs/Transfrags are on average less stable than regions not detected by TARs (Wilcox, P = 2.5 × 10−7). In addition, we previously observed several examples in which highly stable ncRNAs (both predicted ones and known microRNAs) result in a negative signal (“holes”) in tiling-array data (Cheng et al. 2005). These results suggest that tiling arrays have a reduced sensitivity for strongly structured ncRNAs.
While much of the ENCODE region is alignable at least with the genomic DNA of closely related species, and hence used as input in the computational screens detailed above, only a subset of these sequences is under stabilizing selection at the sequence level. We therefore compared the structured RNA candidates with the multiple species analysis for sequence-constrained elements. We used the “moderate” set of constrained elements, which comprises regions detected by at least two of three conservation programs in at least two of three alignments prepared by different methods (Margulies et al. 2007). These conserved elements cover 4.9% of the ENCODE regions.
Eight hundred forty-one RNAz hits (22.69%) overlap with conserved regions, 570 (17.2%) without hits in UTRs and coding regions. For EvoFold predictions the overlap is much higher, 3579 (71.78%) including exons and 2130 (60.41%) without exons, in line with the program’s general tendency to predict structures in highly conserved regions. The fact that a large fraction of predicted conserved RNA structures does not correlate with high sequence conservation does not come as a surprise. Indeed, Torarinsson and colleagues reported expressed noncoding RNAs in regions that are not alignable between human and mouse and nevertheless have conserved secondary structures (Torarinsson et al. 2006). It is interesting, furthermore, that structured RNA accounts for <10% of the sequence-constrained parts of the human genome (based on RNAz, which is relatively unbiased with respect to sequence conservation).
It seems noteworthy that all but one16 of the few known ncRNAs in the ENCODE regions overlap with constrained elements and TARs/Transfrags. This might be special for this set of snoRNAs and miRNAs, which are presumably abundantly expressed as kind of “housekeeping ncRNAs” and have well known reasons for sequence constraints.
The 114 and 142 intergenic/intronic RNAz and EvoFold hits, respectively, that overlap both conserved elements and TARs/Transfrags are of special interest. Twenty-one of these are detected by both EvoFold and RNAz, while 12 of these have an AlifoldZ Z-score <−3.5. These numbers demonstrate that there is only a relatively small, but nonnegligible, number of structured ncRNAs that are similar to the “classical” ncRNA families in terms of high sequence conservation, highly stabilized and well-conserved secondary structures, and high expression levels.
Overlap with GENCODE annotations
The goal of the GENCODE project (Harrow et al. 2006) is the delineation of one complete mRNA sequence for at least one splice isoform of each protein-coding gene in the ENCODE regions, and often, but not systematically, the inference of a number of additional alternative splice forms of these genes. We mapped the predicted structured RNAs in comparison to all scored input regions to this set of annotations (Fig. 5). Extrapolating from our knowledge of described functional RNAs, we have to expect signals in all fractions (intergenic, introns, UTRs, coding sequences), and for RNAz, we can observe only moderate trends of relative enrichment. We see the strongest enrichment for RNAz in 3′-UTRs. This is remarkable given that 3′-UTRs are generally very AU rich (GC content only 44%) and that RNAz has limited sensitivity in AU-rich regions. In contrast, there is no enrichment in the 5′-UTR, which is again interesting given that 5′-UTRs are the fraction with the highest GC content (60%). This result is consistent not only with the EvoFold predictions, which have higher enrichment in 3′-UTRs than 5′-UTRs, but also with previous results from Siepel et al. (2005), who found that highly conserved regions in 3′-UTRs of vertebrates have a significantly increased propensity to form secondary structures, while in 5′-UTRs, this effect is not that pronounced.
Figure 5.
Genomic location of predicted RNAs (high significance level) relative to the GENCODE protein gene annotation. For comparison, the annotation of the input alignments is shown for both RNAz and EvoFold (they differ slightly because of the different filtering steps used for each program; see Methods). “Distal” and “Proximal” refer to a distance boundary of 5 kb away from the next gene (intergenic fraction) or coding exon (intronic fraction). Some hits fall within more than one annotation category, thus the sums of the fractions are slightly >100%.
RNAz predictions are depleted in coding regions despite the high GC content (53%). This is in keeping with the expectation that functional ncRNAs in coding regions should be rare. However, functional RNA structures do occur within coding regions, and thus these predictions are also of interest. As mentioned above, there are a few well-known functions assigned to hairpin structures within coding regions. In addition, there is recent evidence that secondary structures are much more widespread in coding regions of both prokaryotes (Katz and Burge 2003) and eukaryotes (Chamary and Hurst 2005; Meyer and Miklós 2005) than previously thought. EvoFold predictions are highly enriched in coding regions. However, the method has previously been shown to have above average rates of false positives in coding regions (Pedersen et al. 2006), presumably because of the high level of sequence conservation. The interpretation of these coding predictions is thus challenging and often requires additional evidence, such as conservation of synonymous codon positions (Pedersen et al. 2004a, b) or overlapping predictions from several methods. There are 41 overlapping RNAz/EvoFold hits from the high-significance sets in coding exons, 18 of which are particularly stable with AlifoldZ scores Z < −3.5.
In general, we do not see any trend of noncoding structures favoring intronic over intergenic fractions. For RNAz, however, one can observe that “proximal” intergenic and intronic fractions are slightly enriched while distal fractions are depleted, i.e., we see more structures near genes and exons. For EvoFold, both intergenic and intronic fractions are depleted in favor of the more conserved UTR and coding regions.
An interesting result of the GENCODE annotation project is the transcriptional complexity of protein-coding gene loci. For the 487 loci in the ENCODE regions, 2608 different transcripts were identified, 1511 of them noncoding. Two hundred twenty-nine and 940 RNAz and EvoFold hits, respectively, overlap with a noncoding GENCODE transcript. Some of these transcripts are extensively structured (Fig. 7F,G, see below).
Figure 7.
Selected high scoring examples. (Left) UCSC Genome Browser screenshots featuring conserved RNA predictions and additional ENCODE analysis tracks are shown. The significance levels of RNAz and EvoFold hits are color coded (see legend). (*) Significant AlifoldZ hits; the Z-score is shown. In addition, the results of the RACE/microarray experiments, TARs/Transfrags, constrained elements, phastCons scores, and GENCODE annotations are shown. For details on these tracks, refer to Methods. (Right) Consensus structure models generated by RNAalifold are shown for selected hits (marked by gray, dashed boxes; in example G, the first three hits and the sixth hit are shown). In the consensus structures, variable positions are circled indicating compensatory and consistent mutations supporting the structure. The color indicates the number of different nucleotide combinations forming one base pair. Inconsistent mutations lead to pale colors. Examples A–C show predicted structures in intergenic regions. Examples D and E are located in introns of protein-coding regions. Examples F and G show structures associated with alternative spliced transcripts of protein-coding loci detected by the GENCODE project. For further information, refer to the text.
Experimental verification of selected predictions
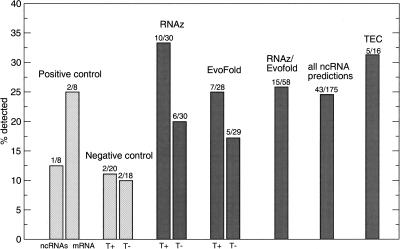
The high false discovery rates clearly show the limitations of the methods used here, indicating that reliable and fully automatic annotation is still out of reach. However, to demonstrate that selection of high-scoring predictions aided by visual inspection (see Methods) can result in high-quality predictions, we have performed verification experiments on selected candidates. We performed 245 RT-PCR experiments on total RNA of six tissues (175 ncRNA predictions, 16 positive controls, 38 negative controls, and 16 nonspliced ESTs clusters) (Harrow et al. 2006). The latter were named “to be experimentally confirmed” (TEC) by the GENCODE annotation. They have poly(A) features and are potentially protein coding (Harrow et al. 2006). Only one (U70) of the eight known ncRNAs (12%) (Fig. 6) but five of the TECs (31%) were recovered by RT-PCR, indicating that this protocol (see Methods) is probably not optimal for small, highly structured RNAs. Overall, we recovered 43 of the 175 predictions (25%). Thus the fraction of verified ncRNA predictions exceeds the amplification rate of randomly selected sequences by a factor of 2–3. Furthermore, we find that predictions that are supported by TARs or Transfrags are more likely to yield positive RT-PCR results (29% compared to 19% without support from tiling arrays). The sequenced fragments of verified ncRNA predictions and TEC were deposited to GenBank under accessions numbers EF212232–EF212281 and EF212282–EF212289, respectively.
Figure 6.
RT-PCR verification of ncRNA predictions. Positive controls include the known small ncRNAs listed in Table 3 as well as eight randomly chosen mRNAs of GENCODE protein-coding genes. Negative controls are randomly selected intergenic and intronic regions. Sets of RNAz and EvoFold predictions were manually selected both overlapping (T+) and not overlapping (T−) with TARs/transfrags. In addition, we selected a set of overlapping RNAz/EvoFold predictions (see Methods).
Examples of selected predictions
Figure 7 shows some examples of predicted RNAs in different genomic contexts. A series of criteria supports the prediction of these regions as functional RNA: (1) several independent RNAz and/or EvoFold hits in close vicinity; (2) overlapping hits of EvoFold/RNAz; (3) additional support from AlifoldZ; (4) support from compensatory/consistent mutations in the predicted structures; (5) overlap with predictions of sequence constrained elements. Evidence for transcription of these regions comes from TARs/Transfrags, ESTs, or GENCODE transcripts (Harrow et al. 2006). In addition, we have performed 5′-RACE/microarray experiments (see Methods).
Examples A, B, and C (in Fig. 7A–C) are located within intergenic regions, all of them >50 kb away from any GENCODE annotation. There are also no “putative” or “pseudogene” GENCODE annotations or any predicted protein-coding genes close by. Nevertheless, we observe sequence-constrained elements. In all cases, the sequences are conserved across eutherian mammals, B is also conserved in chicken, and in C there is a sequence from opossum. We observe several RNAz and EvoFold hits in these regions. In A, for example, we have two independent RNAz hits, one overlapping with an EvoFold hit. This example illustrates the different “sweet spots” of the two programs. The significant RNAz hit is in the region of moderate conservation, while the overlapping hit with EvoFold within the highly conserved region is only of borderline significance. In all three examples there is additional support from AlifoldZ, which is particularly impressive for B and C with Z-scores of −9.5 and −7.0. We want to recall that this Z-score means standard deviations from the expected random background score for a given alignment. The transcription of these RNAs was confirmed by 5′-RACE/array analysis.
Examples D and E (in Fig. 7D,E) show two sequence-constrained “islands” in introns of well-known protein-coding genes. They do not overlap with any predicted coding exons, but show clear signs of conserved RNA structures detected by both RNAz and EvoFold with additional support of AlifoldZ. The structure models show a series of consistent/compensatory mutations, and the RNA was detected by the RACE experiments. In the case of example D, further support for the intronic region to be part of a stable ncRNA comes from TARs/Transfrags as well as a short EST mapping nearby and overlapping with two additional RNAz and EvoFold hits.
Examples F and G (in Fig. 7F,G) show alternative splicing products of two protein loci detected and confirmed by the GENCODE annotation project. In F, we observe an internal transcription start (further supported by a CpG island), which gives rise to a transcript without clear coding potential but that is highly structured: There are five independent RNAz hits, two of which overlap with EvoFold hits and two with significant AlifoldZ scores (−5.0 and −6.4). A similar situation can be observed in G, where high densities of RNAz hits and overlapping EvoFold hits coincide with noncoding transcripts that arise from an alternatively spliced protein–gene locus.
Discussion
RNA secondary structures can provide important clues that a given locus is probably transcribed and that this transcript is functional at the RNA level. Here we attempted to comprehensively detect functional structures. Due to the lack of generic sequence signals that would imply RNA function, at present the only way toward this goal (apart from functional studies of individual transcripts) is comparative analysis. As the ENCODE regions are deeply sequenced, they provide an ideal proving ground for such an endeavor.
In contrast to previous genome-wide screens for structured RNA, which were restricted to very well-conserved regions of the genome, here we screened all alignable sequences. Indeed, high sequence conservation is not necessarily needed for function (Bentwich et al. 2005; Pang et al. 2006). In fact, most known ncRNAs that were missed in the previous RNAz screen of the human genome (Washietl et al. 2005a) were not detectable because they were not present in the highly conserved input set. Here we want to extend the spectrum and screen medium conserved as well as highly conserved regions. There is even a nonnegligible part of ncRNAs that are not alignable at all. For such cases, other methods (Hull Havgaard et al. 2005; Torarinsson et al. 2006; Uzilov et al. 2006) would be necessary that we do not cover here.
Using our highest threshold level and considering our estimates of false positives on shuffled alignments, we estimate ∼1800 and 1500 local RNA secondary structure elements using RNAz and EvoFold, respectively, in the ENCODE regions. We observed a fairly small overlap of predicted structures between RNAz and EvoFold. While surprising and at first sight discouraging, this discrepancy is explained by the fact that both methods are sensitive to dramatically different GC contents and levels of sequence conservation. Since known functional RNAs exist and are detected in the sensitivity ranges of both programs, the methods, in fact, yield complementary results, indicating that the number of structured RNAs is larger than predicted by any one of the programs alone. Furthermore, one should keep in mind that comparative approaches are by construction limited to evolutionarily relatively old sequences: They are bound to miss recent lineage-specific innovations as there is no conserved sequence with which to compare. It is thus likely that the number of functional RNAs in the human genome is even higher than the estimates arising from EvoFold and RNAz.
Despite the rich comparative sequence data in the ENCODE regions, both RNAz and EvoFold exhibit fairly high false discovery rates of 50%–70% as estimated from randomized input data and correction for dinucleotide frequencies. Also, this high noise level reduces the observed overlap. The overlap for previous screens restricted to phastCons conserved regions, for example, resulted in a twofold higher overlap. Substantial noise levels, however, also plague the experimental approaches. For example, tiling arrays, CAGE, and PET diTags techniques show excellent recovery rates and overlap on annotated coding transcripts, but elsewhere result in large numbers of other signals with moderate overlap and of uncertain relevance. The same is true for protein-coding gene prediction, which yields excellent results on known protein-coding exons but also predict thousands of additional exons incorrectly (Guigo et al. 2006). Despite such limitations inherent to all high-throughput methods, the output of such methods can be of high value if sensibly interpreted.
About 25% of a manual selection of ncRNA candidates were verified by means of RT-PCR, indicating that our computational approach detects a significant number of verifiable transcripts. Small and highly structured known ncRNAs are poorly recovered, indicating that the RT-PCR data most likely underestimate the true extent of transcription. In line with the observation from the ENCODE Pilot Project (The ENCODE Project Consortium 2007), we furthermore expect that most noncoding transcripts have a specific spatiotemporal expression pattern; our screen of six tissues is thus a priori expected to have only limited sensitivity.
One can consider various modes of function for noncoding transcripts like transcriptional interference (Martens et al. 2004) or antisense interactions (Katayama et al. 2005). Since we successfully predict ncRNA transcripts based on evolutionary conservation of RNA secondary structure in the presence of sequence variation, our data strongly suggest that a large number of noncoding RNAs require specific well-defined secondary structure or structured regions for their biological function. Current methods are not capable of distinguishing whether or not these structures are required for autonomous actions of the RNAs or whether or not they are part of binding motifs for specific interaction partners.
We found evidence for functional RNA structures in all regions of the genome. A fraction of these signals is likely to correspond to small ncRNAs in the classical sense, which are processed from introns or transcribed from intergenic regions with dedicated promoters, as is known for snoRNAs or miRNAs. We also found many signals in UTRs (particularly enriched in 3′-UTRs) of well-known protein-coding genes, suggesting regulatory functions of these signals at the mRNA level.
Our computational data, as well as the results from high-throughput experiments and the evidence from individual experimental results, strongly suggest that the functional spectrum of ncRNAs is much broader than previously expected. For example, we have convincing evidence for functional RNA structures in a few dozen coding exons. These might have regulatory roles for the mRNA, but it is also conceivable that they serve a double role as mRNA and ncRNA. Indeed, there is one example with such a dual role described in the literature, the steroid receptor activator (SRA) (Lanz et al. 2002; Chooniedass-Kothari et al. 2004). We also observed that alternative transcripts derived from protein loci give rise to transcripts with compelling evidence for functional RNA structures but little coding potential. This further blurs the difference of coding and noncoding genes. There is also a recent example of an enhancer element, which is transcribed and forms a spliced and polyadenylated ncRNA (Evf-2) that binds to the transcription factor as a coactivator that in turn binds to the enhancer element (Feng et al. 2006). This shows that functional RNAs can overlap with various other functional elements. In general, the abundance of predicted functional RNA structures associated with protein genes supports the notion of a “hidden regulatory layer” that exists on the RNA level in complex organisms (Mattick 2003, 2004).
Our data in combination with other ENCODE data and aided by visualization methods (Kent et al. 2002) allow a new way of seeing things and help in directing rationally devised experiments. These data open a perspective on the genome, which we hope will help to better understand the “modern RNA world.”
Methods
Multiple sequence alignments
We used 28-way TBA/MultiZ alignments with human (hg17) as the reference sequence, which were provided by the ENCODE alignment group (Margulies et al. 2007). We chose the TBA/MultiZ method alignments mainly because all previous applications of the programs used were done on TBA/MultiZ alignments or other alignments constructed from BLASTZ-based local comparisons. None of the three programs used for our analysis can handle unprocessed genome-wide alignments as presented by TBA/MultiZ. A series of preprocessing and filtering steps was necessary. The analysis pipeline varies in detail to meet the specific requirements of the three programs.
RNAz predictions
For the RNAz screen, alignments were sliced in overlapping windows of size 120 and slide 40. Each series of windows was started at the beginning of a TBA block. For windows reaching over the end of a block, we tried to append the adjacent block to the current one. Two blocks were only merged if all sequences were exactly or almost consecutive (up to 10 bases were allowed to be missing). Furthermore, sequences with >25% gaps with respect to the human sequence were discarded. Only alignments with more than four sequences, a minimum size of 50 columns, and at most 1% repeat-masked letters were considered. RNAz can only handle alignments with up to six sequences. From alignments with more than six sequences, we chose a subset of six: We used a greedy algorithm and iteratively selected sequences optimizing the set for a mean pairwise identity of ∼80%. In cases of alignments with more than 10 sequences, we sampled three different from such subsets. The windows were finally scored with RNAz version 0.1.1 in the forward and reverse complement directions. Overlapping hits with at least one sampled alignment with P > 0.5 were combined to a single genomic region (“cluster”). Clusters were assigned two significance levels: “P > 0.5” means that there is at least one window in the cluster with a mean P of at least 0.5. “P > 0.9” means that there is at least one window in the cluster with mean P of all samples >0.5 and at least one hit with P > 0.9.
AlifoldZ predictions
The preprocessing steps were the same as for RNAz. However, we only scored one sample per window. If there were less than 10 sequences in the alignment, all sequences were used. If there were more than 10 sequences, a sample of 10 sequences optimized for a mean pairwise identity of 80% was chosen. It does not seem reasonable to score alignments with too many sequences using AlifoldZ because the efficiency of alignment shuffling and alignment errors becomes limiting. In fact, the larger number of sequences per alignment may have contributed to the low number of hits produced by AlifoldZ in comparison to RNAz. We only scored alignments with an RNAalifold consensus MFE better than −15, using the same version of AlifoldZ that was originally published with the paper (Washietl and Hofacker 2004). A sample size of N = 100 was chosen to estimate the Z-scores for both the forward and reverse complement directions. Overlapping hits were clustered as described above for RNAz predictions and assigned two significance levels using Z < −3.5 and Z < −4 as cutoffs.
EvoFold predictions
For the EvoFold analysis, sequences with >20% gaps relative to human were first removed. Second, alignments with sequence from less than six species were eliminated. Third, TBA alignment blocks consecutive relative to human were concatenated. Fourth, nonsyntenic sequences that include segments from disparate genomic regions (more than twice the length of the human reference sequence apart) were removed; however, if the resulting alignment had less than six sequences, none were removed. EvoFold 1.1 was then applied to the concatenated alignments, and their reverse complements, in 120 long overlapping windows each offset by 40. Weak predictions (<10 pairing bases or an average stem length of less than 3) as well as predictions overlapping repeats or retrogenes (as defined by tracks of the UCSC browser) were eliminated. Finally, the set was reduced to single coverage by removing the lowest-scoring candidates if overlap occurred, and ranked according to score. Two prediction sets were defined based on the final score: all predictions and the top 50%.
Randomization of alignments
All three screens were repeated on randomized TBA alignments. The alignments were shuffled as described previously (Washietl and Hofacker 2004), resulting in random alignments of the same base composition, sequence conservation, and gap patterns. We could not exactly preserve local conservation patterns since this would have been limiting in the case of large alignments. However, the adapted shuffling method we used retains a coarse-grained pattern of conservation (only columns with mean pairwise identity >0.5 and <0.5 were shuffled with each other, respectively).
Comparison with other ENCODE data
We used the ENCODE data from December 2005 provided at the Galaxy2ENCODE Web site (Blankenberg et al. 2007). This includes the GENCODE annotation (Harrow et al. 2006), the “moderate” set of constrained elements (Margulies et al. 2007), and the union of Yale and Affymetrix TARs/Transfrags signals from all 11 tissues and RNA extractions [Poly(A)+ and complete RNA] (The ENCODE Project Consortium 2007). Overlap calculations, partition into the different annotation types, and calculating phastCons scores were accomplished using the tools of the Galaxy2ENCODE system and the UCSC table browser (Kent et al. 2002).
Selection of candidates for RT-PCR verification
We manually selected 175 candidates for RT-PCR verification in three sets: RNAz hits (60), EvoFold hits (57), and overlapping RNAz/EvoFold hits (58). For the first two sets, we explicitly chose half of the targets with overlap to TARs/Transfrags and the other half without. The third set of overlapping RNAz/EvoFold hits was chosen without regard to TAR/Transfrag overlap (35 of the 58 have overlap). RNA predictions shorter than 200 nt were extended to target regions of at least 200 nt length (limiting our detection performance of small RNAs, e.g., we cannot detect mature miRNAs).
The criteria that were used for selecting candidates include high RNAz/AlifoldZ and/or EvoFold scores, absence of any indication of alignment errors or other alignment artifacts, presence of compensatory mutations, genomic location in either introns of protein-coding transcripts, or unannotated intergenic regions.
We routinely generated structure annotated and colorized alignments of all hits visualizing the predicted structure together with the mutational pattern. Inspection of the alignments can help to select more reasonable candidates mainly by weeding out obvious false positives. For example, unusual gap patterns or low complexity runs of single letters indicate an artifactual hit. Currently, the programs themselves cannot efficiently recognize such artifacts, and there is still much room for improvement (e.g., by using an explicit indel model in EvoFold).
Negative controls were obtained by randomizing the set of ncRNA target regions using the “Random Intervals” tool of the Galaxy2ENCODE system. From the resulting randomized locations, we chose 38 targets: 19 in intergenic regions (nine overlapping TARs/Transfrags) and 19 in intronic regions (nine overlapping TARs/Transfrags). As positive controls, we randomly chose eight regions in exons of mRNAs of known protein-coding genes and the eight ncRNAs from Table 3.
RT-PCR
Brain, heart, kidney, liver, lung, and testis total RNA (0.1 μg each) was mixed and reversed-transcribed in 25 μL with AMV Reverse Transcriptase XL in the presence of dNTP nucleotide analogs to avoid amplification of genomic DNA contaminants; RNase Inhibitor; and MgCl2 (mRNA Selective PCR kit; Takara). The reaction was carried out in 1× Selective buffer II with 0.4 μM specific primer (see below) following the manufacturer’s instructions, that is, 30°C for 10 min, 42°C for 30 min, and 5°C for 5 min. The PCR amplification was performed in 25 μL with one-fifth of the RT-reaction and primers at a final concentration of 0.4 μM at 85°C for 1 min, 50°C for 1 min, and 72°C for 1 min for 30 cycles following the manufacturer’s instructions (mRNA Selective PCR kit; Takara). Amplimers were separated on a 1.8% agarose gel and sequenced. The primers were selected with Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3www.cgi) and default parameters. For all predictions and controls, we tested the forward and reverse strands. The reason for this is that the programs cannot determine the correct reading direction in all cases (strong RNA signals, i.e., base-pairing patterns, usually can be also detected in the reverse complement).
5′-RACE/array analysis
5′-RACE reactions were performed on brain and testis cDNA prepared from both poly(A)+ and total RNA and oligo(dT) and random hexamers, respectively, as described by Denoeud et al. (2007). The mapping of the RACE primers is given in Figure 7. The RACE amplimers were hybridized to ENCODE tiling arrays as described by Kapranov et al. (2005) and modified by Denoeud et al. (2007).
Data availability
The predictions described in this paper are available as annotation tracks in BED format suitable for use with the UCSC Genome Browser and can be downloaded at http://www.tbi.univie.ac.at/papers/SUPPLEMENTS/ENCODE/. Primer sequences and results of the RT-PCR experiments can also be downloaded from this Web site.
Acknowledgments
We thank Lukas Endler for discussion and David Haussler for valuable comments on the manuscript. We acknowledge funding from the Austrian GEN-AU projects “noncoding RNA” and “Bioinformatics Integration Network” (to I.L.H.), the DFG Bioinformatics Initiative BIZ-6/1-2 (to P.F.S.), the Danish Research Council [#272-05-0319], the National Cancer Institute (both to J.S.P.), a Marie Curie Outgoing International Fellowship (to J.O.K.), ENCODE grants from National Human Genome Research Institute (NHGRI)/National Institutes of Health (NIH) (especially to the following ENCODE subgroups: Yale [#U01HG03156], Affymetrix, Inc. [#U01HG03147], and GENCODE [#U01HG03150]), the Swiss National Science Foundation (to S.E.A. and to A.R.), the NCCR Frontiers in Genetics and the European Union (to S.E.A.), the Jérôme Lejeune (to S.E.A. and A.R.), the Childcare (to S.E.A.), and the Novartis (to A.R.) Foundations.
Footnotes
[The sequenced fragments of verified ncRNA predictions and TEC were deposited to GenBank under accession nos. EF212232–EF212281 and EF212282–EF212289, respectively.]
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.5650707
This approach is also similar in spirit to QRNA, a program that detects conserved RNA structures in pairwise alignments by comparing an SCFG-based RNA model to a background model (Rivas and Eddy 2001).
MIRN483 does not overlap with TARs/Transfrags. It might be specific in fetal liver tissue, which is not among the 11 tissues tested.
References
- Bentwich I., Avniel A.A., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Avniel A.A., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Barzilai A., Einat P., Einav U., Meiri E., Einat P., Einav U., Meiri E., Einav U., Meiri E., Meiri E., et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- Bertone P., Stoc V., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Stoc V., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Tongprasit W., Samanta M., Weissman S., Samanta M., Weissman S., Weissman S., et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Rosenbloom K., Clawson H., Green E.D., Clawson H., Green E.D., Green E.D., et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D., Taylor J., Schenck I., He J., Zhang Y., Ghent M., Veeraraghavan N., Albert I., Miller W., Makova K., Taylor J., Schenck I., He J., Zhang Y., Ghent M., Veeraraghavan N., Albert I., Miller W., Makova K., Schenck I., He J., Zhang Y., Ghent M., Veeraraghavan N., Albert I., Miller W., Makova K., He J., Zhang Y., Ghent M., Veeraraghavan N., Albert I., Miller W., Makova K., Zhang Y., Ghent M., Veeraraghavan N., Albert I., Miller W., Makova K., Ghent M., Veeraraghavan N., Albert I., Miller W., Makova K., Veeraraghavan N., Albert I., Miller W., Makova K., Albert I., Miller W., Makova K., Miller W., Makova K., Makova K., et al. A framework for collaborative analysis of ENCODE data: Making large-scale analyses biologist-friendly. Genome Res. 2007 doi: 10.1101/gr.5578007. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bompfünewerer A.F., Flamm C., Fried C., Fritzsch G., Hofacker I.L., Lehmann J., Missal K., Mosig A., Müller B., Prohaska S.J., Flamm C., Fried C., Fritzsch G., Hofacker I.L., Lehmann J., Missal K., Mosig A., Müller B., Prohaska S.J., Fried C., Fritzsch G., Hofacker I.L., Lehmann J., Missal K., Mosig A., Müller B., Prohaska S.J., Fritzsch G., Hofacker I.L., Lehmann J., Missal K., Mosig A., Müller B., Prohaska S.J., Hofacker I.L., Lehmann J., Missal K., Mosig A., Müller B., Prohaska S.J., Lehmann J., Missal K., Mosig A., Müller B., Prohaska S.J., Missal K., Mosig A., Müller B., Prohaska S.J., Mosig A., Müller B., Prohaska S.J., Müller B., Prohaska S.J., Prohaska S.J., et al. Evolutionary patterns of non-coding RNAs. Theory Biosci.. 2005;123:301–369. doi: 10.1016/j.thbio.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Buratti E., Baralle F.E., Baralle F.E. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol. Cell. Biol. 2004;24:10505–10514. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P., Kasukawa T., Katayama S., Gough J., Frith M., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Kasukawa T., Katayama S., Gough J., Frith M., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Katayama S., Gough J., Frith M., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Gough J., Frith M., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Frith M., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Oyama R., Ravasi T., Lenhard B., Wells C., Ravasi T., Lenhard B., Wells C., Lenhard B., Wells C., Wells C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Chamary J.V., Hurst L.D., Hurst L.D. Evidence for selection on synonymous mutations affecting stability of mRNA secondary structure in mammals. Genome Biol. 2005;6:R75. doi: 10.1186/gb-2005-6-9-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Patel S., Long J., Stern D., Tammana H., Helt G., Long J., Stern D., Tammana H., Helt G., Stern D., Tammana H., Helt G., Tammana H., Helt G., Helt G., et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- Chooniedass-Kothari S., Emberley E., Hamedani M.K., Troup S., Wang X., Czosnek A., Hube F., Mutawe M., Watson P.H., Leygue E., Emberley E., Hamedani M.K., Troup S., Wang X., Czosnek A., Hube F., Mutawe M., Watson P.H., Leygue E., Hamedani M.K., Troup S., Wang X., Czosnek A., Hube F., Mutawe M., Watson P.H., Leygue E., Troup S., Wang X., Czosnek A., Hube F., Mutawe M., Watson P.H., Leygue E., Wang X., Czosnek A., Hube F., Mutawe M., Watson P.H., Leygue E., Czosnek A., Hube F., Mutawe M., Watson P.H., Leygue E., Hube F., Mutawe M., Watson P.H., Leygue E., Mutawe M., Watson P.H., Leygue E., Watson P.H., Leygue E., Leygue E., et al. The steroid receptor RNA activator is the first functional RNA encoding a protein. FEBS Lett. 2004;566:43–47. doi: 10.1016/j.febslet.2004.03.104. [DOI] [PubMed] [Google Scholar]
- Clote P., Ferre F., Kranakis E., Krizanc D., Ferre F., Kranakis E., Krizanc D., Kranakis E., Krizanc D., Krizanc D. Structural RNA has lower folding energy than random RNA of the same dinucleotide frequency. RNA. 2005;11:578–591. doi: 10.1261/rna.7220505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoeud F., Kapranov P., Ucla C., Frankish A., Castelo R., Drenkow J., Lagarde J., Alioto T., Manzano C., Chrast J., Kapranov P., Ucla C., Frankish A., Castelo R., Drenkow J., Lagarde J., Alioto T., Manzano C., Chrast J., Ucla C., Frankish A., Castelo R., Drenkow J., Lagarde J., Alioto T., Manzano C., Chrast J., Frankish A., Castelo R., Drenkow J., Lagarde J., Alioto T., Manzano C., Chrast J., Castelo R., Drenkow J., Lagarde J., Alioto T., Manzano C., Chrast J., Drenkow J., Lagarde J., Alioto T., Manzano C., Chrast J., Lagarde J., Alioto T., Manzano C., Chrast J., Alioto T., Manzano C., Chrast J., Manzano C., Chrast J., Chrast J., et al. Prominent use of distal 5′ transcription start sites and discovery of a large number of additional exons in ENCODE regions. Genome Res. 2007 doi: 10.1101/gr.5660607. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007 doi: 10.1038/nature05874. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Bi C., Clark B.S., Mady R., Shah P., Kohtz J.D., Bi C., Clark B.S., Mady R., Shah P., Kohtz J.D., Clark B.S., Mady R., Shah P., Kohtz J.D., Mady R., Shah P., Kohtz J.D., Shah P., Kohtz J.D., Kohtz J.D. The evf-2 noncoding RNA is transcribed from the dlx-5/6 ultraconserved region and functions as a dlx-2 transcriptional coactivator. Genes & Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabory A., Ripoche M.A., Yoshimizu T., Dandolo L., Ripoche M.A., Yoshimizu T., Dandolo L., Yoshimizu T., Dandolo L., Dandolo L. The H19 gene: Regulation and function of a noncoding RNA. Cytogenet. Genome Res. 2006;113:188–193. doi: 10.1159/000090831. [DOI] [PubMed] [Google Scholar]
- Gardner P.P., Giegerich R., Giegerich R. A comprehensive comparison of comparative RNA structure prediction approaches. BMC Bioinformatics. 2004;5:140. doi: 10.1186/1471-2105-5-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glusman G., Qin S., El-Gewely M.R., Siegel A.F., Roach J.C., Hood L., Smit A.F., Qin S., El-Gewely M.R., Siegel A.F., Roach J.C., Hood L., Smit A.F., El-Gewely M.R., Siegel A.F., Roach J.C., Hood L., Smit A.F., Siegel A.F., Roach J.C., Hood L., Smit A.F., Roach J.C., Hood L., Smit A.F., Hood L., Smit A.F., Smit A.F. A third approach to gene prediction suggests thousands of additional human transcribed regions. PLoS Comput. Biol. 2006;2:e18. doi: 10.1371/journal.pcbi.0020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Moxon S., Marshall M., Khanna A., Eddy S.R., Bateman A., Moxon S., Marshall M., Khanna A., Eddy S.R., Bateman A., Marshall M., Khanna A., Eddy S.R., Bateman A., Khanna A., Eddy S.R., Bateman A., Eddy S.R., Bateman A., Bateman A. Rfam: Annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guigo R., Flicek P., Abril J.F., Reymond A., Lagarde J., Denoeud F., Antonarakis S., Ashburner M., Bajic V.B., Birney E., Flicek P., Abril J.F., Reymond A., Lagarde J., Denoeud F., Antonarakis S., Ashburner M., Bajic V.B., Birney E., Abril J.F., Reymond A., Lagarde J., Denoeud F., Antonarakis S., Ashburner M., Bajic V.B., Birney E., Reymond A., Lagarde J., Denoeud F., Antonarakis S., Ashburner M., Bajic V.B., Birney E., Lagarde J., Denoeud F., Antonarakis S., Ashburner M., Bajic V.B., Birney E., Denoeud F., Antonarakis S., Ashburner M., Bajic V.B., Birney E., Antonarakis S., Ashburner M., Bajic V.B., Birney E., Ashburner M., Bajic V.B., Birney E., Bajic V.B., Birney E., Birney E., et al. EGASP: The human ENCODE Genome Annotation Assessment Project. Genome Biol. 2006;(Suppl 1) 7:S2.1–S2.31. doi: 10.1186/gb-2006-7-s1-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J., Denoeud F., Frankish A., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Denoeud F., Frankish A., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Frankish A., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Gilbert J.G., Storey R., Swarbreck D., Storey R., Swarbreck D., Swarbreck D., et al. GENCODE: Producing a reference annotation for ENCODE. Genome Biol. 2006;(Suppl 1) 7:S4.1–S4.9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker I.L., Fekete M., Stadler P.F., Fekete M., Stadler P.F., Stadler P.F. Secondary structure prediction for aligned RNA sequences. J. Mol. Biol. 2002;319:1059–1066. doi: 10.1016/S0022-2836(02)00308-X. [DOI] [PubMed] [Google Scholar]
- Hubert N., Walczak R., Sturchler C., Myslinski E., Schuster C., Westhof E., Carbon P., Krol A., Walczak R., Sturchler C., Myslinski E., Schuster C., Westhof E., Carbon P., Krol A., Sturchler C., Myslinski E., Schuster C., Westhof E., Carbon P., Krol A., Myslinski E., Schuster C., Westhof E., Carbon P., Krol A., Schuster C., Westhof E., Carbon P., Krol A., Westhof E., Carbon P., Krol A., Carbon P., Krol A., Krol A. RNAs mediating cotranslational insertion of selenocysteine in eukaryotic selenoproteins. Biochimie. 1996;78:590–596. doi: 10.1016/s0300-9084(96)80005-8. [DOI] [PubMed] [Google Scholar]
- Hull Havgaard J.H., Lyngsø R., Stormo G.D., Gorodkin J., Lyngsø R., Stormo G.D., Gorodkin J., Stormo G.D., Gorodkin J., Gorodkin J. Pairwise local structural alignment of RNA sequences with sequence similarity less than 40% Bioinformatics. 2005;21:1815–1824. doi: 10.1093/bioinformatics/bti279. [DOI] [PubMed] [Google Scholar]
- Kapranov P., Drenkow J., Cheng J., Long J., Helt G., Dike S., Gingeras T.R., Drenkow J., Cheng J., Long J., Helt G., Dike S., Gingeras T.R., Cheng J., Long J., Helt G., Dike S., Gingeras T.R., Long J., Helt G., Dike S., Gingeras T.R., Helt G., Dike S., Gingeras T.R., Dike S., Gingeras T.R., Gingeras T.R. Examples of the complex architecture of the human transcriptome revealed by race and high-density tiling arrays. Genome Res. 2005;15:987–997. doi: 10.1101/gr.3455305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., Kasukawa T., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., Nishida H., Yap C.C., Suzuki M., Kawai J., Yap C.C., Suzuki M., Kawai J., Suzuki M., Kawai J., Kawai J., et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Katz L., Burge C.B., Burge C.B. Widespread selection for local RNA secondary structure in coding regions of bacterial genes. Genome Res. 2003;13:2042–2051. doi: 10.1101/gr.1257503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Pringle T.H., Zahler A.M., Haussler D., Zahler A.M., Haussler D., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen B., Hein J., Hein J. RNA secondary structure prediction using stochastic context-free grammars and evolutionary history. Bioinformatics. 1999;15:446–454. doi: 10.1093/bioinformatics/15.6.446. [DOI] [PubMed] [Google Scholar]
- Knudsen B., Hein J., Hein J. Pfold: RNA secondary structure prediction using stochastic context-free grammars. Nucleic Acids Res. 2003;31:3423–3428. doi: 10.1093/nar/gkg614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz R.B., Razani B., Goldberg A.D., O’Malley B.W., Razani B., Goldberg A.D., O’Malley B.W., Goldberg A.D., O’Malley B.W., O’Malley B.W. Distinct RNA motifs are important for coactivation of steroid hormone receptors by steroid receptor RNA activator (SRA) Proc. Natl. Acad. Sci. 2002;99:16081–16086. doi: 10.1073/pnas.192571399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies E.H., Cooper G.M., Asimenos G., Thomas D.J., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Cooper G.M., Asimenos G., Thomas D.J., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Asimenos G., Thomas D.J., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Thomas D.J., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Birney E., Keefe D., Schwartz A.S., Hou M., Keefe D., Schwartz A.S., Hou M., Schwartz A.S., Hou M., Hou M., et al. Analyses of deep mammalian sequence alignments and constraint predictions for 1% of the human genome. Genome Res. 2007 doi: 10.1101/gr.6034307. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens J.A., Laprade L., Winston F., Laprade L., Winston F., Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- Mattick J.S. Challenging the dogma: The hidden layer of non-protein-coding RNAs in complex organisms. Bioessays. 2003;25:930–939. doi: 10.1002/bies.10332. [DOI] [PubMed] [Google Scholar]
- Mattick J.S. RNA regulation: A new genetics? Nat. Rev. Genet. 2004;5:316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- Meyer I.M., Miklós I., Miklós I. Statistical evidence for conserved, local secondary structure in the coding regions of eukaryotic mRNAs and pre-mRNAs. Nucleic Acids Res. 2005;33:6338–6348. doi: 10.1093/nar/gki923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignone F., Gissi C., Liuni S., Pesole G., Gissi C., Liuni S., Pesole G., Liuni S., Pesole G., Pesole G. Untranslated regions of mRNAs. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-3-reviews0004. REVIEWS0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O., Rousset J.P., Napthine S., Brierley I., Rousset J.P., Napthine S., Brierley I., Napthine S., Brierley I., Brierley I. Reprogrammed genetic decoding in cellular gene expression. Mol. Cell. 2004;13:157–168. doi: 10.1016/s1097-2765(04)00031-0. [DOI] [PubMed] [Google Scholar]
- Okazaki Y., Furuno M., Kasukawa T., Adachi J., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., Furuno M., Kasukawa T., Adachi J., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., Kasukawa T., Adachi J., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., Adachi J., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., Nikaido I., Osato N., Saito R., Suzuki H., Osato N., Saito R., Suzuki H., Saito R., Suzuki H., Suzuki H., et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- Pang K.C., Frith M.C., Mattick J.S., Frith M.C., Mattick J.S., Mattick J.S. Rapid evolution of noncoding RNAs: Lack of conservation does not mean lack of function. Trends Genet. 2006;22:1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Pedersen J.S., Forsberg R., Meyer I.M., Hein J., Forsberg R., Meyer I.M., Hein J., Meyer I.M., Hein J., Hein J. An evolutionary model for protein-coding regions with conserved RNA structure. Mol. Biol. Evol. 2004a;21:1913–1922. doi: 10.1093/molbev/msh199. [DOI] [PubMed] [Google Scholar]
- Pedersen J.S., Meyer I.M., Forsberg R., Simmonds P., Hein J., Meyer I.M., Forsberg R., Simmonds P., Hein J., Forsberg R., Simmonds P., Hein J., Simmonds P., Hein J., Hein J. A comparative method for finding and folding RNA secondary structures within protein-coding regions. Nucleic Acids Res. 2004b;32:4925–4936. doi: 10.1093/nar/gkh839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen J.S., Bejerano G., Siepel A., Rosenbloom K., Lindblad-Toh K., Lander E.S., Kent J., Miller W., Haussler D., Bejerano G., Siepel A., Rosenbloom K., Lindblad-Toh K., Lander E.S., Kent J., Miller W., Haussler D., Siepel A., Rosenbloom K., Lindblad-Toh K., Lander E.S., Kent J., Miller W., Haussler D., Rosenbloom K., Lindblad-Toh K., Lander E.S., Kent J., Miller W., Haussler D., Lindblad-Toh K., Lander E.S., Kent J., Miller W., Haussler D., Lander E.S., Kent J., Miller W., Haussler D., Kent J., Miller W., Haussler D., Miller W., Haussler D., Haussler D. Classification of conserved RNA secondary structures in the human genome. PLoS Comput. Biol. 2006;2:e33. doi: 10.1371/journal.pcbi.0020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas E., Eddy S.R., Eddy S.R. Noncoding RNA gene detection using comparative sequence analysis. BMC Bioinformatics. 2001;2:8. doi: 10.1186/1471-2105-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semon M., Duret L., Duret L. Evidence that functional transcription units cover at least half of the human genome. Trends Genet. 2004;20:229–232. doi: 10.1016/j.tig.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Clawson H., Spieth J., Hillier L.W., Richards S., Spieth J., Hillier L.W., Richards S., Hillier L.W., Richards S., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torarinsson E., Sawera M., Havgaard J., Fredholm M., Gorodkin J., Sawera M., Havgaard J., Fredholm M., Gorodkin J., Havgaard J., Fredholm M., Gorodkin J., Fredholm M., Gorodkin J., Gorodkin J. Thousands of corresponding human and mouse genomic regions unalignable in primary sequece contain common RNA structure. Genome Res. 2006;16:885–889. doi: 10.1101/gr.5226606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzilov A.V., Keegan J.M., Mathews D.H., Keegan J.M., Mathews D.H., Mathews D.H. Detection of noncoding RNAs on the basis of predicted secondary structure formation free energy change. BMC Bioinformatics. 2006;7:173. doi: 10.1186/1471-2105-7-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S., Hofacker I.L., Hofacker I.L. Consensus folding of aligned sequences as a new measure for the detection of functional RNAs by comparative genomics. J. Mol. Biol. 2004;342:19–39. doi: 10.1016/j.jmb.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Washietl S., Hofacker I.L., Lukasser M., Hüttenhofer A., Stadler P.F., Hofacker I.L., Lukasser M., Hüttenhofer A., Stadler P.F., Lukasser M., Hüttenhofer A., Stadler P.F., Hüttenhofer A., Stadler P.F., Stadler P.F. Mapping of conserved RNA secondary structures predicts thousands of functional non-coding RNAs in the human genome. Nat. Biotechnol. 2005a;23:1383–1390. doi: 10.1038/nbt1144. [DOI] [PubMed] [Google Scholar]
- Washietl S., Hofacker I.L., Stadler P.F., Hofacker I.L., Stadler P.F., Stadler P.F. Fast and reliable prediction of noncoding RNAs. Proc. Natl. Acad. Sci. 2005b;102:2454–2459. doi: 10.1073/pnas.0409169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman C., Krogh A., Krogh A. No evidence that mRNAs have lower folding free energies than random sequences with the same dinucleotide distribution. Nucleic Acids Res. 1999;27:4816–4822. doi: 10.1093/nar/27.24.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The predictions described in this paper are available as annotation tracks in BED format suitable for use with the UCSC Genome Browser and can be downloaded at http://www.tbi.univie.ac.at/papers/SUPPLEMENTS/ENCODE/. Primer sequences and results of the RT-PCR experiments can also be downloaded from this Web site.