Abstract
Genomic tiling microarrays have become a popular tool for interrogating the transcriptional activity of large regions of the genome in an unbiased fashion. There are several key parameters associated with each tiling experiment (e.g., experimental protocols and genomic tiling density). Here, we assess the role of these parameters as they are manifest in different tiling-array platforms used for transcription mapping. First, we analyze how a number of published tiling-array experiments agree with established gene annotation on human chromosome 22. We observe that the transcription detected from high-density arrays correlates substantially better with annotation than that from other array types. Next, we analyze the transcription-mapping performance of the two main high-density oligonucleotide array platforms in the ENCODE regions of the human genome. We hybridize identical biological samples and develop several ways of scoring the arrays and segmenting the genome into transcribed and nontranscribed regions, with the aim of making the platforms most comparable to each other. Finally, we develop a platform comparison approach based on agreement with known annotation. Overall, we find that the performance improves with more data points per locus, coupled with statistical scoring approaches that properly take advantage of this, where this larger number of data points arises from higher genomic tiling density and the use of replicate arrays and mismatches. While we do find significant differences in the performance of the two high-density platforms, we also find that they complement each other to some extent. Finally, our experiments reveal a significant amount of novel transcription outside of known genes, and an appreciable sample of this was validated by independent experiments.
Mapping transcribed regions of the human genome in an unbiased fashion is a crucial step toward understanding at a molecular level the organization of hereditary information and the specific functions of each human cell or tissue type. To this end, a number of approaches using genomic tiling microarrays have been tested and published over the last few years, including key studies by Kapranov et al. (2002), Rinn et al. (2003), Bertone et al. (2004), Schadt et al. (2004), and Cheng et al. (2005). While the strategies differ substantially in most of their details, they all share a basic array design concept: to construct an array whose probes (the molecules attached to the microarray at the manufacturing) cover all of the nonrepetitive sequence of the genome or genomic region under investigation.
Kapranov et al. (2002) used a high-density oligonucleotide array design containing perfect match probes of length 25 bp and corresponding mismatch probes. The arrays were synthesized in situ (directly on the supporting array material) using physical masks (Lipshutz et al. 1999) and covered chromosomes 21 and 22 with probe starting positions spaced every 35 bp (genomic distance). They were hybridized with samples representing 11 cell lines. The data was later reanalyzed (Kampa et al. 2004) and a more sophisticated approach to genomic segmentation was introduced. We refer to this setup as the Affymetrix tiling-array platform.
Rinn et al. (2003) mapped transcribed regions of chromosome 22 with an array of PCR products (amplicons), tiled end-to-end with a probe size range of 300–1400 bp. This array represents the PCR tiling-array platform and was hybridized with placenta poly(A)+ RNA (Rinn et al. 2003) and later with RNA from two cell lines (White et al. 2004).
Schadt et al. (2004) used tiling arrays where the probes were synthesized on the array using the Agilent ink-jet technology (Shoemaker et al. 2001). They tiled chromosomes 20 and 22 with 60-mers uniformly spaced every 30 bp. The statistical treatment of the data was presented in Ying et al. (2003).
Bertone et al. (2004) used oligonucleotide microarrays with 36-bp probes spaced every 46 bp to map transcribed regions of the entire nonrepetitive portion of the human genome. The arrays are synthesized in situ using maskless technologies developed by NimbleGen Systems. We refer to this as the MAS (maskless array synthesis) tiling array platform (Singh-Gasson et al. 1999; Nuwaysir et al. 2002).
Cheng et al. (2005) used an updated version of the Affymetrix platform with a tighter spacing of the probes, every 5 bp, and covering 10 chromosomes of the human genome. Transcript maps were generated for polyadenylated cytosolic RNA from eight cell lines (and for one of these cell lines, also nonpolyadenylated RNA).
These different studies produced a wealth of data. However, the experiments represent very different choices in array design and manufacturing, RNA extraction and hybridization conditions, and data processing methods. As such, comparing the results from these studies is not trivial.
Here, we outline some of the key parameters differentiating the various studies. The array design parameters include the length and genomic spacing of the probes, the use of mismatch probes, and whether to cover one or both genomic strands. For the oligonucleotide tiling experiments referenced above, the probe length varies between 25 and 70 bases. The genomic spacing of the probes is measured between probe initiation points and can range from the smallest possible distance of one single base up to the length of the probe, or even further. At the design stage it is important to minimize potential cross-hybridization, self-pairing, and other probe sequence artifacts such as DNA secondary structure formation (SantaLucia Jr. and Hicks 2004). Genomic regions considered as repeats (by, e.g., RepeatMasker [A.F.A. Smit and P. Green, unpubl.]) are usually omitted from the design due to potential cross-hybridization. If some flexibility is allowed in the design process, probes may be chosen so as to achieve better probe thermodynamics. This is possible for arrays interrogating genes (Mathews et al. 1999; Hughes et al. 2001; Rouillard et al. 2003), but for tiling arrays with high genomic density probe optimization options are limited (Bertone et al. 2006).
The experimental protocols for extraction, labeling, and hybridization of the RNA sample to the array vary considerably. Choosing the type of target RNA (i.e., tissue or cell line, poly(A)+ or total RNA), and the reactions and conditions to use in the hybridization will affect the results. The number of technical and biological replicates is an additional crucial parameter, more replicates potentially enables greater certainty and detail in the interpretation of the results.
Once the tiling arrays have been designed, manufactured, hybridized with labeled RNA, and the hybridization intensities have been extracted, there are a number of ways to transform the raw intensities into a score for each probe. This is usually done using statistical methods such as a sign test or the t-test. Exactly what methods are available depends on the design features of the array, such as the presence of mismatch probes. The segmentation of the genome into transcribed and nontranscribed regions is then performed based on the scores.
Our goal is to assess different tiling microarrays that are currently used for transcription mapping, an area where no detailed comparison thus far has been performed, and ultimately to aid the ENCODE Consortium when choosing strategy for the multiple tissue whole-genome transcription mapping of the human genome (The ENCODE Project Consortium 2004). Previous work on comparing gene-based microarrays include studies by Tan et al. (2003), Jarvinen et al. (2004), Mah et al. (2004), Park et al. (2004), and Yauk et al. (2004). Most of these indicate differences in the gene expression results from different microarray platforms, which have been attributed to differences in data processing or inadequate choice of comparison metrics (Larkin et al. 2005).
We start our microarray comparison by analyzing a set of already published chromosome 22 transcription experiments. Overall, this study indicated that high-density oligonucleotide arrays perform significantly better than amplicon (PCR) arrays.
We then describe a direct comparison of the two in situ-synthesized oligonucleotide-based platforms MAS (Bertone et al. 2004) and Affymetrix (Affy) (Kapranov et al. 2002) on the manually picked part of the ENCODE regions of the human genome (http://www.genome.gov/10005107). We hybridized identical biological samples to the arrays and developed a unified data-processing scheme based on statistical treatment of the data. Using this approach, we compare the results from the two platforms with each other and with the recently generated GENCODE gene annotation (Guigo et al. 2003; Ashurst et al. 2005; http://genome.imim.es/gencode/).
Results and Discussion
Pilot study: Comparison of public chromosome 22 tiling data
We carried out an initial comparison of previously published transcription maps of chromosome 22 generated from PCR-based tiling arrays (Rinn et al. 2003; White et al. 2004) and two oligonucleotide tiling-array platforms, MAS and Affymetrix (Kapranov et al. 2002; Bertone et al. 2004) (Fig. 1). These maps were generated from 15 separate experiments (tissues or cell lines). We used the RefSeq annotation (Pruitt et al. 2005) as a benchmark, since GENCODE annotation is not yet available for the entire chromosome 22. For each experiment we measured the consistency between gene annotation and transcribed regions identified by individual studies. The transcription data from oligonucleotide arrays agrees better with the RefSeq exon annotation than the data from PCR arrays, an observation that holds true across all experiments. The results have to be interpreted with some care since they were not obtained with the same biological samples or scoring schemes. Nonetheless, we conclude that PCR-based arrays are clearly less useful for a detailed transcription mapping study, possibly because of their lower genomic resolution. Spotted arrays (e.g., PCR-based) also have a significantly lower feature resolution on the array compared with arrays with in-situ synthesized probes. Therefore, we focus our subsequent experimental and analysis efforts on the oligonucleotide tiling microarrays.
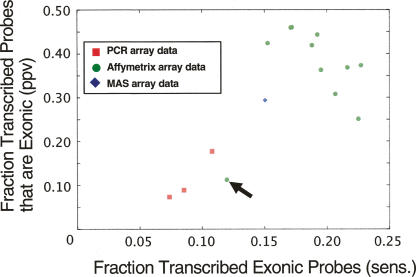
Figure 1.
Comparing human chromosome 22 transcription data sets with gene annotation. Transcription data sets were derived from previously published studies. They were generated from three different microarray platforms: PCR (red squares), MAS (blue diamond), and Affymetrix (green circles); in a total of 15 separate experiments (tissues or cell lines), each represented by a point in the figure. We used the RefSeq annotation as a benchmark to assess the quality of the data from each experiment. (x-axis) The fraction of exonic probes that were identified to be transcribed in individual experiments (sensitivity). (y-axis) The fraction of transcribed probes overlapping with an exon (PPV). The PCR tiling-array data were from placenta, fibroblast, and B-cells (Rinn et al. 2003; White et al. 2004), the MAS data from liver (Bertone et al. 2004), and the Affymetrix sets were collected from Kapranov et al. (2002) representing 11 different cell lines. (Arrow) The Affymetrix data from the U87 cell line is not representative since a long section of chromosome 22 is identified as transcriptionally silent, suggesting that this particular experiment probably did not work or something is unusual about U87.
Approach
Oligonucleotide array designs and hybridizations
An oligonucleotide array containing 36 bp oligonucleotides that tile both strands of the nonrepetitive sequence of the ENCODE regions end-to-end (allowing some positional shifts to reduce self-complementarity) was prepared using maskless photolithography, MAS (maskless array synthesis). The MAS arrays cover both strands of the ENCODE regions ENm001–ENm011 (11.6 Mb). An Affymetrix ENCODE array, which covers one strand of the entire ENCODE region on one array, tiled with 25-mer oligonucleotides with an average distance between oligonucleotide starts of 21 bases was obtained from the manufacturer. This array has both perfect match (PM) and mismatch (MM) probes.
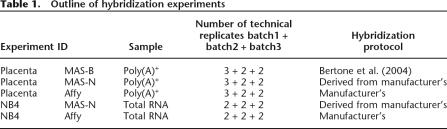
As outlined in Table 1, five different hybridization experiments were carried out: two different RNA targets (placenta poly(A)+ RNA and NB4 total RNA) were hybridized to the two different array types. (We follow the nomenclature of Royce et al. [2006, i.e., target or sample is the RNA extracted from a biological entity [tissue or cell line], which is hybridized to the probes on the microarray.) The Affymetrix arrays were hybridized according to the manufacturer’s recommendation. The MAS arrays were hybridized using two different experimental protocols, MAS-B, described in Bertone et al. (2004), and MAS-N, a variant of the manufacturer’s recommended protocol. The placental RNA was hybridized using both MAS protocols, the NB4 RNA only with MAS-N.
Table 1.
Outline of hybridization experiments
Generating comparable maps of transcriptionally active regions (TARs)
Development of consistent scoring schemes
To bring the outcomes from the two technologies MAS and Affymetrix into a comparable form, we developed ways of scoring them similarly. For each spot on the microarrays, a hybridization intensity was collected. For oligonucleotide tiling arrays, it is usually advantageous to aggregate the intensities from probes that are adjacent to each other in genomic space (Kampa et al. 2004; Cheng et al. 2005; Royce et al. 2005). This is done by applying a sliding genomic window encompassing multiple probes and converting the intensities within the window into a score, which is assigned to the middle probe. The windowed approach is logical since we are ultimately interested in obtaining a set of regions whose intensities are significantly higher than the background, and we expect those regions to be of the same length as exons (150–200 bp on average, depending on exon type) rather than of single probes (25–36 bp in this study).
We developed new ways of scoring the MAS arrays and describe these in terms of three levels of scoring: single probe intensities, robust statistics within a sliding window, and robust statistics using paired data within a sliding window (Cawley et al. 2004).
Single-probe intensities
Single-probe intensity scoring uses the raw intensities from the arrays. By wisely choosing methods and parameters to deal with the genomic segmentation (see below) it is possible to obtain reasonable results from this approach (Bertone et al. 2004). In this approach, both intra- and interarray normalization of the microarray data may be particularly important (Royce et al. 2005).
Robust nonparametric statistics within a sliding window
We used the sign test for scoring MAS array data. The sign test is attractive since it is statistically robust and does not assume normally distributed data. Comparing each intensity within a sliding genomic window of a specified size with the array median yields a measure or a score of the significance of the intensities (see Methods for details). It is easy to include multiple replicates in this scheme: Each probe is simply compared with the median intensity of its own array, and no interarray normalization is necessary. The number of available score levels is restricted; however, due to the discrete values introduced by the counting (it is a binomial), it may not be sufficient in situations in which discerning the top scores (say, top 5%) from near-top scores is important. With an average genomic spacing of 36 bp between the starts of two adjacent probes, the window (160 bp) encompasses five probes. We also applied the sign test on the Affymetrix data as a part of our comparison.
Robust nonparametric statistics using paired data within a sliding window
When paired data is available, such as the PM and MM probe intensities on Affymetrix arrays, the paired Wilcoxon signed rank test is a more powerful option than the standard sign test. It was first used with tiling microarrays by Cawley et al. (2004) to score ChIP-chip data (Horak and Snyder 2002), and it is also immediately applicable to transcription data as is shown in Kampa et al. (2004) and Cheng et al. (2005). All pairwise PM–MM differences within the window are calculated and a P-value, which essentially measures how significantly the distribution of PM–MM differences is skewed to either side around zero, is calculated, along with the corresponding point estimate (the pseudomedian). While this approach is analogous to the standard sign test, it has considerably greater statistical power.
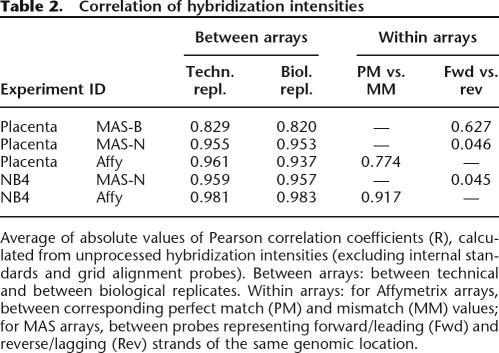
The MAS arrays did not contain proper mismatch probes. Instead, we tried to simulate these using the complementary strand oligonucleotide of the MAS arrays as the “mismatch” probe. We call this approach the Fwd-Rev scoring, and it is justified on the MAS-B (placenta) data, since the correlation between forward and reverse-strand probes is close to the correlation between PM and MM probes for the Affy placenta data (Table 2).
Table 2.
Correlation of hybridization intensities
Average of absolute values of Pearson correlation coefficients (R), calculated from unprocessed hybridization intensities (excluding internal standards and grid alignment probes). Between arrays: between technical and between biological replicates. Within arrays: for Affymetrix arrays, between corresponding perfect match (PM) and mismatch (MM) values; for MAS arrays, between probes representing forward/leading (Fwd) and reverse/lagging (Rev) strands of the same genomic location.
Segmentation of genomic regions
After obtaining one score value per oligonucleotide probe, the next step is to construct a transcription map based on these scores, i.e., to segment the genomic regions into transcribed and nontranscribed regions. We call the transcribed regions TARs (Transcriptionally Active Regions) (Rinn et al. 2003), regardless of overlap with genes, exons, or other genomic features. (Note, an alternate term, transfrag [Transcriptional Fragment], was introduced by Kampa et al. [2004).
Maxgap/minrun segmentation
In Bertone et al. (2004), TARs were generated by requiring at least five adjacent probes with a raw intensity in the top 10% of all intensities of that slide. Thus, the threshold above which to consider a probe “positive” was the intensity value corresponding to the 90th percentile, and any probe that was below the threshold immediately terminated the transcribed region. In the Affymetrix series of publications (Kapranov et al. 2002; Cheng et al. 2005), the threshold for generating TARs was based on setting a maximum false-positive rate of the hybridization levels of negative bacterial controls, thus enabling an optimized percentile cutoff for each array set and biological sample. Furthermore, gaps were allowed, such that a maximum stretch of a certain number of nucleotides (called maximal gap, or maxgap for short) with a score below the threshold was allowed between probes whose scores were above the cutoff. Typically, the maxgap parameter allows one or two probes to be below the cutoff while still being incorporated into the TAR. The total length of a TAR is then required to be of at least a certain length (a minimal run, or minrun), usually corresponding to at least two probes.
HMM segmentation
As an alternative to the maxgap/minrun segmentation, a hidden Markov model (HMM) (Rabiner 1989; Ji and Wong 2005; Li et al. 2005) was used to predict TARs, given the derived probe scores (above). Each probe can be in one of four HMM states (TAR, non-TAR, and two intermediate transition states), emitting the assigned score (i.e., the emission spectrum is continuous). The parameters of the HMM can be estimated by learning from the sequences of probes that fall into regions with known transcription characteristics (e.g., according to gene annotation). The HMM can then be applied to sequences of probes bearing the same scoring protocol to determine the most likely corresponding state sequence, in order to identify TARs (Viterbi decoding).
Platform comparison
We have analyzed the five microarray tiling experiments, representing the MAS and Affymetrix platforms, introduced in Table 1 at multiple stages throughout the data processing.
First, we calculate a correlation coefficient between the raw hybridization intensities of technical replicates within each platform to assess the level of basic experimental reproducibility. We also assess the overlap of preliminary TAR sets generated from technical replicates using single-probe intensities.
Before proceeding to the next level, the TAR sets to be compared must be determined. This includes decisions about what scoring algorithm, what segmentation method, and what corresponding parameter settings to use for each of the five experimental data sets. In summary, the input data type (e.g., PM only or PM–MM), the number of replicates, the scoring algorithm and, if applicable, its corresponding genomic window size define the scoring scheme, which together with the segmentation algorithm and its parameters specify a particular TAR set.
The resulting sets of transcribed regions are compared with each other, both within and across microarray platforms and biological samples, and the degree of overlap between detected transcription and gene annotation is measured. For the annotation comparison, we have chosen to use the GENCODE annotation, which aims at finding and verifying all protein-coding genes in the ENCODE regions. There are two main measurements: how much of the known annotated exons are covered by a detected transcribed region (sensitivity) and the degree to which the detected transcription falls within known exons (positive predictive value [PPV]). The PPV is defined as the number of nucleotides in TARs that overlap with exonic regions, divided by the total number of nucleotides in the TAR set (this is sometimes referred to as “specificity” [Burge and Karlin 1997]). The sensitivity is defined as the number of nucleotides in annotated exons that overlap with TARs divided by the total number of nucleotides in annotated exons. We do not expect a sensitivity of 100% since in any given tissue or cell line at any given time point, far from all annotated genes will be expressed. Also, we do not expect a PPV of 100% since GENCODE, although arguably the most comprehensive and accurate gene annotation available, is incomplete.
The transcription status is assessed for all 1342 annotated splice variants of all 264 known genes in the GENCODE annotation of regions ENm001–ENm01. For each splice variant, the scores of all exon overlapping probes are collected and the transcription status assessed using the sign test (by comparing each individual score with the median score) (Bertone et al. 2004). A gene is considered transcribed if at least one of its splice variants is deemed transcribed at the chosen significance level. We also assess for each transcript (with at least two exons) how many of its exons are considered transcribed based on the median intensity score of each exon within the transcript. Ideally, either all exons or no exons should be transcribed. We call this concept the multi-exon coherence, and a suitable quantitative measure is the percentage of transcribed transcripts that display all of their exons as “on.”
Finally, we perform experimental validation of the results in placenta (MAS-B and Affy), using RT–PCR on a subset of the novel TARs (including both TARs unique to a platform and TARs present in both). We also perform experimental validation of a number of genes with differing transcription status in the two platforms including, as a negative control, a set of genes that are considered off in both platforms.
The results are available at http://tiling.gersteinlab.org/platformcmp.
Outcomes
Conclusions about optimal scoring and segmentation systems
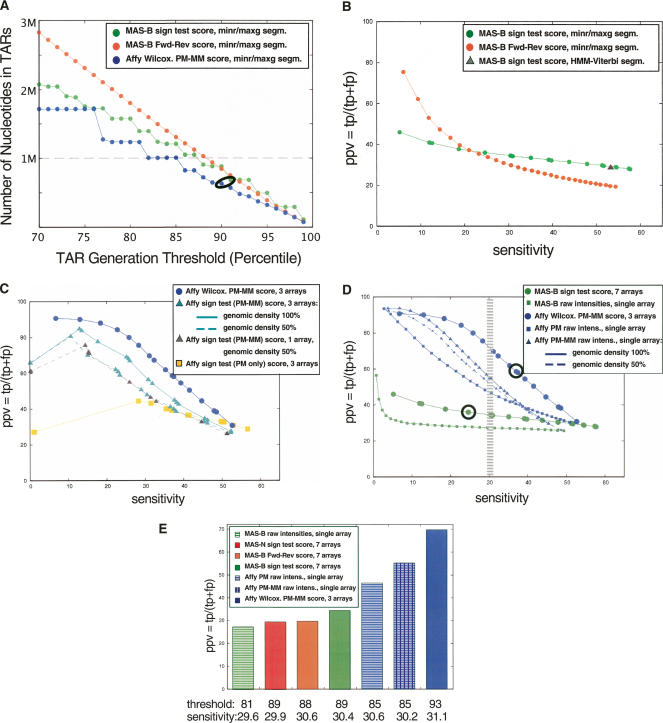
We first examined the effect of the segmentation threshold on the size of the resulting TAR sets. The results are in Figure 2A and Supplemental Figure S1, and, as expected, the lower the segmentation threshold the greater the size of the TAR sets. The step-like pattern in these figures is because of the finite number of available scores. For the minrun/maxgap segmentation, the maxgap parameter was set to 50 for the Affymetrix data and 80 for the MAS data, thus including in a TAR a probe whose score is below the score threshold if it is flanked on both sides by probes with scores above the threshold. Other maxgap settings were tested, but the results did not improve in terms of gene annotation agreement (data not shown). The minrun parameter was set to 50 bp, i.e., the minimum length of a TAR is 50 bp. Thus, at least four probes have to be included in an Affymetrix TAR and three for the MAS array TARs.
Figure 2.
(A) Number of nucleotides in placental TARs as a function of segmentation threshold (percentiles). TARs were generated with the maxgap/minrun algorithm based on the scored hybridization intensity data using a genomic window and technical replicates: MAS-B scored with the standard sign test (green); MAS Fwd-Rev scoring using reverse strand as “mismatch” (orange), pseudomedian; Affymetrix scored using pseudomedian from PM-MM (blue). The data points corresponding to the data sets used in the Comparison section are circled: Thresholds are 90th percentile for Affy and 91st percentile for MAS-B (sign test scoring). (x-axis) The percentile score threshold for calling a probe “positive.” (y-axis) The number of nucleotides in TARs (in megabase pairs). The dashed line corresponds to the number of nucleotides in exons in the analyzed region (1,001,238 nt). (B) Positive predictive value (PPV) versus sensitivity for three different ways of scoring and segmenting the MAS-B data, varying the segmentation threshold from 70th percentile (to the right in the figure) to 99th percentile (to the left) for the MAS-B set scored with the standard sign test (green); scored using reverse strand as “mismatch” (Fwd-Rev scoring, orange); and the result from HMM segmentation (Viterbi decoding) of sign test-scored data (gray triangle). Sensitivity (x-axis), defined as the percentage of bases in GENCODE exonic regions that are covered by a TAR. PPV (y-axis), defined as the percentage of bases in the TARs that overlap with a GENCODE exonic region. (C) PPV versus sensitivity for two different ways of scoring the placenta Affy data, using three replicates (six array features) unless otherwise stated: Wilcoxon signed rank test (blue circles), and standard sign test (using PM–MM values: cyan triangles; using PM-only values: yellow squares). The result from reducing the genomic density of the Affy array to 50% (i.e., removing the data from every second probe) is also shown using PM–MM values (three replicates: cyan triangles, dashed line; single replicate only: gray triangles, dashed line). (D) PPV versus sensitivity for MAS-B and Affy placenta data, varying the segmentation threshold from the 70th percentile (right) to the 99th percentile (left). The average results of TARs generated from raw intensities from single arrays for Affy (PM only [blue squares], and PM–MM [blue triangles; solid line actual genomic density, dashed line 50% genomic density]) and MAS-B (green squares) are plotted, as well as scored results for Affy (blue circles) and MAS-B (green circles). Sensitivity (x-axis), and PPV (y-axis), defined as above. The data points corresponding to the data sets used in the Comparison section are circled. The hatched area marks where a sensitivity of 30% is achieved for the various sets. (E) PPV for placental TAR sets when choosing a segmentation threshold that yields ∼30% sensitivity (hatched area in B). Note that the actual sensitivity varies slightly between the sets.
Different scoring schemes give different results and also differ from the results obtained using single-probe intensity scores when comparing to the gene annotation. As is clear from Figure 2B (and Supplemental Fig. S2a), the standard sign test provides the best performance for MAS-B (placenta) if a sensitivity at or above 25% is required, but its improvement in terms of PPV when increasing the segmentation threshold is modest. The Fwd-Rev scoring is more sensitive to the choice of threshold, and it performs better than the sign test scoring for segmentation thresholds above the 94th percentile. For the standard sign test, we also tried applying different weights to the probes within a window, e.g., multiplying each score with a discretized Gaussian, but no improvement in performance was recorded (Supplemental Fig. S2).
For the Affymetrix data, Figure 2C (placenta) and Supplemental Figure S3 (NB4) reveal that the use of mismatch probes improves performance, in particular for the NB4 total RNA experiment, and that the Wilcoxon scoring performs very well. Figure 2C shows that for sensitivities up to 35%, using mismatch probes is a better strategy than doubling the genomic density of the probes, and also that it is better to use a single array with a PM–MM setup than two replicates with PM probes only.
Figure 2D and Supplemental Figure S3 show that the more elaborate scoring models using replicates outperform the single-probe intensity scoring in terms of sensitivity and PPV for both MAS and Affy.
These two points (elaborate scoring with replicates and the advantage of using mismatches) are further illustrated in Figure 2E, where the PPV of the TAR sets when choosing a segmentation threshold that corresponds to a sensitivity of 30% have been plotted.
The analysis of different segmentation algorithms reveals that the TAR sets generated by the nonparametric HMM segmentation (Viterbi decoding) are biased toward a high sensitivity (Fig. 2B; Supplemental Fig. S2) where it performs on par with the minrun/maxgap algorithm.
Results from comparison pipeline
Replicate comparison of unprocessed hybridization intensities
As is shown in Table 2, we obtained Pearson correlation coefficients of 0.83 and 0.96 for placenta MAS-B and MAS-N data, respectively, measured on pairwise comparison of the raw hybridization intensities of the arrays. The figure for Affymetrix was 0.96 and NB4 results were similar. We also note that the correlation of PM and MM probes for Affy placenta is close to the correlation of Fwd and Rev probes for MAS-B. Comparing the preliminary TAR sets, generated from single arrays, across technical replicates (Supplemental Table S1) again indicates that the MAS-B data is the most variable.
Choosing TAR sets to include in comparison
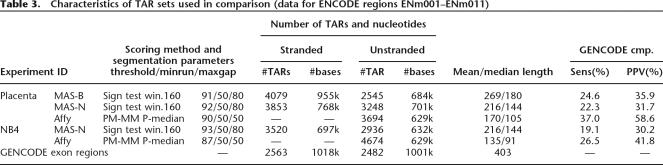
For each of the five experiments, the best-performing scoring and segmentation algorithm was chosen, and the segmentation threshold was tuned to generate TAR sets of roughly equal size, as measured in number of bases. The chosen sets are presented in Table 3 (and its extended version, Supplemental Table S2) and the points corresponding to these sets in Figure 2A and Supplemental Figure S1 have been circled. To enable a comparison of the TAR sets, MAS array TARs on the two strands were merged into one set of unstranded TARs for each biological sample (Affymetrix TARs do not have strand information). For MAS, the standard sign test scoring was chosen, and for Affymetrix, the Wilcoxon signed rank test (pseudo-median). The segmentation thresholds range from the 87th to the 93rd percentiles for the various sets. The resulting sizes of the sets included in the subsequent comparison range from 629 to 701 kb (between 2545 and 4674 TARs). The total number of bases in exons in the analyzed regions (ENm001–ENm011) is 1001 kb, which means that the chosen sets can reach a sensitivity of 63%–70% at most (as discussed above). The length distributions of all five TAR sets are unimodal and decay roughly exponentially (Supplemental Fig. S6).
Table 3.
Characteristics of TAR sets used in comparison (data for ENCODE regions ENm001–ENm011)
Compare TAR sets with each other and to GENCODE annotation and conserved regions
Figure 2D and Supplemental Figure S3 reveal that the agreement with annotation is better for the Affy sets than for the MAS sets, both placenta and NB4. While similar sensitivity levels are achievable, the Affy TAR sets reach significantly higher PPVs. Likewise, the agreement is larger for the placenta sets (MAS-B, MAS-N, and Affy) than for the NB4 sets (MAS-N and Affy). This is also summarized in Table 3.
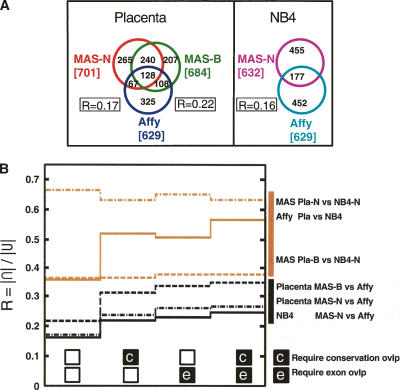
Figure 3A shows the overlap between the placenta and NB4 TAR sets from the different experiments. As a measure of the overlap between the sets, we calculate a ratio R = |∩|/|U| for each pairwise comparison, where the numerator represents the size of the intersection and the denominator represents the size of the union of the two sets under comparison. For two sets that agree completely, R = 1. We find that the MAS-B placenta TAR set agrees better with the Affy placenta TAR set (R = 0.22) than the MAS-N TAR sets do with Affy (R is 0.16–0.17). Between 62% and 72% of the nucleotides in the placenta sets are exclusive to a particular experiment (pairwise comparison MAS-B vs. Affy and MAS-N vs. Affy). For the NB4 total RNA TAR sets (MAS-N and Affy), more than 70% of the nucleotides in either TAR set are exclusive to that set.
Figure 3.
TAR set agreement. (A) Overlap of TAR sets, measured in number of overlapping nucleotides (kilobases). All three placenta TAR sets (MAS-B, MAS-N, Affy) and both NB4 TAR sets (MAS-N and Affy). R is a measure of the size of the overlap. R = |∩|/|U| (calculated pairwise for the three placenta TAR sets). (B) Size of TAR set overlap, expressed in R, for comparisons within biological samples but across different array platforms (black lines), and comparisons within array platforms but across the biological samples (brown lines). Values in the leftmost column of the graph are calculated with no further constraints. Second column, only TARs overlapping with conserved regions are included. Third column, only TARs overlapping with GENCODE exons are included. Fourth column, only TARs overlapping with both conserved and exon regions are included.
As shown in Figure 3B, the overlap across the different biological samples within each experimental technology is larger than the overlap within the same biological sample between the two experimental technologies. An extreme example is the MAS-N placenta and NB4 sets which agree much better (dashed-dotted brown line; R = 0.67) than NB4 MAS-N and Affy (solid black line; R = 0.16). Restricting the overlap calculations to the subset of TARs that overlap conserved or exonic regions, or both (i.e., moving to the right in Fig. 3B; Supplemental Fig. S8) yields higher values of R for the within-biological sets comparisons (black) and for the within-Affy comparisons (solid brown), but not for the within-MAS comparisons (nonsolid brown). Consequently, there is no enrichment for conserved regions or known genes within the common parts of the MAS TAR sets.
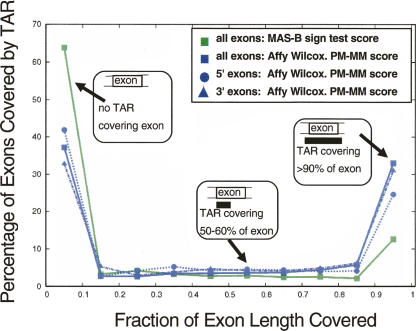
The bimodal distribution in Figure 4 shows that most GENCODE unique exons are either fully covered by a TAR (>90% of exon nucleotides overlap with a TAR) or not covered at all (<10%). This is true for all TAR sets (Supplemental Fig. S9). We notice a slight 3′ bias for the Affymetrix poly(A)+ data (but not for the MAS data), detecting 32% of the 3′ exons and 25% of the 5′ exons entirely.
Figure 4.
Distribution of GENCODE exon coverage by placenta TARs: all exons (MAS-B, green squares, and Affy, blue squares); 5′ exons (Affy, blue circles); 3′ exons (Affy, blue triangles). (x-axis) The fraction to which an exon is covered by a TAR; 0.0–1.0 split up in 10 bins. (y-axis) The percentage of exons covered by a TAR to the fraction represented on the x-axis.
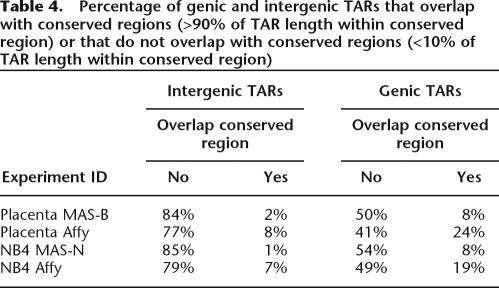
Table 4 and Supplemental Figure S10 show a comparison of each of the five TAR sets to a set of conserved elements, generated from the union of conserved regions called by the Threader Blockset Aligner (TBA) (Blanchette et al. 2004) and MLagan (Brudno et al. 2003). The union set of conserved elements covers ∼10% of the ENCODE regions. We find that most of the novel (intergenic) TARs do not overlap with conserved regions. Only 7%–8% of Affymetrix novel TARs and 1%–2% of MAS novel TARs overlap fully (>90%) with conserved regions.
Table 4.
Percentage of genic and intergenic TARs that overlap with conserved regions (>90% of TAR length within conserved region) or that do not overlap with conserved regions (<10% of TAR length within conserved region)
Transcription status of known genes and exons
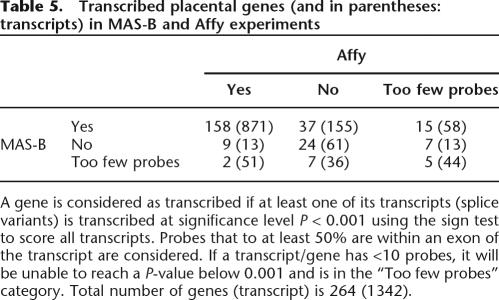
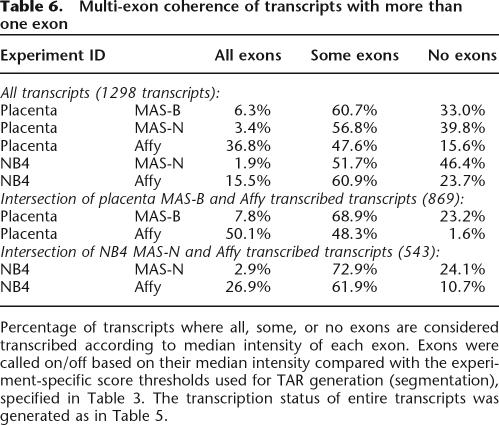
The transcription status of all known splice variants (transcripts) of the 264 GENCODE genes in the regions ENm001–ENm011 was assessed, and the results are shown in Table 5. A gene is considered as “transcribed” if at least one of its transcripts is detected at significance level P < 0.001. If a transcript has <10 probes, it will be unable to reach a P-value below 0.001, and if this is true for all splice variants of a gene, that gene is in the “Too few probes” category. In total, 158 (69.3%) genes are considered transcribed according to both platforms and 221 (83.7%) according to at least one platform (similar percentages on the transcript level). In Table 6, the multi-exon coherence (either all exons on or all exons off) is assessed and found to be higher for Affy. For both placenta and NB4 there is an enrichment of multi-exon coherence in transcripts that are considered as transcribed in both platforms. One example is the Affy NB4 set, for which, in total, 15.5% of all transcripts have all their exons transcribed, while 26.9% of the transcripts that are on in both NB4 sets have all their exons transcribed. The difference in score distribution between exons and introns is assessed (Supplemental Fig. S7) and for all five sets exons are indeed overrepresented at the high end of the score spectrum, but also many introns have high scores.
Table 5.
Transcribed placental genes (and in parentheses: transcripts) in MAS-B and Affy experiments
A gene is considered as transcribed if at least one of its transcripts (splice variants) is transcribed at significance level P < 0.001 using the sign test to score all transcripts. Probes that to at least 50% are within an exon of the transcript are considered. If a transcript/gene has <10 probes, it will be unable to reach a P-value below 0.001 and is in the “Too few probes” category. Total number of genes (transcript) is 264 (1342).
Table 6.
Multi-exon coherence of transcripts with more than one exon
Percentage of transcripts where all, some, or no exons are considered transcribed according to median intensity of each exon. Exons were called on/off based on their median intensity compared with the experiment-specific score thresholds used for TAR generation (segmentation), specified in Table 3. The transcription status of entire transcripts was generated as in Table 5.
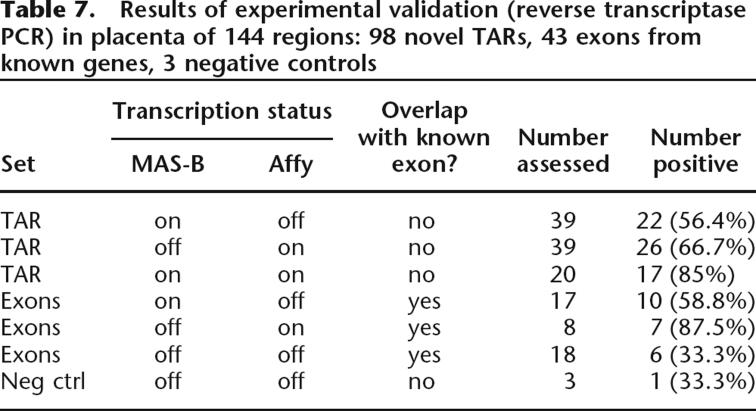
Experimental validation of novel TARs and known genes
Experimental validation of the microarray transcription data is crucial to the interpretation of the results. Table 7 shows that we used RT–PCR to assess, in total, 144 regions experimentally in placenta. Of these, 98 were novel TARs (no overlap with known genes). The experiments verified the presence of 56.4% (22/39) of the assayed novel TARs that were exclusively found on the MAS platform (MAS-B), 66.7% (26/39) of the novel TARs that were exclusively found on the Affymetrix platform, and 85% (17/20) of the assessed novel TARs that were common to both. In total, 66.3% of all assessed novel TARs were verified.
Table 7.
Results of experimental validation (reverse transcriptase PCR) in placenta of 144 regions: 98 novel TARs, 43 exons from known genes, 3 negative controls
Forty-three known genes were also validated. Genes that were completely off (i.e., none of their splice variants were considered transcribed) according to one of the platforms, but not the other, were assessed. In total, 58.8% (10/17) of the MAS-B exclusive genes were verified and 87.5% (7/8) of the Affymetrix exclusive genes were verified. For genes that were considered “off” in both platforms, 33.3% (6/18) were found in our experimental validation.
Discussion
In this work we have attempted to assess the suitability of two oligonucleotide tiling microarray strategies for transcription mapping in human. We tried to overcome the inherent differences between the approaches through using the same biological samples and a unified scoring and TAR generation procedure, and we have produced, compared, and validated several sets of transcribed regions. We conclude that many factors are significant for the outcome of the experiments. Here, we elaborate on some key findings.
Arrays are noisy
In the comparison between the two microarray tiling platforms, the Affymetrix platform yielded TARs that better agreed with the GENCODE annotation (Figs. 2D,E, 4; Supplemental Fig. S3). A simple explanation for this would be a higher noise level for MAS arrays. Moreover, given that the Pearson’s correlation coefficients between raw intensities of technical replicates of MAS-N and Affymetrix arrays (Table 2; see also Supplemental Table S1) are similar, it is likely that the MAS-N noise was rather systematic than random, while the MAS-B data seem to have a larger component of random noise. The systematic noise hypothesis is further supported by the observation that the overlap of TARs is larger within platform than within biological sample (Fig. 3). The noise could result, e.g., from probe sequence artifacts, sample contamination (after it was split into different aliquots for the experiments), suboptimal hybridization parameters, or protocol-dependent labeling artifacts (Nazarenko et al. 2002). A related issue is the cross-hybridization. For a transcription experiment, the amount of different RNA species present in the sample is large. Since the target RNA is derived from the entire genome, cross-hybridization is potentially present at high levels. Longer probes can be hybridized at higher temperatures and are thus less sensitive to cross-hybridization, but at a given temperature, these probes are more susceptible to nonspecific binding.
A comparison of the NB4 total RNA and placenta poly(A)+ TAR sets within the two platforms (Tables 2–4, 6; Fig. 3) revealed that the agreement between the platforms, and between the results of each platform and annotation, is larger for the poly(A)+ sets. One possible reason is that for total RNA introns may be labeled. This would explain the worse performance of the NB4 sets compared with placenta, but not the differences in performance between MAS and Affy arrays. The exon and intron score distributions (Supplemental Fig. S7) show that for both placenta poly(A)+ and NB4 total RNA, exons in general have higher scores than introns. For both MAS and Affy experiments there is a slight shift of the total RNA intron score distribution toward higher scores, as compared with poly(A)+ distribution, indicating the presence of intron labeling.
Counteract the noise: More data, appropriate scoring
Figure 2D,E show that using more replicates enables TAR sets that better agree with annotation. Figure 2C shows that the genomic probe density also is important—reducing the genomic density of the Affy array to 50% (excluding every other probe on the arrays) worsens the performance, specifically for sensitivities below 40%–45%. Furthermore, the density determines how well the endpoints of the TARs can be defined. The theoretical uncertainty of where a transcribed region starts and ends has an upper limit in the genomic distance between two adjacent probes. It also influences the results through the scoring procedure, where often a genomic window is used for the statistical calculations. A window that is significantly larger than the average size of an exon is not desired, since it would likely contain both probes that represent actual transcription (exons) and probes that belong to truly nontranscribed regions (introns). Taken together, we conclude that the number of recorded data points per genomic unit is a crucial parameter for tiling microarray transcription mapping—the more data the better results.
To take advantage of the data, appropriate probe scoring procedures are needed. We tried several scoring schemes for our array data and found, in Figure 2D,E, that statistically based scoring using replicates and a genomic window can significantly improve the results compared with using single-probe intensities. Using the standard sign test scoring was ultimately deemed the best way to score the MAS data (in particular the more noisy MAS-B data), while the best way to score the Affy data was to use the pseudomedian from the Wilcoxon signed rank test. For MAS-B data, using a Fwd-Rev scoring algorithm as a surrogate for a true PM–MM scoring improved agreement with annotation for high-segmentation thresholds in the maxgap/minrun algorithm (Fig. 2B). Exploring the Affy data showed that the sign test did not perform particularly well using PM-only data as input, but quite well using PM–MM (Fig. 2C). In fact, the PM-only Affy data scored with the standard sign test (i.e., identical scoring as the MAS sign test) resulted in a sensitivity/PPV behavior very similar to that of MAS—a relatively low agreement with annotation, and reduced impact of increasing the segmentation threshold (PPV insensitive to threshold increases). These results indicate that mismatches can be very useful. Altogether, we conclude that regardless of array design, statistically based scoring, taking into account the available data in an appropriate way, is indispensable in the analysis of tiling microarray data.
In Figure 2D we also analyzed the trade-off between increasing the genomic density of probes versus using the array space for MM probes. We found that in the investigated genomic density and sensitivity ranges, it is better to use half of the array features for MM probes (one PM/MM probe pair every 42 nt) than to double the genomic density (to 21 nt) and use PM probes only. This is true for both scored (sign test) (Fig. 2C) and unscored (Fig. 2D) data. From Figure 2C we also observe that using a single-array PM–MM setup is actually preferable to using technical replicates of PM-only data for sensitivities up to 40% (retaining the genomic density and using sign test scoring). These findings suggest that true mismatch probes is a straightforward way to significantly improving the signal-to-noise ratio of oligonucleotide tiling arrays.
Conclusions from the array platform comparison
According to our study, the current form of the Affymetrix tiling microarray platform is better suited than the MAS platform for detailed transcription mapping of the human genome. This is true in the sense that the agreement of the TARs with known annotation is larger (Fig. 2), and also in the sense that the exons in multiple-exon transcripts are more coherently transcribed (Table 6). From our study, we attribute this foremost to the higher genomic density of the probes and the presence of mismatch probes and how these can be used to reduce the impact of nonspecific hybridization. However, we cannot entirely exclude the effects of the differing labeling and hybridization protocols. On the other hand, the two technologies are almost equal in their ability to detect novel transcription, as indicated by our experimental validation of novel TARs: In total 66.1% of the novel MAS-B and 72.9% of the novel Affymetrix placenta TARs are validated using RT–PCR (Table 7). TARs supported by both platforms are even more reliable and 85% of these are validated. The overlap of genes that are considered transcribed by the different platforms is substantial. Experimental validation of a subset of the genes considered transcribed by only one of the two platforms indicated that the Affymetrix setup is ahead of MAS in this respect as well, although the sample sizes are relatively small. It is also clear that if a gene is not detected by either platform, it is less likely to actually be transcribed. Our validation study shows that the two technologies are complementary, since much transcription detected by only one array platform is in fact verified as transcribed. They also reinforce each other, in the sense that array-based transcriptional evidence (or lack thereof) from both platforms yields more reliable results.
While the results obtained from the Affy arrays agree better with the annotation and the validation results, the advantage of the MAS technology is that it allows for rapid manufacturing of customized designs and cost-effective production of small array series. Using true mismatches in the MAS design may improve the results for MAS arrays as well, but there are currently no results publicly available. We conclude that oligonucleotide tiling microarrays are suitable to detect novel transcribed regions, and that the use of replicates and statistically based scoring schemes significantly improves the performance for all investigated oligonucleotide-tiling microarray-based transcription-mapping experiments
Methods
Array designs
Affymetrix arrays
Arrays were designed and manufactured by Affymetrix, Inc., using a physical mask. Probes are 25-bp long with an average genomic spacing of 21 bp, and they cover one genomic strand, with the exception of repeat regions, as defined by RepeatMasker (A.F.A. Smit and P. Green, unpubl.). Each probe is present in a “perfect match” and a “mismatch” version. The mismatch probe contains a single substitution at the middle probe position (A→T, T→A, C→G, G→C). Each array contains in total ∼1,400,000 features.
MAS arrays
Arrays were designed by us and manufactured by NASA using a NimbleGen maskless array synthesizer. Probes are 36-bp long with an average genomic spacing of 36 bp. Positional shifts were allowed to avoid self-complementarity at the probe ends (defined as at least four consecutive complementary nucleotides within the six 5′/3′ nucleotides). The probes cover both genomic strands, with the exception of repeat regions. The design was done on the NCBI v34 of the human genome build, and each array contains almost 390,000 features.
RNA extraction and array hybridization
Cell culture
The human NB4 cells were cultured in RPMI medium containing 20 mM L-glutamine (Media Tech) and supplemented with 10% fetal bovine serum (Invitrogen), 100 IU/mL penicillin (Media Tech) and 100 μg/mL streptomycin (Media Tech). Cells were maintained at 37°C under 5% CO2/95% air in a humidified incubator.
RNA samples
Total RNA from the human NB4 cells was extracted using a Qiagen RNA extraction kit according to the manufacturer’s instructions. Human placental poly(A)+ mRNA (obtained from total RNA) was purchased from Ambion.
Protocols
The Supplemental material contains a detailed description of all three experimental protocols (MAS-B, MAS-N, Affy). The MAS-N protocol yields in-vitro transcribed, biotin-labeled, single-stranded cRNA (Van Gelder et al. 1990), fragmented to an average size of 50–200 bp before hybridization. The MAS-B protocol yields Cy3-aminoallyl-labeled unfragmented single-stranded cDNA. The Affymetrix protocol yields end-labeled (bio-ddATP) double-stranded cDNA, fragmented to an average size of 50–100 bp before hybridization.
Scoring schemes
To obtain the desired statistical resolution, MAS array scoring was done pooling the data from all three biological samples (for both placenta and NB4). For placenta, this corresponds to seven measurements for each probe and six for NB4. For Affymetrix, three technical replicates were used, corresponding to six measurements for each probe (three PM and three MM probes).
Sign test using array median intensity
The intensity of every probe within the window is compared with the median intensity of the slide and assigned a “1” if it is above and “0” otherwise. The number of ones within the window is counted and the probability P of finding at least this number of 1’s under the null hypothesis that half of the probes should be above the median is calculated. The score assigned to the probe in the middle of the window is then defined as score = −log(P). Window sizes of 90–240 bp were tried, choosing 160 (five probes) for the MAS data in this study. No inter-array normalization is performed, since each intensity is compared with the median intensity on its own array only. A variant of this scoring approach is to weight the probes within the window differently, such that the central probe(s) becomes more important. For instance, the intensities within the window can be multiplied with a discretized Gaussian envelope. Several parameter settings were tried (Supplemental Figs. S2–S5).
Paired Wilcoxon signed rank sum test
Inter-array normalization is undertaken through dividing each intensity with the array median (median normalization). Within a window, all pairwise differences between the intensities of a perfect match probe and its corresponding mismatch probe are calculated and ranked. A sign is assigned to each rank number depending on whether the PM or the MM intensity was greater, and a P-value is calculated from the sum of this signed ranking (keeping track of the rank sum of all negative ranks and the rank sum of all positive ranks). The P-value, which is a measure of how significantly the distribution of PM–MM differences is skewed to either side around zero, can then be used to compute the final score for the probe in the middle of the window (Kampa et al. 2004; Royce et al. 2005). The corresponding point estimate, the pseudomedian, is obtained by taking the median value of all the pairwise averages of PM–MM values within the window. The Affymetrix scores used in this study were calculated by Affymetrix using a window size of 101 nt, corresponding to on average five probes in the window. For MAS arrays, the paired Wilcoxon signed rank test was applied using the probe corresponding to the reverse strand of the exact same genomic locus as mismatch probe (instead of a designed mismatch probe).
Segmentation of genomic regions
Maxgap/minrun segmentation
The transcribed regions were generated from scored data. The maxgap parameter was set to 50 for Affymetrix data and 80 for MAS data. The minrun parameter was set to 50 for both approaches. Other maxgap/minrun parameter settings were also tested (data not shown). We evaluated segmentation thresholds of the 70–99th percentile.
HMM segmentation
The emission and transition probability distributions of the four-state HMM for each data set were learned according to the scores of those probes that fall into known gene regions, where the score characteristics in the exon regions were used to estimate the parameters for the TAR state, and those in the intron regions for the non-TAR state. The parameters for the two intermediate transition states were obtained by investigating those probes containing both exon and intron regions. These emission distributions were fitted with mixed-Gaussian distributions to generate a continuous model. The Viterbi algorithm was utilized to identify TARs.
Assessing transcription of annotated genes
The transcription status was assessed using the sign test as described for all annotated splice variants of all known genes in the GENCODE annotation of regions ENm001–ENm011, accepting the exons with labels “VEGA_known,” “VEGA_Novel_CDS,” “VEGA_Novel_transcript_gencode_conf,” and “VEGA_Putative_ gencode_conf.” For the exon/intron-based investigations (Table 6; Supplemental Fig. S7), the median probe score for each feature was used, with a percentile threshold for on/off calls as defined for each experiment in Table 3.
Choosing primer pairs for validation
Primer pairs were generated using Primer3 (Rozen and Skaletsky 2000). Primers assessing novel TARs were required to define a genomic region with no overlap with any GENCODE gene. When assessing known genes, the exon with the highest P-value-based transcription score was chosen. Primer3 settings were as default or more stringent, e.g., GC content within 35%–65%, primer size was forced to be between 20 and 28 nt, and the resulting PCR products to be between 100 and 200 bp. Validation candidates were checked using UCSC In Silico PCR (http://genome.ucsc.edu/cgi-bin/hgPcr) against the NCBI v35 human genome build to ensure that exactly one PCR product was possible; those that generated no or multiple hits were discarded. Three regions that did not contain any verified or predicted transcription were chosen to act as negative controls. The experimental protocol of the PCR validation is in the Supplemental material.
Accessing data and results
The MAS ENCODE array platform has GEO (Gene Omnibus Expression, http://www.ncbi.nlm.nih.gov/geo/) accession number GPL2105; the corresponding data series has GEO accession number GSE2720 (placenta and untreated NB4). The Affymetrix anti-sense ENCODE array platform has GEO accession number GPL1789; the corresponding data series has accession number GSE2671 (placenta) and GSE2679 (untreated NB4). The TAR sets, the gene/transcript/exon transcription status, the validation results, and the raw data are available at or from http://tiling.gersteinlab.org/platformcmp.
Acknowledgments
O.E. is supported by a Knut and Alice Wallenberg Foundation postdoctoral fellowship. We acknowledge support from the NIH (1U01HG003156-01).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.5014606
References
- Ashurst J.L., Chen C.K., Gilbert J.G., Jekosch K., Keenan S., Meidl P., Searle S.M., Stalker J., Storey R., Trevanion S., Chen C.K., Gilbert J.G., Jekosch K., Keenan S., Meidl P., Searle S.M., Stalker J., Storey R., Trevanion S., Gilbert J.G., Jekosch K., Keenan S., Meidl P., Searle S.M., Stalker J., Storey R., Trevanion S., Jekosch K., Keenan S., Meidl P., Searle S.M., Stalker J., Storey R., Trevanion S., Keenan S., Meidl P., Searle S.M., Stalker J., Storey R., Trevanion S., Meidl P., Searle S.M., Stalker J., Storey R., Trevanion S., Searle S.M., Stalker J., Storey R., Trevanion S., Stalker J., Storey R., Trevanion S., Storey R., Trevanion S., Trevanion S., et al. The vertebrate genome annotation (Vega) database. Nucleic Acids Res. 2005;33:D459–D465. doi: 10.1093/nar/gki135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertone P., Stolc V., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Stolc V., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Tongprasit W., Samanta M., Weissman S., Samanta M., Weissman S., Weissman S., et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- Bertone P., Trifonov V., Rozowsky J.S., Schubert F., Emanuelsson O., Karro J., Kao M.-Y., Snyder M., Gerstein M., Trifonov V., Rozowsky J.S., Schubert F., Emanuelsson O., Karro J., Kao M.-Y., Snyder M., Gerstein M., Rozowsky J.S., Schubert F., Emanuelsson O., Karro J., Kao M.-Y., Snyder M., Gerstein M., Schubert F., Emanuelsson O., Karro J., Kao M.-Y., Snyder M., Gerstein M., Emanuelsson O., Karro J., Kao M.-Y., Snyder M., Gerstein M., Karro J., Kao M.-Y., Snyder M., Gerstein M., Kao M.-Y., Snyder M., Gerstein M., Snyder M., Gerstein M., Gerstein M. Design optimization methods for genomic DNA tiling arrays. Genome Res. 2006;16:271–281. doi: 10.1101/gr.4452906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Kent W.J., Riemer C., Elnitski L., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Riemer C., Elnitski L., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Elnitski L., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Rosenbloom K., Clawson H., Green E.D., Clawson H., Green E.D., Green E.D., et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudno M., Do C., Cooper G., Kim M.F., Davydov E., Green E.D., Sidow A., Batzoglou S., Do C., Cooper G., Kim M.F., Davydov E., Green E.D., Sidow A., Batzoglou S., Cooper G., Kim M.F., Davydov E., Green E.D., Sidow A., Batzoglou S., Kim M.F., Davydov E., Green E.D., Sidow A., Batzoglou S., Davydov E., Green E.D., Sidow A., Batzoglou S., Green E.D., Sidow A., Batzoglou S., Sidow A., Batzoglou S., Batzoglou S. LAGAN and Multi-LAGAN: Efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 2003;13:721–731. doi: 10.1101/gr.926603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C., Karlin S., Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Cawley S., Bekiranov S., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Bekiranov S., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Sementchenko V., Cheng J., Williams A.J., Cheng J., Williams A.J., Williams A.J., et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- Cheng J., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Patel S., Long J., Stern D., Tammana H., Helt G., Long J., Stern D., Tammana H., Helt G., Stern D., Tammana H., Helt G., Tammana H., Helt G., Helt G., et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, The ENCODE (ENCyclopedia Of DNA Elements) project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- Guigo R., Dermitzakis E.T., Agarwal P., Ponting C.P., Parra G., Reymond A., Abril J.F., Keibler E., Lyle R., Ucla C., Dermitzakis E.T., Agarwal P., Ponting C.P., Parra G., Reymond A., Abril J.F., Keibler E., Lyle R., Ucla C., Agarwal P., Ponting C.P., Parra G., Reymond A., Abril J.F., Keibler E., Lyle R., Ucla C., Ponting C.P., Parra G., Reymond A., Abril J.F., Keibler E., Lyle R., Ucla C., Parra G., Reymond A., Abril J.F., Keibler E., Lyle R., Ucla C., Reymond A., Abril J.F., Keibler E., Lyle R., Ucla C., Abril J.F., Keibler E., Lyle R., Ucla C., Keibler E., Lyle R., Ucla C., Lyle R., Ucla C., Ucla C., et al. Comparison of mouse and human genomes followed by experimental verification yields an estimated 1,019 additional genes. Proc. Natl. Acad. Sci. 2003;100:1140–1145. doi: 10.1073/pnas.0337561100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horak C., Snyder M., Snyder M. ChIP-chip: A genomic approach for identifying transcription factor binding sites. Methods Enzymol. 2002;350:469–483. doi: 10.1016/s0076-6879(02)50979-4. [DOI] [PubMed] [Google Scholar]
- Hughes T.R., Mao M., Jones A.R., Burchard J., Marton M.J., Shannon K.W., Lefkowitz S.M., Ziman M., Schelter J.M., Meyer M.R., Mao M., Jones A.R., Burchard J., Marton M.J., Shannon K.W., Lefkowitz S.M., Ziman M., Schelter J.M., Meyer M.R., Jones A.R., Burchard J., Marton M.J., Shannon K.W., Lefkowitz S.M., Ziman M., Schelter J.M., Meyer M.R., Burchard J., Marton M.J., Shannon K.W., Lefkowitz S.M., Ziman M., Schelter J.M., Meyer M.R., Marton M.J., Shannon K.W., Lefkowitz S.M., Ziman M., Schelter J.M., Meyer M.R., Shannon K.W., Lefkowitz S.M., Ziman M., Schelter J.M., Meyer M.R., Lefkowitz S.M., Ziman M., Schelter J.M., Meyer M.R., Ziman M., Schelter J.M., Meyer M.R., Schelter J.M., Meyer M.R., Meyer M.R., et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 2001;19:342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- Jarvinen A.-K., Hautaniemi S., Edgren H., Auvinen P., Saarela J., Kallioniemi O.P., Monni O., Hautaniemi S., Edgren H., Auvinen P., Saarela J., Kallioniemi O.P., Monni O., Edgren H., Auvinen P., Saarela J., Kallioniemi O.P., Monni O., Auvinen P., Saarela J., Kallioniemi O.P., Monni O., Saarela J., Kallioniemi O.P., Monni O., Kallioniemi O.P., Monni O., Monni O. Are data from different gene expression microarray platforms comparable? Genomics. 2004;83:1164–1168. doi: 10.1016/j.ygeno.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Ji H., Wong W.H., Wong W.H. TileMap: Create chromosomal map of tiling array hybridizations. Bioinformatics. 2005;21:3629–3636. doi: 10.1093/bioinformatics/bti593. [DOI] [PubMed] [Google Scholar]
- Kampa D., Cheng J., Kapranov P., Yamanaka M., Brubaker S., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Cheng J., Kapranov P., Yamanaka M., Brubaker S., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Kapranov P., Yamanaka M., Brubaker S., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Yamanaka M., Brubaker S., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Brubaker S., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Piccolboni A., Bekiranov S., Helt G., Bekiranov S., Helt G., Helt G., et al. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 2004;14:331–342. doi: 10.1101/gr.2094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P., Cawley S.E., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Cawley S.E., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Strausberg R.L., Fodor S.P., Gingeras T.R., Fodor S.P., Gingeras T.R., Gingeras T.R. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- Larkin J.E., Frank B.C., Gavras H., Sultana R., Quackenbush J., Frank B.C., Gavras H., Sultana R., Quackenbush J., Gavras H., Sultana R., Quackenbush J., Sultana R., Quackenbush J., Quackenbush J. Independence and reproducibility across microarray platforms. Nat. Methods. 2005;2:337–343. doi: 10.1038/nmeth757. [DOI] [PubMed] [Google Scholar]
- Li W., Meyer C.A., Liu X.S., Meyer C.A., Liu X.S., Liu X.S. A hidden Markov model for analyzing ChIP-chip experiments on genome tiling arrays and its application to p53 binding sequences. Bioinformatics. 2005;21:i274–i282. doi: 10.1093/bioinformatics/bti1046. [DOI] [PubMed] [Google Scholar]
- Lipshutz R.J., Fodor S.P., Gingeras T.R., Lockhart D.J., Fodor S.P., Gingeras T.R., Lockhart D.J., Gingeras T.R., Lockhart D.J., Lockhart D.J. High density synthetic oligonucleotide arrays. Nat. Genet. 1999;21:20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- Mah N., Thelin A., Lu T., Nikolaus S., Kuhbacher T., Gurbuz Y., Eickhoff H., Kloppel G., Lehrach H., Mellgard B., Thelin A., Lu T., Nikolaus S., Kuhbacher T., Gurbuz Y., Eickhoff H., Kloppel G., Lehrach H., Mellgard B., Lu T., Nikolaus S., Kuhbacher T., Gurbuz Y., Eickhoff H., Kloppel G., Lehrach H., Mellgard B., Nikolaus S., Kuhbacher T., Gurbuz Y., Eickhoff H., Kloppel G., Lehrach H., Mellgard B., Kuhbacher T., Gurbuz Y., Eickhoff H., Kloppel G., Lehrach H., Mellgard B., Gurbuz Y., Eickhoff H., Kloppel G., Lehrach H., Mellgard B., Eickhoff H., Kloppel G., Lehrach H., Mellgard B., Kloppel G., Lehrach H., Mellgard B., Lehrach H., Mellgard B., Mellgard B., et al. A comparison of oligonucleotide and cDNA-based microarray systems. Physiol. Genomics. 2004;16:361–370. doi: 10.1152/physiolgenomics.00080.2003. [DOI] [PubMed] [Google Scholar]
- Mathews D.H., Burkard M.E., Freier S.M., Wyatt J.R., Turner D.H., Burkard M.E., Freier S.M., Wyatt J.R., Turner D.H., Freier S.M., Wyatt J.R., Turner D.H., Wyatt J.R., Turner D.H., Turner D.H. Predicting oligonucleotide affinity to nucleic acid targets. RNA. 1999;5:1458–1469. doi: 10.1017/s1355838299991148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarenko I., Pires R., Lowe B., Obaidy M., Rashtchian A., Pires R., Lowe B., Obaidy M., Rashtchian A., Lowe B., Obaidy M., Rashtchian A., Obaidy M., Rashtchian A., Rashtchian A. Effect of primary and secondary structure of oligodeoxyribonucleotides on the fluorescent properties of conjugated dyes. Nucleic Acids Res. 2002;30:2089–2095. doi: 10.1093/nar/30.9.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuwaysir E.F., Huang W., Albert T.J., Singh J., Nuwaysir K., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., Huang W., Albert T.J., Singh J., Nuwaysir K., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., Albert T.J., Singh J., Nuwaysir K., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., Singh J., Nuwaysir K., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., Nuwaysir K., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., Richmond T., Gorski T., Berg J.P., Ballin J., Gorski T., Berg J.P., Ballin J., Berg J.P., Ballin J., Ballin J., et al. Gene expression analysis using oligonucleotide arrays produced by maskless photolithography. Genome Res. 2002;12:1749–1755. doi: 10.1101/gr.362402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park P.J., Cao Y.A., Lee S.Y., Kim J.W., Chang M.S., Hart R., Choi S., Cao Y.A., Lee S.Y., Kim J.W., Chang M.S., Hart R., Choi S., Lee S.Y., Kim J.W., Chang M.S., Hart R., Choi S., Kim J.W., Chang M.S., Hart R., Choi S., Chang M.S., Hart R., Choi S., Hart R., Choi S., Choi S. Current issues for DNA microarrays: Platform comparison, double linear amplification, and universal RNA reference. J. Biotechnol. 2004;112:225–245. doi: 10.1016/j.jbiotec.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Pruitt K.D., Tatusova T., Maglott D.R., Tatusova T., Maglott D.R., Maglott D.R. NCBI Reference Sequence (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabiner L. A tutorial on hidden Markov models and selected applications in speech recognition. Proc. IEEEE. 1989;77:257–286. [Google Scholar]
- Rinn J.L., Euskirchen G., Bertone P., Martone R., Luscombe N.M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Euskirchen G., Bertone P., Martone R., Luscombe N.M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Bertone P., Martone R., Luscombe N.M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Martone R., Luscombe N.M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Luscombe N.M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Nelson F.K., Miller P., Gerstein M., Miller P., Gerstein M., Gerstein M., et al. The transcriptional activity of human Chromosome 22. Genes & Dev. 2003;17:529–540. doi: 10.1101/gad.1055203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillard J.M., Zuker M., Gulari E., Zuker M., Gulari E., Gulari E. OligoArray 2.0: Design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res. 2003;31:3057–3062. doi: 10.1093/nar/gkg426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royce T.E., Rozowsky J.S., Bertone P., Samanta M., Stolc V., Weissman S., Snyder M., Gerstein M., Rozowsky J.S., Bertone P., Samanta M., Stolc V., Weissman S., Snyder M., Gerstein M., Bertone P., Samanta M., Stolc V., Weissman S., Snyder M., Gerstein M., Samanta M., Stolc V., Weissman S., Snyder M., Gerstein M., Stolc V., Weissman S., Snyder M., Gerstein M., Weissman S., Snyder M., Gerstein M., Snyder M., Gerstein M., Gerstein M. Issues in the analysis of oligonucleotide tiling microarrays for transcript mapping. Trends Genet. 2005;21:466–475. doi: 10.1016/j.tig.2005.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royce T.E., Rozowsky J.S., Luscombe N.M., Emanuelsson O., Yu H., Zhu X., Snyder M., Gerstein M., Rozowsky J.S., Luscombe N.M., Emanuelsson O., Yu H., Zhu X., Snyder M., Gerstein M., Luscombe N.M., Emanuelsson O., Yu H., Zhu X., Snyder M., Gerstein M., Emanuelsson O., Yu H., Zhu X., Snyder M., Gerstein M., Yu H., Zhu X., Snyder M., Gerstein M., Zhu X., Snyder M., Gerstein M., Snyder M., Gerstein M., Gerstein M. Extrapolating traditional DNA microarray statistics to the tiling and protein microarrays technologies. Methods Enzymol. 2006;411:282–311. doi: 10.1016/S0076-6879(06)11015-0. [DOI] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H.J., Skaletsky H.J. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S., Misener S., Misener S., editors. Bioinformatics methods and protocols: Methods in molecular biology. Humana Press; Totowa, N.J: 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- SantaLucia J. , Jr., Hicks D., Hicks D. Annu. Rev. Biophys. Biomol. Struct. Vol. 33. 2004. The thermodynamics of DNA structural motifs; pp. 415–440. [DOI] [PubMed] [Google Scholar]
- Schadt E.E., Edwards S.W., GuhaThakurta D., Holder D., Ying L., Svetnik V., Leonardson A., Hart K.W., Russell A., Li G., Edwards S.W., GuhaThakurta D., Holder D., Ying L., Svetnik V., Leonardson A., Hart K.W., Russell A., Li G., GuhaThakurta D., Holder D., Ying L., Svetnik V., Leonardson A., Hart K.W., Russell A., Li G., Holder D., Ying L., Svetnik V., Leonardson A., Hart K.W., Russell A., Li G., Ying L., Svetnik V., Leonardson A., Hart K.W., Russell A., Li G., Svetnik V., Leonardson A., Hart K.W., Russell A., Li G., Leonardson A., Hart K.W., Russell A., Li G., Hart K.W., Russell A., Li G., Russell A., Li G., Li G., et al. A comprehensive transcript index of the human genome generated using microarrays and computational approaches. Genome Biol. 2004;5:R73. doi: 10.1186/gb-2004-5-10-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker D.D., Schadt E.E., Armour C.D., He Y.D., Garrett-Engele P., McDonagh P.D., Loerch P.M., Leonardson A., Lum P.Y., Cavet G., Schadt E.E., Armour C.D., He Y.D., Garrett-Engele P., McDonagh P.D., Loerch P.M., Leonardson A., Lum P.Y., Cavet G., Armour C.D., He Y.D., Garrett-Engele P., McDonagh P.D., Loerch P.M., Leonardson A., Lum P.Y., Cavet G., He Y.D., Garrett-Engele P., McDonagh P.D., Loerch P.M., Leonardson A., Lum P.Y., Cavet G., Garrett-Engele P., McDonagh P.D., Loerch P.M., Leonardson A., Lum P.Y., Cavet G., McDonagh P.D., Loerch P.M., Leonardson A., Lum P.Y., Cavet G., Loerch P.M., Leonardson A., Lum P.Y., Cavet G., Leonardson A., Lum P.Y., Cavet G., Lum P.Y., Cavet G., Cavet G., et al. Experimental annotation of the human genome using microarray technology. Nature. 2001;409:922–927. doi: 10.1038/35057141. [DOI] [PubMed] [Google Scholar]
- Singh-Gasson S., Green R.D., Yue Y., Nelson C., Blattner F., Sussman M.R., Cerrina F., Green R.D., Yue Y., Nelson C., Blattner F., Sussman M.R., Cerrina F., Yue Y., Nelson C., Blattner F., Sussman M.R., Cerrina F., Nelson C., Blattner F., Sussman M.R., Cerrina F., Blattner F., Sussman M.R., Cerrina F., Sussman M.R., Cerrina F., Cerrina F. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat. Biotechnol. 1999;17:974–978. doi: 10.1038/13664. [DOI] [PubMed] [Google Scholar]
- Tan P.K., Downey T.J., Spitznagel E.L., Jr., Xu P., Fu D., Dimitrov D.S., Lempicki R.A., Raaka B.M., Cam M.C., Downey T.J., Spitznagel E.L., Jr., Xu P., Fu D., Dimitrov D.S., Lempicki R.A., Raaka B.M., Cam M.C., Spitznagel E.L., Jr., Xu P., Fu D., Dimitrov D.S., Lempicki R.A., Raaka B.M., Cam M.C., Xu P., Fu D., Dimitrov D.S., Lempicki R.A., Raaka B.M., Cam M.C., Fu D., Dimitrov D.S., Lempicki R.A., Raaka B.M., Cam M.C., Dimitrov D.S., Lempicki R.A., Raaka B.M., Cam M.C., Lempicki R.A., Raaka B.M., Cam M.C., Raaka B.M., Cam M.C., Cam M.C. Evaluation of gene expression measurements from commercial microarray platforms. Nucleic Acids Res. 2003;31:5676–5684. doi: 10.1093/nar/gkg763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gelder R.N., von Zastrow M.E., Yool A., Dement W.C., Barchas J.D., Eberwine J.H., von Zastrow M.E., Yool A., Dement W.C., Barchas J.D., Eberwine J.H., Yool A., Dement W.C., Barchas J.D., Eberwine J.H., Dement W.C., Barchas J.D., Eberwine J.H., Barchas J.D., Eberwine J.H., Eberwine J.H. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc. Natl. Acad. Sci. 1990;87:1663–1667. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E.J., Emanuelsson O., Scalzo D., Royce T., Kosak S., Oakeley E.J., Weissman S., Gerstein M., Groudine M., Snyder M., Emanuelsson O., Scalzo D., Royce T., Kosak S., Oakeley E.J., Weissman S., Gerstein M., Groudine M., Snyder M., Scalzo D., Royce T., Kosak S., Oakeley E.J., Weissman S., Gerstein M., Groudine M., Snyder M., Royce T., Kosak S., Oakeley E.J., Weissman S., Gerstein M., Groudine M., Snyder M., Kosak S., Oakeley E.J., Weissman S., Gerstein M., Groudine M., Snyder M., Oakeley E.J., Weissman S., Gerstein M., Groudine M., Snyder M., Weissman S., Gerstein M., Groudine M., Snyder M., Gerstein M., Groudine M., Snyder M., Groudine M., Snyder M., Snyder M., et al. DNA replication-timing analysis of human chromosome 22 at high resolution and different developmental states. Proc. Natl. Acad. Sci. 2004;101:17771–17776. doi: 10.1073/pnas.0408170101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauk C.L., Berndt M.L., Williams A., Douglas G.R., Berndt M.L., Williams A., Douglas G.R., Williams A., Douglas G.R., Douglas G.R. Comprehensive comparison of six microarray technologies. Nucleic Acids Res. 2004;32:e124. doi: 10.1093/nar/gnh123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying L., Schadt E.E., Holder S.V.D., Edwards S., Guhathakurtka D., Schadt E.E., Holder S.V.D., Edwards S., Guhathakurtka D., Holder S.V.D., Edwards S., Guhathakurtka D., Edwards S., Guhathakurtka D., Guhathakurtka D. 2003 Proc. of the American Statistical Association. 2003. Identification of chromosomal regions containing transcribed sequences using microarray expression data; pp. 4672–4677. [Google Scholar]