Abstract
Recent progress in mapping transcription factor (TF) binding regions can largely be credited to chromatin immunoprecipitation (ChIP) technologies. We compared strategies for mapping TF binding regions in mammalian cells using two different ChIP schemes: ChIP with DNA microarray analysis (ChIP-chip) and ChIP with DNA sequencing (ChIP-PET). We first investigated parameters central to obtaining robust ChIP-chip data sets by analyzing STAT1 targets in the ENCODE regions of the human genome, and then compared ChIP-chip to ChIP-PET. We devised methods for scoring and comparing results among various tiling arrays and examined parameters such as DNA microarray format, oligonucleotide length, hybridization conditions, and the use of competitor Cot-1 DNA. The best performance was achieved with high-density oligonucleotide arrays, oligonucleotides ≥50 bases (b), the presence of competitor Cot-1 DNA and hybridizations conducted in microfluidics stations. When target identification was evaluated as a function of array number, 80%–86% of targets were identified with three or more arrays. Comparison of ChIP-chip with ChIP-PET revealed strong agreement for the highest ranked targets with less overlap for the low ranked targets. With advantages and disadvantages unique to each approach, we found that ChIP-chip and ChIP-PET are frequently complementary in their relative abilities to detect STAT1 targets for the lower ranked targets; each method detected validated targets that were missed by the other method. The most comprehensive list of STAT1 binding regions is obtained by merging results from ChIP-chip and ChIP-sequencing. Overall, this study provides information for robust identification, scoring, and validation of TF targets using ChIP-based technologies.
Identification of transcription factor binding sites is essential for understanding the regulatory circuits that control cellular processes such as cell division and differentiation as well as metabolic and physiological balance. Traditionally the pursuit of transcription factor targets has exposed only a few binding regions at a time. However, recent years have witnessed several new approaches for the global mapping of transcriptional regulatory regions. Such approaches include computational methods (Bailey and Elkan 1995; Liu et al. 2001, 2002; Wasserman and Sandelin 2004) as well as more direct in vivo methods that require isolation of target DNA through chromatin immunoprecipitation (ChIP) of the transcription factor of interest. These ChIP-based strategies identify target binding regions by using the immunoprecipitated DNA to either probe a DNA microarray that tiles significant regions of the human genome (ChIP-chip) (Horak et al. 2002; Ren et al. 2002; Weinmann et al. 2002; Martone et al. 2003; Cawley et al. 2004; Euskirchen et al. 2004; Odom et al. 2004) or for direct DNA sequencing (ChIP sequencing) (Impey et al. 2004; Chen and Sadowski 2005; Kim et al. 2005a; Roh et al. 2005; Wei et al. 2006). In ChIP-chip experiments, the DNA associated with a transcription factor of interest is compared to a reference sample, generally either genomic DNA or any DNA that might be immunoprecipitated with a negative control antibody. ChIP-chip experiments entail the use of DNA tiling microarrays that are prepared either by deposition of PCR products or by oligonucleotide synthesis. These arrays may tile promoter regions, large genomic segments, entire chromosomes, or in some cases an entire genome (Martone et al. 2003; Cawley et al. 2004; Boyer et al. 2005; Kim et al. 2005b; Lee et al. 2006). ChIP sequencing experiments, on the other hand, do not require the use of a reference sample. Sequencing is performed from individually cloned ChIP fragments (Weinmann et al. 2001; Hug et al. 2004); from concatenations of single “tags,” where each tag is a signature derived from a ChIP DNA fragment (STAGE) (Impey et al. 2004; Chen and Sadowski 2005; Kim et al. 2005a; Roh et al. 2005); or from concatenations of Paired-End diTags cloned from the 5′- and 3′-ends of each ChIP DNA fragment (ChIP-PET) (Loh et al. 2006; Wei et al. 2006).
Although ChIP-based technologies have demonstrated widespread utility, many experimental parameters important for enhancing the performance of ChIP have not been adequately explored for mammalian cells. Moreover, a direct comparison of ChIP-chip and ChIP sequencing has not been performed. Such information is crucial for the large number of experiments that are performed on subsets of mammalian genomes and will become even more crucial as these experiments expand to cover entire genomes.
While many microarray parameters for ChIP-chip appear to translate well from previously established microarray protocols (see, for example, Hegde et al. 2000; Oberley and Farnham 2003; Buck and Lieb 2004; Wu et al. 2006), other variables are more tenuous. In particular, we focused on addressing oligonucleotide length and array format, the presence or absence of Cot-1 DNA, and the number of replicas required to obtain the maximum of data. Currently there is considerable variation in the use of each of these.
We explored parameters for ChIP-chip using the sequence-specific transcription factor STAT1 (Signal Transducer and Activator of Transcription). STAT1 is a cytoplasmic protein that translocates to the nucleus when cells encounter interferons or other peptide signals (for review, see Boehm et al. 1997; Bromberg and Chen 2001; Levy and Darnell 2002). STAT1-dependent transcription is important for immune and inflammatory responses, antiviral effects, proliferation, apoptosis, and differentiation (Boehm et al. 1997; Levy and Darnell 2002). STAT1 was selected by The ENCODE Project Consortium (The ENCODE Project Consortium 2004) as an ideal factor to test the performance of ChIP DNA across platforms and is a model factor for two main reasons: (1) STAT1 ChIP experiments show less enrichment than those with chromatin modifications or more general DNA-binding proteins such as RNA polymerase II and hence would be expected to more thoroughly test the performance of various platforms, and (2) STAT1 is inducible, and therefore it offers a direct biologic control in the form of STAT1 ChIP samples prepared from control cells not treated with interferon-gamma (IFNG).
STAT1 ChIP-chip studies have been conducted previously on a Chromosome 22 PCR product tiling array (Hartman et al. 2005). In the study presented herein, ChIPs were performed to find many previously unidentified binding regions for STAT1 under IFNG stimulation.
Our STAT1 mapping studies focus on the ENCODE regions, which represent 1% (30 Mb) of the human genome (The ENCODE Project Consortium 2004). The ENCODE regions are comprised of 44 subregions that range in length from 500 kb to 1.9 Mb and were selected to include loci of biological interest and regions that stratify both gene density and nonexonic conservation with mouse. The final results from the data sets described here have also been included in the meta-analysis conducted by the ENCODE Transcriptional Regulatory Elements Subgroup (The ENCODE Project Consortium 2004). Our studies are expected to provide useful information for comparing and integrating data generated from the ENCODE group as well as for future genome-scale studies that map transcription factor binding regions using ChIP-based methods.
Results
Exploring ChIP-chip performance: Longer oligonucleotides yield better signals
In the first phase of these studies, we investigated ChIP-chip performance on oligonucleotide arrays synthesized by maskless photolithography (Nuwaysir et al. 2002). For these studies we used chromatin-immunoprecipitated STAT1 DNA, which produces modest signal enrichments relative to ChIP DNA isolated to study other transcription factors and chromatin modifications. HeLa S3 cells were treated with IFNG to induce STAT1 binding, and then incubated briefly with 1% formaldehyde to cross-link protein to DNA. Nuclei were prepared, and chromatin was sheared to ∼1 kb final DNA size. STAT1 and its associated DNA were immunoprecipitated using an anti-STAT1 antibody. Cross-links were reversed, and the success of each immunoprecipitation was examined by PCR analysis using primers to a known STAT1-binding region in the promoter of IRF1 (Interferon Regulatory Factor 1) (Hartman et al. 2005), whose locus is included in the ENCODE regions.
Using this assay, we investigated the effects of varying a number of parameters on the performance of ChIP-chip. These parameters included the type of beads used in the immunoprecipitation step (magnetic or Sepharose), various labeling technologies, and array hybridization conditions. The final ChIP and microarray conditions selected are reported in Methods. No difference in immunoprecipitation efficiency was observed using magnetic as opposed to Sepharose beads. However, signal enrichment and array uniformity were significantly improved when the hybridization solution was continuously circulated over the array surface using microfluidic chambers; thus all arrays were subjected to this procedure. We also included unlabeled Cot-1 competitor DNA in all hybridizations except as noted below.
Arrays with oligonucleotides of different lengths (25−60 bases [b]) are currently used for ChIP-chip experiments (Cawley et al. 2004; Boyer et al. 2005; Kim et al. 2005b). We systematically examined the contribution of array oligonucleotide length to ChIP-chip performance. Custom arrays with 36-, 50-, 60-, or 70-b oligonucleotides tiling most or all of the ENCODE regions were synthesized by maskless photolithography. The oligonucleotides were designed to comprehensively cover nonrepetitive regions and are tiled end-to-end such that immediately adjacent genomic DNA segments are represented on the arrays. Thus the short oligonucleotide arrays have more probes per region, but are expected to exhibit lower signals and increased cross-hybridization relative to arrays with longer oligonucleotides, depending on the exact conditions used. Lower signals will reduce accuracy when enrichment ratios are determined (see Discussion). STAT1 ChIP DNA prepared from nuclear extracts of IFN-γ treated cells was labeled with Alexa647 and hybridized to the arrays along with Alexa555-labeled STAT1 ChIP DNA isolated in parallel from the nuclear extracts of uninduced (STAT1-nuclear excluded) cells. Each biological replicate was labeled and hybridized independently. The 36-b array data set contained two biological replicates; all other ChIP-chip data sets contained three or more biological replicates (see Supplemental Table 1). Array signals representing enrichments in ChIP DNA samples from IFN-γ-treated cells relative to those ChIP DNA samples prepared from untreated control cells were scored using a sliding window approach (see Supplemental Methods). As shown in Figure 1, significantly higher signal enrichments for STAT1 target regions were observed from the 50-b arrays relative to the 36-b arrays. Increases in oligonucleotide lengths to 60 and 70 b improved array performances only marginally compared to the 50-b arrays (data not shown). The reduced performance of the 36-b arrays was not due to the use of two replicates; a comparison of signal enrichments from data sets comprised of two biological replicates hybridized to each of two 36- and 50-b arrays yielded a similar outcome (see section and Fig. 6 below). The lower signal enrichments observed with the 36-b arrays were also not likely because of suboptimal hybridization affinities as several different hybridization conditions were tested for the arrays at each oligonucleotide length and improved signal enrichments were not apparent with any of these alternative conditions (see Supplemental Methods). Moreover, the expected difference in hybridization temperature for the 36-b array relative to the 50-b array is calculated to be 4°C or less (Bertone et al. 2006). Thus, longer oligonucleotides enhance performance, and 50-b arrays were used for all remaining experiments.
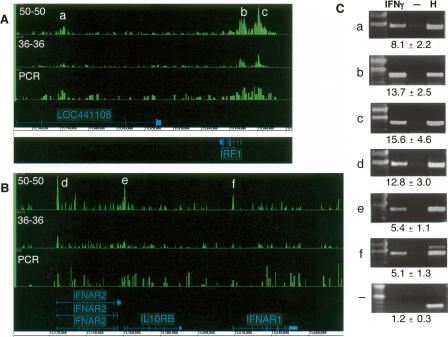
Figure 1.
Comparison of signal tracks. Signal enrichment tracks are plotted for the 50-b, 36-b, and PCR product array platforms for two different loci. Signals of STAT1 bound regions in IFNG-stimulated cells relative to untreated cells were derived from multiple biological replicates with one replicate hybridized per array (Methods; Supplemental Table 1). Annotations above the coordinate axis are for genes on the forward strand, and those below are for reverse-strand genes. Signal enrichment tracks are plotted to the same scale for the platforms displayed, from 0 to 2.5 in panel A and from 0 to 1.3 in panel B. (A) The IRF1 locus and flanking regions on Chromosome 5 (coordinates 131,770,000 to 131,870,000 from build NCBIv35 [hg17]). (B) The loci on Chromosome 21, which contain the cytokine receptors, IFNAR2, IL10RB, and IFNAR1 (coordinates 33,500,000 to 33,680,000). (C) Targets that have been validated by ChIP-PCR (shown) are indicated by symbols a through f. The lanes are labeled for ChIP DNA from IFNG-stimulated (IFNγ) cells, ChIP DNA from unstimulated cells, and for HeLa S3 genomic DNA. Fold enrichments, as calculated for several biological replicates (see Methods), are indicated for each target (a–f) and for a negative control region (−).
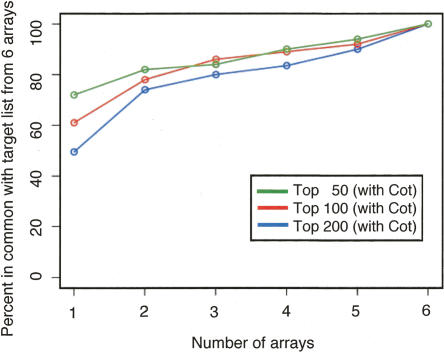
Figure 6.
The value of adding biological replicates to a ChIP-chip data set. For the six 50 b every 38-b arrays that were hybridized in the presence of Cot DNA, the reproducibility of target lists for the top 50 (green), 100 (red), and 200 (blue) binding regions was examined as a function of the number of biological replicates analyzed. Each biological replicate is hybridized to a separate array. The agreement is compared against the target list identified by using all six arrays. We see that greater than 80% agreement is obtained when three or more biological replicates are used.
Validation of targets from the 50-b oligonucleotide array data
Signal enrichment maps are suggestive of binding regions, but in order for array performance to be properly assessed, it is essential to validate the targets identified from the ChIP-chip experiments. Therefore, we devised a scheme to measure the sensitivity and specificity of the experiments. STAT1 targets were ranked according to their signal enrichments, and a subset of targets was sampled across the rankings and tested for enrichment in STAT1 ChIP DNA by ChIP-PCR analysis. A twofold or greater enrichment in each of at least two STAT1 biological replicate ChIP-PCR experiments was chosen as a threshold for enrichment. Target validation was plotted as a function of rank order for each ChIP-chip data set. As shown in Figure 2A, targets at the top of the rank list validated as true positives, and the frequency of target validation diminishes further down the rank list. Thus, most of the first 75 targets are expected to be bona fide targets, whereas most of the regions below 100 on the rank list are negative. Extrapolation of the confirmed positives as a function of rank order for the entire list suggests that there are ∼124 positives in the top 200 targets listed (Table 1). This figure is expected to be an overestimation because many targets lie immediately adjacent to one another and likely represent enrichments from a single common target region. If targets are combined into 10-kb regions, then the total number of STAT1 targets is ∼67 for the ChIP-chip data set using the arrays with 50-b end-to-end tiling.
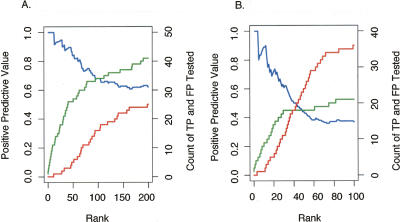
Figure 2.
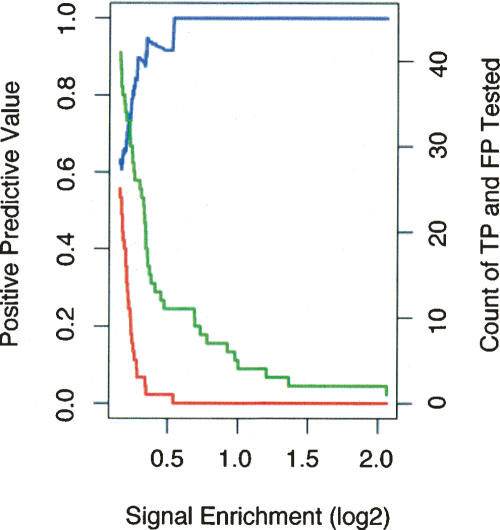
Validation frequency as a function of rank order for the 50 b every 50-b array and PCR product array platforms. For each ChIP-chip data set (derived from multiple biological replicates with one replicate hybridized per array) (Supplemental Table 1), we identified the target regions above a threshold. The targets were tested and divided into true positives (TP) and false positives (FP) based on a ChIP-PCR validation assay (as described in Methods). Sensitivity [defined as TP/(TP + FN), where FN are the number of false negatives] and specificity [defined as TN/(TN + FP), where TN are the number of true negatives] of the target list at this threshold are difficult to accurately estimate since the total number of actual binding sites (TP + FN) as well as the number of true negatives (TN) are not known, and other methods for direct, in vivo identification binding independent of ChIP methods do not exist. Nonetheless, the positive predictive value [defined as TP/(TP + FP)] for ChIP-chip experiments can be estimated using data from ChIP-PCR validations. (A) The number of targets confirmed by validation, true positives (green line), as well as the number targets that did not validate, false positives (red line), is plotted as a function of target rank (ordered by signal enrichment) for the 50 b every 50-b array platform (from three biological replicate arrays). The positive predictive value (blue line) is also shown as a function of rank. (B) The number of true positives (green line), the number of false positives (red line), and the positive predictive value (blue line) are shown as a function of rank for the PCR product array platform (data from six biological replicate arrays).
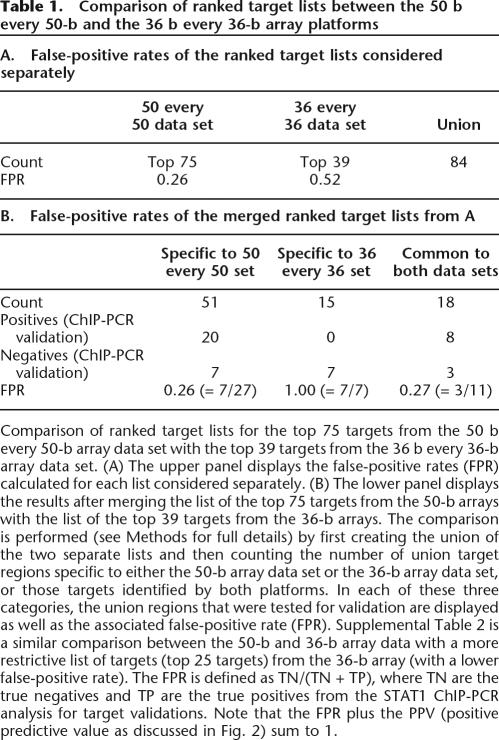
Table 1.
Comparison of ranked target lists between the 50 b every 50-b and the 36 b every 36-b array platforms
Comparison of ranked target lists for the top 75 targets from the 50 b every 50-b array data set with the top 39 targets from the 36 b every 36-b array data set. (A) The upper panel displays the false-positive rates (FPR) calculated for each list considered separately. (B) The lower panel displays the results after merging the list of the top 75 targets from the 50-b arrays with the list of the top 39 targets from the 36-b arrays. The comparison is performed (see Methods for full details) by first creating the union of the two separate lists and then counting the number of union target regions specific to either the 50-b array data set or the 36-b array data set, or those targets identified by both platforms. In each of these three categories, the union regions that were tested for validation are displayed as well as the associated false-positive rate (FPR). Supplemental Table 2 is a similar comparison between the 50-b and 36-b array data with a more restrictive list of targets (top 25 targets) from the 36-b array (with a lower false-positive rate). The FPR is defined as TN/(TN + TP), where TN are the true negatives and TP are the true positives from the STAT1 ChIP-PCR analysis for target validations. Note that the FPR plus the PPV (positive predictive value as discussed in Fig. 2) sum to 1.
We also compared the accuracy of target detection for the 50-b ChIP-chip data set as a function of signal enrichment. As shown in Figure 3, the fraction of validated positives decreases and the fraction of false positives increases at a very sharp signal enrichment threshold. Thus there is a very sharp transition at a particular signal enrichment (∼0.25 on a log2 scale) above which most targets validate as positives.
Figure 3.
Validation frequency as a function of signal for the 50 b every 50-b array data set. The data from Figure 2A were analyzed as a function of array signal enrichment for the 50 b every 50-b array platform. Signal enrichment is defined as the log2 ratio of signal intensity of the ChIP DNA over the signal intensity of the reference DNA sample (for STAT1, this is the log2 ratio of intensities for IFNG-stimulated against unstimulated ChIP DNA samples). A target region is identified as a “peak” in a signal enrichment track (see Fig. 1; for details, see Supplemental Methods) and is assigned its maximal signal enrichment, the height of the peak. The number of true positives is the green line, the number of false positives is the red line, and the positive predictive value is the blue line as in Figure 2A. The horizontal scale in this figure is in the opposite orientation to the horizontal scale displayed in Figure 2; high signal enrichment, which appears to the right-hand side here, corresponds to higher rank, which is to the left in Figure 2. At a log2 signal of ∼0.25, the number of false positives increases sharply to the left.
We next compared the accuracy of STAT1 targets identified from the 50-base array ChIP-chip data set to those identified from the 36-b array ChIP-chip data set. We selected the highest 75 ranked targets from the 50-b array data, corresponding to a false-positive rate of 0.26, and cross-referenced these with the entire list of 39 targets identified from the 36-b array data. For the 36-b arrays, only the top-ranked 39 regions had positive signals at a statistically significant cutoff. We suspect this low number is due to diminished signal on the 36-b arrays. The targets of the 50- and 36-b arrays combined into 84 distinct target regions (see “Comparison of Target Lists” in Methods); 18 were common to both lists, and most of these (eight of the 11 tested by ChIP-PCR analysis) validated as bona fide targets. The 36-b oligonucleotide array failed to identify 68% (51/75) of the targets detected with the 50-b oligonucleotide array. Of these 51 targets, 27 were tested by ChIP-PCR analysis, and the majority of these (20/27) could be validated. In contrast, 15 targets were unique to the 36-b array. Seven of these 15 were tested by ChIP-PCR analysis, and none showed enrichment. If we restrict analysis of the 36-b array to the top 25 targets, thereby reducing its false-positive rate from 0.52 to 0.38, a similar trend is observed (Supplemental Table 2) and fewer targets specific to the 36-b array are identified, indicating a greater overlap of the top-ranked targets between the 50- and 36-b lists. In conclusion, based on chromosomal maps of signal enrichments (Fig. 1) and target validations, the 50-b arrays outperformed the shorter 36-b arrays under the conditions we used.
Comparison of oligonucleotide and PCR product arrays
Both oligonucleotide arrays and PCR product arrays are used extensively for ChIP-chip experiments (e.g., Martone et al. 2003). PCR product arrays have features of longer length than oligonucleotide arrays and could in principle perform better in mammalian ChIP-chip experiments. Six independent biological replica STAT1 ChIP samples were isolated and hybridized to six PCR product arrays (Supplemental Table 1) and compared to those targets obtained from a data set using three biological replicates hybridized to three 50-b oligonucleotide arrays; in many cases, the same ChIP samples were used. As shown in Figure 1, the signal enrichments appeared better for the oligonucleotide array data set relative to the PCR product array data set.
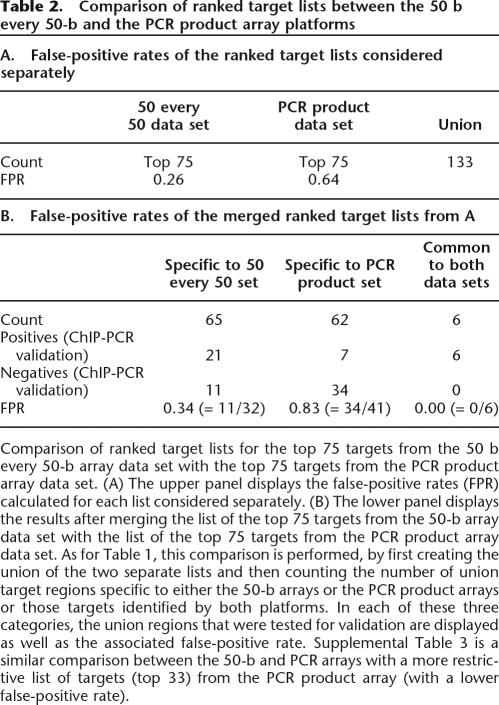
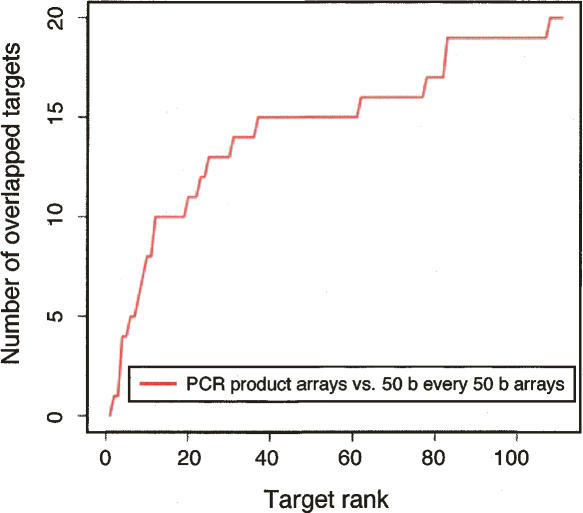
The top 75 ranked targets from the 50-b oligonucleotide array data and the top 75 ranked targets from the PCR product array data were then merged to form a union of regions that could be used as the basis for comparing the two ChIP-chip data sets (Table 2A; see “Comparison of Target Lists” in Methods). Six targets overlapped between the 50-b oligonucleotide array and the PCR product array target lists (Table 2B). The different platforms were compared as a function of their rank order on the target lists. As shown in Figure 4, the positives at the very top of the rank order lists usually agree, and less concurrence is observed for targets with lower rankings. If we restrict analysis of the PCR product array data set to the top 33 targets, thereby reducing its false-positive rate from 0.64 to 0.40, a similar trend is observed (Supplemental Table 3).
Table 2.
Comparison of ranked target lists between the 50 b every 50-b and the PCR product array platforms
Comparison of ranked target lists for the top 75 targets from the 50 b every 50-b array data set with the top 75 targets from the PCR product array data set. (A) The upper panel displays the false-positive rates (FPR) calculated for each list considered separately. (B) The lower panel displays the results after merging the list of the top 75 targets from the 50-b array data set with the list of the top 75 targets from the PCR product array data set. As for Table 1, this comparison is performed, by first creating the union of the two separate lists and then counting the number of union target regions specific to either the 50-b arrays or the PCR product arrays or those targets identified by both platforms. In each of these three categories, the union regions that were tested for validation are displayed as well as the associated false-positive rate. Supplemental Table 3 is a similar comparison between the 50-b and PCR arrays with a more restrictive list of targets (top 33) from the PCR product array (with a lower false-positive rate).
Figure 4.
Agreement between the ranked target lists for the 50 b every 50-b array and the PCR product array platforms. Each data set is comprised of multiple biological replicates with one replicate hybridized per array (Supplemental Table 1). The vertical axis is the number of targets common between the two rank lists up to a certain rank (the horizontal axis). The agreement increases steeply for the highest-ranked targets and then starts to plateau.
To ascertain if targets from the PCR product arrays and the 50-b oligonucleotide arrays validate at similar rates, and to determine if the two platforms exhibit similar sensitivities and specificities, targets were selected and tested for validation across a wide range of rank orders using ChIP-PCR analysis. As shown in Figure 2B, the frequency of validated targets (i.e., the positive predictive value) from the PCR product array data was diminished relative to the 50-b oligonucleotide array data (Fig. 2A), indicating that the PCR product array data set contains more false positives. In addition, the sensitivity of the PCR product array format was lower.
To investigate these differences in array performance, we examined regions that were specific to one of the target lists and that were tested for enrichment by ChIP-PCR (Table 2B). Seven targets that were identified by the PCR product array data set and validated by ChIP-PCR analysis were not present on the target list from the 50-b oligonucleotide array data set. Inspection of these regions revealed six of the seven targets contained a combination of repetitive elements and AT-rich sequences that likely resulted in low signal enrichments on the oligonucleotide arrays. In contrast, 21 targets identified from the oligonucleotide array data set and validated by ChIP-PCR analysis were not found using the PCR product arrays. Two of the 21 were adjacent to positive regions detected by the PCR product arrays, but we could not identify aspects of sequence composition that might cause the other 19 targets to escape detection in the ChIP-chip experiments performed with the PCR product arrays.
The presence of competitor Cot-1 DNA in the hybridization improves signal-to-noise
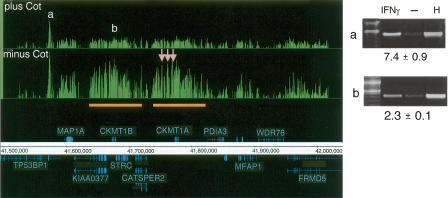
Highly repetitive sequences comprise 50% of mammalian genomes and can be potential targets as well as a source of noise. We therefore investigated the value of including unlabeled Cot-1 repetitive DNA in the hybridizations because the addition of Cot-1 DNA might be expected to decrease nonspecific hybridization (DeRisi et al. 1996) and improve the accurate detection of transcription factor targets. To make this comparison, six biological replicates were divided after labeling and hybridized on 12 arrays in the presence and absence of Cot-1 DNA, using 50-b arrays with 38-b spacing. The addition of an excess of Cot-1 DNA produced a modest improvement in signal-to-noise. Figure 5 illustrates this point for a region on Chromosome 15, where several peaks and often the overall background were noticeably reduced. The three false positives in this region (pink arrows) had high signal enrichments in the experiment lacking Cot-1 DNA, but had low signal enrichments in the experiment containing Cot-1 DNA. Target b in Figure 5 (which lies in a region containing a gene duplication; orange bars) was confirmed by ChIP-PCR analysis. It was ranked 22nd on the target list for STAT1 ChIP DNA hybridized in the absence of Cot-1 DNA and slipped just below the threshold on the ranked target list (to 95th) when the matching sample pairs were hybridized in the presence of Cot-1 DNA.
Figure 5.
The effect of Cot-1 DNA in determining STAT1 targets. Signal enrichment tracks are shown for data sets of paired samples (see Methods; Supplemental Table 1) that were hybridized either in the presence of Cot-1 DNA (top track) and or in the absence of Cot-1 DNA (lower track), both on the 50 b every 38-b array platform. Annotations above the coordinate axis are for genes on the forward strand, and annotations below the coordinate axis are for genes on the reverse strand. Signal enrichment tracks are plotted to the same scale from 0 to 3.3 for the Cot-present and Cot-absent data sets. Targets with labels “a” and “b” are identified by both experiments (ChIP-PCR gel images are shown). Three targets (pink arrows) appeared only in the Cot-absent experiment and were identified as false positives by ChIP-PCR validation (gel images not shown). The orange bars indicate a region of segmental duplication, which is a potential cause of the false positives (due to cross-hybridization with confirmed target “b”). For the ChIP-PCR validations displayed, the lanes are labeled for ChIP DNA from IFNG-stimulated cells, ChIP DNA from unstimulated cells, and for HeLa S3 genomic DNA. The fold enrichments are indicated and were calculated for several biological replicates (see Methods).
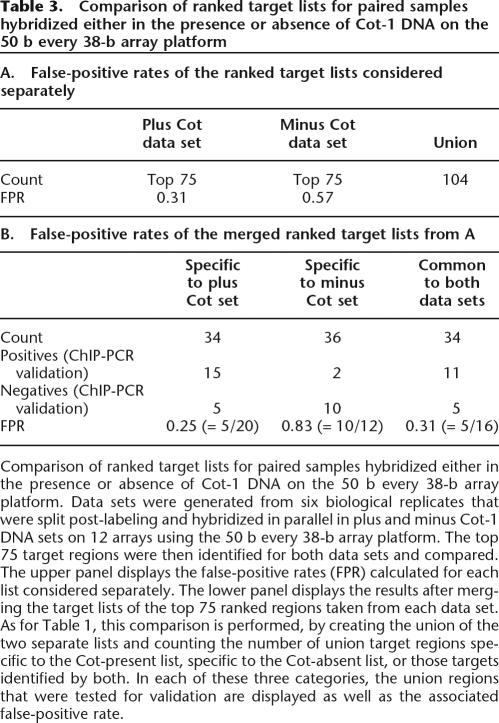
An inspection of 22 targets specific to the Cot-absent data set revealed that 13 targets had highly repetitive elements in their regions and eight targets had segmental duplications. When the same sliding window scoring method was applied to the Cot-absent and Cot-present data sets, a significant number of additional targets was found in the Cot-absent ranked target list (181 targets) relative to the Cot-present ranked target list (three targets) at the equivalent threshold of 3.5-fold enrichment. Importantly, validation of targets revealed a much higher accuracy for the STAT1-associated regions identified in the presence of Cot-1 DNA than in the absence of Cot-1 DNA. Targets specific to either the Cot-present or Cot-absent data sets were sampled from among the top 75 ranked targets identified (Table 3) and tested for enrichment by ChIP-PCR analysis. The experiment containing Cot-1 DNA detected 15 validated positive regions specific to that data set at a false-positive rate of 0.25, whereas the experiment lacking Cot-1 DNA detected only two validated positive regions specific to that data set at a false-positive rate of 0.83 (Table 3). Thus, more accurate results can be obtained through inclusion of Cot-1 DNA in ChIP-chip hybridizations.
Table 3.
Comparison of ranked target lists for paired samples hybridized either in the presence or absence of Cot-1 DNA on the 50 b every 38-b array platform
Comparison of ranked target lists for paired samples hybridized either in the presence or absence of Cot-1 DNA on the 50 b every 38-b array platform. Data sets were generated from six biological replicates that were split post-labeling and hybridized in parallel in plus and minus Cot-1 DNA sets on 12 arrays using the 50 b every 38-b array platform. The top 75 target regions were then identified for both data sets and compared. The upper panel displays the false-positive rates (FPR) calculated for each list considered separately. The lower panel displays the results after merging the target lists of the top 75 ranked regions taken from each data set. As for Table 1, this comparison is performed, by creating the union of the two separate lists and counting the number of union target regions specific to the Cot-present list, specific to the Cot-absent list, or those targets identified by both. In each of these three categories, the union regions that were tested for validation are displayed as well as the associated false-positive rate.
The value of adding more biological replicate experiments
Researchers typically perform multiple biological replicate experiments for microarray data sets, although a systematic analysis of how replicas improve accuracy and reproducibility of targets has not been previously investigated. We therefore examined the value of performing multiple experiments. The top 50, 100, and 200 targets were taken from six biological replicates hybridized with Cot-1 DNA to six arrays with 50 b every 38-b spacing (Supplemental Table 1). As noted above, the top 50 targets have the highest frequency of enrichment in ChIP-PCR validations, and those near the bottom of the list (e.g., ranked 150–200) have the lowest frequency of positive validation. The efficiency of target detection from among all targets identified in this Cot-present data set was determined using a single biological replicate on one array, and then progressively increasing the number of biological replicates, with each replicate hybridized to a separate array. As shown in Figure 6, 50%–70% of all targets from the six-array Cot-present data set can be identified even with a single array. As expected, a higher fraction of the targets are identified using the top 50 target list relative to the top 200 target list since the largest fraction of positive regions resides at the highest rankings as shown in the ChIP-PCR validation studies. The analysis of three independent biological replicates, which is typical for most published ChIP-chip experiments, identified most (80%–86%) of the final targets included in the six-array data set.
Comparison of ChIP-chip to ChIP-PET
In ChIP sequencing, a ChIP-enriched fragment is represented by either a single internal 20-base-pair (bp) tag sequence (ChIP-STAGE) or a 36-bp paired-end ditag (ChIP-PET in which the ditag is constructed from 18-bp 5′ and 3′ signature sequences extracted from each end of the ChIP DNA fragment, thus demarcating the full length of the sonicated ChIP fragment). The binding sites are then deduced by the frequency with which tags are extracted from ChIP DNA fragments relative to the background expectation. The advantage of using paired-end-ditags over single tags is that the PETs mark the start and end of each ChIP fragment. When PET fragments are mapped to the reference genome (e.g., the NCBIv35 [hg17] build of the human genome sequence), the identity of each individual ChIP fragment can be inferred by the PET mapping location, and binding sites can be accurately defined by the common regions within clusters of overlapping PETs. Furthermore, duplicate PET fragments arising from fragment amplification events during cloning can be easily distinguished and removed by treating these multiple PETs that map to an identical location as a single fragment.
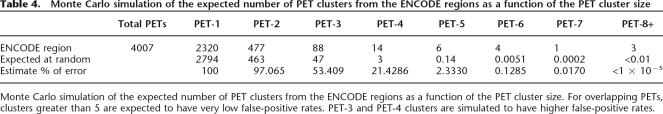
In all, 725,877 PETs were sequenced from STAT1 ChIP DNA isolated from IFNG-induced cells. Sixty-six percent of the PETs map to unique locations in the genome and represent 327,838 distinct ChIP DNA fragments ranging from 0.1 to 6 kb. Of these unique paired-end diTags, only those PET fragments with 5′- and 3′-ends <6 kb apart were considered. The PET-defined ChIP fragments that overlapped with each other were grouped into clusters: clusters of two overlapping fragments are termed as PET-2, clusters of three overlapping fragments as PET-3, and clusters of three or more overlapping fragments as PET3+, and so on. The frequency of each cluster throughout the ENCODE regions is shown in Table 4. The ENCODE region with the most overlapping fragments lies upstream of IRF1 and is a PET-33 cluster (Fig. 7A). Monte Carlo simulation was performed to determine the frequency of clusters expected by random chance (Table 4; see Supplemental Methods). Based on the frequency of PET clusters generated at random, more than 46% of PET-3 clusters and more than 88% of PET4+ clusters are likely to represent bona fide binding targets.
Table 4.
Monte Carlo simulation of the expected number of PET clusters from the ENCODE regions as a function of the PET cluster size
Monte Carlo simulation of the expected number of PET clusters from the ENCODE regions as a function of the PET cluster size. For overlapping PETs, clusters greater than 5 are expected to have very low false-positive rates. PET-3 and PET-4 clusters are simulated to have higher false-positive rates.
Figure 7.
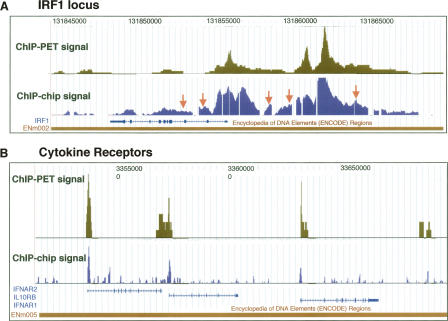
Comparison of ChIP-chip and ChIP-PET. Signal tracks for the ChIP-chip data set from the 50 b every 50-b platform are shown and compared to ChIP-PET signals (the vertical axis corresponds to the number of overlapping ditags at a given genomic coordinate). (A) The IRF1 locus on Chromosome 5 (coordinates 131,842,000 to 131,865,000 from build NCBIv35 [hg17]). The orange arrows indicate the validated ChIP-chip targets from the 50-b array experiment that were on the shoulders of ChIP-PET clusters in the IRF1 region. (B) The region on Chromosome 21 (coordinates 33,500,000 to 33,700,000) containing the cytokine receptors IFNAR2, IL10RB, and IFNAR1. Significant concurrence is observed between the signal readouts from each method.
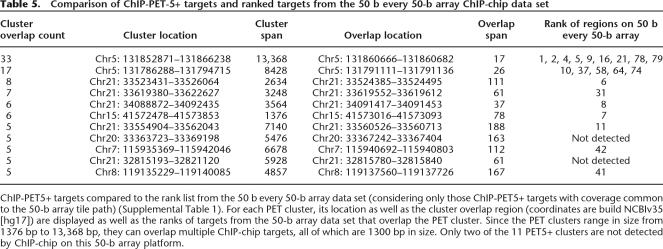
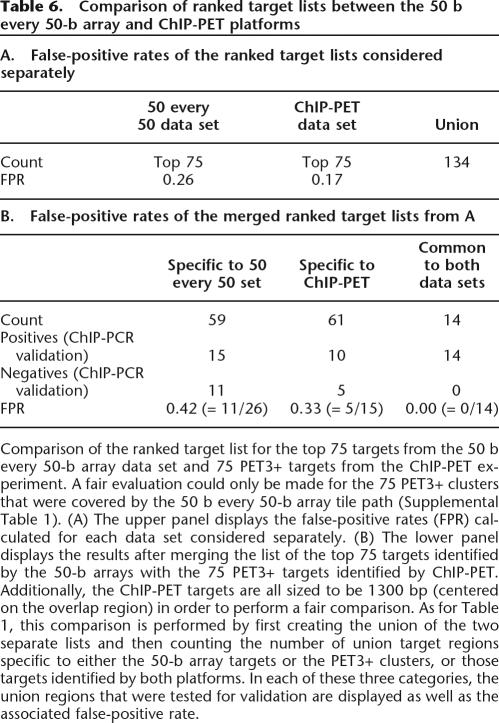
Comparison of signal maps derived from ChIP-chip and Chip-PET data reveals appreciable agreement between the two approaches (Fig. 7), and the concurrence is highest for those targets with the highest signal (Table 5). Since the ChIP-PET sequencing experiment inherently covers all of the ENCODE regions, we only considered those 75 PET3+ clusters whose sequence was represented on the 50 b every 50-b array tile path (Supplemental Table 1) for a comparison between the two platforms. Of these 75 PET3+ clusters, there were 11 PET5+ clusters (those with the highest enrichment), nine of which were also identified in the 50 b every 50-b array data set (Table 5). For the remaining 64 PET-3 and PET-4 clusters, only five overlap the targets lists for the ChIP-chip data set, giving an overall concurrence of 14 targets (Table 6).
Table 5.
Comparison of ChIP-PET-5+ targets and ranked targets from the 50 b every 50-b array ChIP-chip data set
ChIP-PET5+ targets compared to the rank list from the 50 b every 50-b array data set (considering only those ChIP-PET5+ targets with coverage common to the 50-b array tile path) (Supplemental Table 1). For each PET cluster, its location as well as the cluster overlap region (coordinates are build NCBIv35 [hg17]) are displayed as well as the ranks of targets from the 50-b array data set that overlap the PET cluster. Since the PET clusters range in size from 1376 bp to 13,368 bp, they can overlap multiple ChIP-chip targets, all of which are 1300 bp in size. Only two of the 11 PET5+ clusters are not detected by ChIP-chip on this 50-b array platform.
Table 6.
Comparison of ranked target lists between the 50 b every 50-b array and ChIP-PET platforms
Comparison of the ranked target list for the top 75 targets from the 50 b every 50-b array data set and 75 PET3+ targets from the ChIP-PET experiment. A fair evaluation could only be made for the 75 PET3+ clusters that were covered by the 50 b every 50-b array tile path (Supplemental Table 1). (A) The upper panel displays the false-positive rates (FPR) calculated for each data set considered separately. (B) The lower panel displays the results after merging the list of the top 75 targets identified by the 50-b arrays with the 75 PET3+ targets identified by ChIP-PET. Additionally, the ChIP-PET targets are all sized to be 1300 bp (centered on the overlap region) in order to perform a fair comparison. As for Table 1, this comparison is performed by first creating the union of the two separate lists and then counting the number of union target regions specific to either the 50-b array targets or the PET3+ clusters, or those targets identified by both platforms. In each of these three categories, the union regions that were tested for validation are displayed as well as the associated false-positive rate.
To further investigate the targets that were unique to either the ChIP-chip or ChIP-PET target lists, validation experiments were performed. Ten of the targets identified by ChIP-PET3+ cluster regions and missed in the 50 b every 50-b array data set were selected for ChIP-PCR validation and shown to be bona fide targets (Table 6). Repetitive DNA elements appeared to obstruct the identification of six of these 10 targets in the 50 b every 50-b ChIP-chip data set. These repetitive regions had the following characteristics:
Four regions did not have the area of highest PET signal measured on the tiling arrays because highly repetitive elements were centered on the PET overlap spans, and hence these nucleotides were removed from the array tile path. An example of this is shown in Supplemental Figure 1A for a PET-5 target on Chromosome 21.
Repetitive regions were similarly noted for two target regions where a combination of AT-rich and RepeatMasked sequences were congruent with the PET overlap spans. This case includes the other validated PET-5 cluster (Supplemental Fig. 1B).
The remaining four PET3+ targets not detected by the 50-b array were missed for no apparent reason.
Investigation of the 15 confirmed targets that were detected in the 50 b every 50-b array ChIP-chip data set but that were not on the PET3+ list (Table 6B) revealed that seven resided near a ChIP-PET target but were on the shoulder relative to the site of maximal signal. Five of the 7 targets corresponded to the IRF1 locus, which has one of the strongest signals in the genome (Fig. 7A). Thus these array targets correspond to a single common target region. Four of the remaining eight ChIP-chip targets from the 50 b every 50-b array data set intersected PET-2 clusters; we presume increased sequencing depth would have detected these STAT1-binding regions.
We also inspected those regions that did not show enrichment by ChIP-PCR analysis (11 negatives specific to the 50 b every 50-b array data set and five negatives specific to the ChIP-PET experiment) (Table 6B) to ascertain what sequence features might contribute to the identification of these targets as false positives. Of the 11 false positives from the 50 b every 50-b array ChIP-chip data set, six are either largely or entirely comprised of simple repeats, one additional target region occurs as a segmental duplication, another lies near a strong target in the IRF1 5′-non-coding region, and no unusual features that may be uniquely attributable to ChIP-chip performance could be established for the other three. All five ChIP-PETs that were not enriched in ChIP-PCR validation experiments (Table 6) were PET-3 clusters. As indicated by the Monte Carlo simulation (Table 4), ∼50% of PET-3 clusters are expected to be false positives arising from random background. Another possible explanation for the ChIP-PET false positives could be nearby repetitive genomic regions that lead to mapping artifacts. One of the five ChIP-PET false positives does reside in a repetitive region and may have been misassigned during mapping to the hg17 reference sequence. In another example (shown in Supplemental Fig. 2), the false positive in the region Chr5:131963298–131964597 [hg17] was initially called a PET-3, although subsequent analysis revealed that it is more likely to be a PET-2 cluster as two of the DNA fragments in this cluster are almost identical and were likely derived from the same ChIP fragment. In summary, these results indicate that ChIP-chip and ChIP-PET exhibit considerable agreement, particularly on the strongest targets. Each approach is capable of identifying validated targets not found by the other technique.
Discussion
The combination of sequenced genomes and ChIP-based technologies has inspired progress for the comprehensive detection of transcription factor binding regions in vivo. While most efforts have focused on ChIP-chip strategies, ChIP sequencing is gaining popularity as a parallel method. In this study, we performed STAT1 chromatin immunoprecipitations from IFNG-stimulated cells and used the resulting ChIP DNA to map STAT1-binding regions by both microarray hybridizations and DNA sequencing. Based on the outcome of these studies, we determined that reliable ChIP-chip results can be obtained using maskless high-density arrays containing longer rather than shorter oligonucleotides and also by including Cot-1 DNA as a competitor to improve hybridization accuracy. In cross-referencing STAT1 targets obtained by ChIP-chip with those detected by ChIP-PET, we found regions that overlapped between ChIP-chip and ChIP-PET, as well as enriched regions specific to only one of these methods. Thus the sequencing of ChIP DNA fragments is shown to be a valuable and alternative strategy for target identification.
The ChIP-chip conditions applied here for STAT1 can be extended to other DNA-interacting proteins that are constitutively present in the nucleus. In these experiments, the hybridization reference samples are either total genomic DNA or ChIP DNA prepared using normal serum. Examples of other factors we have analyzed by ChIP-chip on 50-b maskless ENCODE tiling arrays include the chromatin remodeling proteins BAF155 and BAF170, as well as the transcription factor c-Jun; the binding profiles of all three of these proteins are part of the ENCODE meta-analyses, and their tracks are available in the UCSC Browser (The ENCODE Project Consortium 2004). As with STAT1, we labeled unamplified ChIP samples of SMARCC1, SMARCC2, and JUN in order to avoid possible biases that may arise during PCR or other amplification methods, and these unamplified ChIP samples exhibited good signal enrichments in our hybridizations.
For the maskless array platforms, longer oligonucleotides most likely improve performance through reduced cross-hybridization and potentially stronger signals. This, in turn, should lead to more accurate measurements and thus more accurate ratios of immunoprecipitated DNA relative to control DNA. Extending this logic, PCR product arrays have even longer DNA fragments as array elements and in theory should provide superior results to oligonucleotide arrays. This is not the case, probably for several reasons. First, multiple probes on high-density oligonucleotide arrays allow for several independent measurements across a region of interest. If any individual probe performs poorly (e.g., because of secondary structure, cross-hybridization, or AT-rich regions), then sampling over multiple probes using a sliding window approach (see Supplemental Methods) can still provide useful signals. Indeed, we have found that signals generated by one or a few oligonucleotides are not usually trustworthy. Second, repetitive sequences on PCR product arrays may reduce signal-to-noise ratios. Finally, a small fraction of PCR products (5%–10%) amplify from regions other than those intended (Rinn et al. 2003). This will lead to misassignment, and the targets will not be validated.
Our validation strategy involved analyzing regions sampled across a range of targets ranked by signal enrichments. By extending the validation frequency as a function of rank, we can extrapolate and determine the sensitivity of the experiment at a particular threshold. It should be noted, however, that positives that are unable to be detected by a specific protocol cannot be assessed for sensitivity using this validation method. Nonetheless, this strategy is expected to provide the best approach available for determining these measurements.
Our study reveals that ChIP-chip and ChIP-PET generally yield similar results, particularly for the strongest signals. However, targets that are uniquely identified by one of these technologies are also captured, and many of these targets could be validated as positives by ChIP-PCR analysis. Targets exclusive to either ChIP-chip or ChIP-PET fall into several classes:
Many unique targets arise from the manner in which positives are scored. Current ChIP-chip scoring methods merge stretches of probes showing signal enrichments into short windows (we used 1.3 kb), and thus adjacent segments are often part of a single larger target region (>1.3 kb), whereas ChIP-PET clusters were connected if the PETs share 1 bp of overlap with no restriction on the length of each cluster region. Grouping adjacent ChIP-chip targets will alleviate this problem, particularly for highly enriched segments where these incidents occur most frequently.
Other targets solely identified by one platform can often be attributed to neighboring repetitive sequences. RepeatMasked sequences are eliminated during the array design process in the ChIP-chip experiments. Consequently, targets that lie within or immediately adjacent to genomic repeats are more likely to be missed by ChIP-chip, but detected by PET sequencing. Conversely, repetitive regions may also lead to false positives through cross-hybridization to real targets (in ChIP-chip) or by the occasional misassignment of a tag containing repetitive DNA elements (in ChIP-PET).
In addition, we would expect that very small targets flanked by large adjacent repeats are also likely to be missed by ChIP-PET but might be detected by ChIP-chip. Since both ChIP-chip and ChIP-PET identify unique, validated targets, the use of both of these technologies in an integrated fashion is anticipated to produce optimal sensitivity and specificity for detecting binding targets.
Our studies suggest that several design parameters can be modified to enhance the performances of ChIP-chip and ChIP-PET. For ChIP-chip, future generations of array design may incorporate the following improvements:
It should be possible to more accurately retrieve targets that lie next to repetitive sequences by increasing the number of oligonucleotide tiles adjacent to repeats.
The judicious choice of nonidentical oligonucleotides should improve array performance.
Finally, the use of isothermal arrays, where the oligonucleotides on the array vary in length to give a more uniform annealing temperature, should improve performance (Urban et al. 2006).
For ChIP-PET, slight modifications to the mapping algorithm should eliminate those few instances in which nearly identical ChIP fragments were double counted in determining the ChIP-PET cluster number (see example in Supplemental Fig. 2).
Another desirable feature of ChIP-PET is that it is inherently whole genome and can theoretically find all targets present in genomic sequences. Currently both ChIP-PET and whole-genome ChIP-chip are expensive because of the considerable cost of high-throughput sequencing and whole-genome oligonucleotide arrays. However, both of these technologies are expected to exhibit dramatic decreases in cost in the near future as new sequencing technologies become available (Margulies et al. 2005; Shendure et al. 2005; Service 2006) and as array densities continue to increase. Thus, both ChIP-chip and ChIP-sequencing technologies will become substantially more cost-effective, and their mutual combination would maximize accuracy.
Methods
STAT1 chromatin immunoprecipitations
STAT1 ChIP samples were prepared from IFNG-stimulated HeLa S3 cells, and ChIP DNA quality was verified as previously described (Hartman et al. 2005). Cultures of 12 × 108 HeLaS3 cells were divided in half and were either induced with 5 ng/mL human recombinant IFNG (R&D Systems #285-IF), or left untreated, for 30 min at 37°C, 5% CO2 and then fixed with 1% formaldehyde final concentration for 10 min at room temperature. Fixations were quenched by addition of glycine to 125 mM final concentration, and cells were washed twice in cold 1× Dulbecco’s PBS. Cells were swelled for 10 min in hypotonic lysis buffer (20 mM HEPES at pH 7.9, 10 mM KCl, 1 mM EDTA at pH 8, 10% glycerol, 1 mM DTT, 0.5 mM PMSF, 0.1 mM sodium orthovanadate, and Roche protease inhibitors #11-697-498-001) and lysed by dounce homogenization (using pestle B). Nuclear pellets were collected and lysed in 1× RIPA buffer (10 mM Tris-Cl at pH 8.0, 140 mM NaCl, 1% Triton X-100, 0.1% SDS, 1% deoxycholic acid, 0.5 mM PMSF, 1 mM DTT, 0.1 mM sodium orthovanadate, and Roche protease inhibitors). Nuclear lysates were sonicated with a Branson 250 Sonifier (output 20%, 100% duty cycle) to shear the chromatin to ∼1 kb in size. Clarified lysates were incubated overnight at 4°C with anti-STAT1 α p91 (C-24) rabbit polyclonal antibody (Santa Cruz Biotechnology #sc-345). Protein–DNA complexes were precipitated with RIPA-equilibrated protein A agarose beads (Upstate #16-156), and immunoprecipitates were washed three times in 1× RIPA, once in 1× PBS, and then eluted from the beads by addition of 1% SDS, 1× TE (10 mM Tris-Cl at pH 7.6, 1 mM EDTA at pH 8), and incubation for 10 min at 65°C. Cross-links were reversed overnight at 65°C. All samples were purified by treatment first with 200 μg/mL RNase A (QIAGEN #19101) for 1 h at 37°C, then with 200 μg/mL Proteinase K (Ambion #2548) for 2 h at 45°C, followed by extraction with phenol:chloroform:isoamyl alcohol and precipitation at −70°C with 0.1 volume of 3 M sodium acetate, 2 volumes of 100% ethanol, and 1.5 μL of pellet paint coprecipitant (Novagen #69049-3). ChIP DNA prepared from 1 × 108 cells was resuspended in 50 μL of ultrapure water (GIBCO-Invitrogen #10977-015).
ChIP sample preparation and labeling
Biological replicates are defined as STAT1 ChIP DNA prepared from distinct cell cultures grown, harvested, and processed on separate days. ChIP DNA samples from individual biological replicates were labeled separately and hybridized separately (without pooling) as one biological replicate per array (Supplemental Table 1). In many cases, the same biological replicates were hybridized to each of the array platforms. For the experiment comparing hybridizations in the presence and absence of Cot-1 DNA, six biological replicates were divided after labeling and hybridized over 12 arrays in plus and minus Cot sets.
For PCR product arrays (gift of Bing Ren, UCSD) and maskless arrays with 50 b every 50-b spacing and 36 b every 36-b spacing (both oligo length arrays manufactured by NASA Ames Research Center), ChIP DNA from 1 × 108 cells was random primed with Klenow (enzyme and primers from BioPrime DNA Labeling System; Invitrogen #18094-011), and Aminoallyl-dUTP (Sigma #A0410) was incorporated. Next Alexa Fluor dyes (Invitrogen #A32755; Alexa647 for ChIP DNA isolated from IFNG-stimulated cells and Alexa555 for ChIP DNA isolated from unstimulated cells) were coupled to the Aminoallyl-dUTP. Coupling reactions were terminated with hydroxylamine. Alexa555- and Alexa647-coupled ChIP DNA samples were combined and recovered using a CyScribe GFX Purification Kit (Amersham #27-9606-02) according to the manufacturer’s protocol. The recovered probe was further purified by ethanol precipitation with 0.1 volume of 3 M sodium acetate (pH 5.2).
For maskless arrays (Nuwaysir et al. 2002) with 50 b every 38-b spacing (NimbleGen Systems of Iceland, LLC), ChIP DNA from 1 × 108 cells was directly labeled (per manufacturer’s protocol) by Klenow random priming with Cy5 nonamers (ChIP DNA isolated from IFNG-stimulated cells) or Cy3 nonamers (ChIP DNA isolated from unstimulated cells).
Microarray hybridizations
All arrays were hybridized with mixing in MAUI hybridization stations from BioMicro Systems for 16−18 h at 42°C. PCR product arrays were prehybridized in 5× SSC/25% formamide/0.05% SDS/1% BSA for 1 h at 42°C. Labeled ChIP DNA was precipitated and resuspended in 60 μL of 5× SSC/25% formamide/0.05% SDS with 5 μg of human Cot-1 DNA (Invitrogen #15279-011) per array. The PCR product arrays were washed in 42°C 2× SSC/0.1% SDS, room temperature 0.1× SSC/0.1% SDS, and 0.1× SSC. Labeled ChIP DNA for maskless arrays (Nuwaysir et al. 2002) with 50 b every 50-b spacing and 36 b every 36-b spacing (both oligo length arrays manufactured by NASA Ames Research Center) was precipitated with 30 μg of human Cot-1 DNA per array, and pellets were resuspended in 45 μL of hybridization buffer (final concentrations: 40% formamide, 5× SSC, 0.1% SDS, and 0.2× TE). Arrays were washed once with 42°C 0.2% SDS/0.2× SSC, once with room temperature NSWB (6× SSPE, 0.01% Tween 20, 1 mM DTT), twice with 0.2× SSC, and twice with 0.05× SSC. For maskless arrays (Nuwaysir et al. 2002) with 50 b every 38-b spacing (NimbleGen Systems of Iceland, LLC), labeled ChIP DNA was hybridized in buffer containing 20% formamide, 1.2 M Betaine, and 0.1 μg/μL herring sperm DNA per the manufacturer’s protocol. The plus Cot-1 experiments included 10 μg of human Cot-1 DNA per array. Arrays were washed in 42°C 0.2% SDS/0.2× SSC, room temperature 0.2× SSC, and 0.05× SSC.
ChIP-PET experiment
The STAT1 ChIP-PET library was constructed as previously described (Wei et al. 2006). PET sequences were extracted from the raw reads and mapped to human genome sequence assembly [hg17]. The process of PET extraction and mapping is essentially the same as previously described for cDNA analysis (Ng et al. 2005). The mapping criteria are that both the 5′ and 3′ signatures must have a minimal 17-bp match, be present on the same chromosome and same strand, in the correct orientation (5′ → 3′), and within 6 kb of genomic distance.
STAT1 target validations
Primers were designed to amplify 200–350-bp fragments from regions throughout the rank-ordered target lists as well as regions where array signals were below cutoff values. ChIP DNA from either 4 × 106 IFNG-stimulated or unstimulated cells was amplified. For each primer pair, parallel reactions were run with 0.2 μg of HeLa S3 genomic DNA to ensure that a sample set would yield a single band of the expected size. The entire completed PCR reactions were loaded on 1.5% agarose gels, and only those primer sets in which entire sample volumes were loaded were analyzed further. Each plate of PCR reactions included positive and negative controls, and all reactions from a plate were loaded on the same gel. Densitometric analyses were made using ImageJ software (http://rsb.info.nih.gov/ij/). For each primer pair, enrichments were calculated for yield from IFNG-stimulated cells relative to yield from unstimulated cells. To qualify as a validated region, enrichments had to be consistently greater than twofold from each of two or more biological replicates. In many cases, more than two biological replicates were tested, and for some regions, validation results were quantified from multiple primer pairs (in separate reactions) to eliminate any primer artifacts. In total, 280 regions were tested for validation. Primer sequences used in the ChIP-PCR assays are available at http://encode.gersteinlab.org/data/Euskirchen_etal/.
Comparison of target lists
As described above, target lists are rank lists of nonoverlapping target regions of uniform size 1300 bp. In order to fairly compare the ChIP-chip data against the ChIP-PET experiment, the ChIP-PET targets were likewise converted into 1300-bp regions centered on the ChIP-PET cluster. Also, comparisons were done for targets identified in regions common to both platforms because the 50 b every 50-b array does not tile all of the ENCODE regions (Supplemental Table 1). When computing the overlap between any two lists of regions (whether the data are from ChIP-chip or ChIP-PET), the number of entries in the first list intersecting the second is not necessarily the same as the number of the second list intersecting the first (this discrepancy typically happens in loci where multiple target sites are located in a short genomic span). In order to avoid this ambiguity, we first merged the two lists under comparison to form a list of union regions. Then using the union set of regions as a basis, we computed the number of regions belonging to only one of the two original lists, or union regions that came from both lists. One important note is that some union target regions occurring in more complicated loci tend to be longer and might only contribute one joint region to the counts of number of union regions shared by both lists, even though the region might correspond to multiple entries on each of the original two lists. Regions that have been tested for validation can also be compared against these union target regions to assess validation rates for union regions that were detected on only one of the two lists or by both data sets. This is how the data in Tables 1, 2, 3, and 6 and Supplemental Tables S2 and S3 were generated.
Acknowledgments
The authors are grateful to Bing Ren (UCSD) for sharing the PCR product arrays. We thank Janine Mok and Mike Hudson for critical reading of the manuscript. Elsa Eysteinsdottir and Chloe Leplar of NimbleGen Systems of Iceland, LLC provided expert microarray support. This work was funded by NIH ENCODE grant HG003156.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.5583007
References
- Bailey T.L., Elkan C., Elkan C. The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1995;3:21–29. [PubMed] [Google Scholar]
- Bertone P., Trifonov V., Rozowsky J.S., Schubert F., Emanuelsson O., Karro J., Kao M.Y., Snyder M., Gerstein M., Trifonov V., Rozowsky J.S., Schubert F., Emanuelsson O., Karro J., Kao M.Y., Snyder M., Gerstein M., Rozowsky J.S., Schubert F., Emanuelsson O., Karro J., Kao M.Y., Snyder M., Gerstein M., Schubert F., Emanuelsson O., Karro J., Kao M.Y., Snyder M., Gerstein M., Emanuelsson O., Karro J., Kao M.Y., Snyder M., Gerstein M., Karro J., Kao M.Y., Snyder M., Gerstein M., Kao M.Y., Snyder M., Gerstein M., Snyder M., Gerstein M., Gerstein M. Design optimization methods for genomic DNA tiling arrays. Genome Res. 2006;16:271–281. doi: 10.1101/gr.4452906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm U., Klamp T., Groot M., Howard J.C., Klamp T., Groot M., Howard J.C., Groot M., Howard J.C., Howard J.C. Cellular responses to interferon-γ. Annu. Rev. Immunol. 1997;15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Kumar R.M., Murray H.L., Jenner R.G., Murray H.L., Jenner R.G., Jenner R.G., et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg J., Chen X., Chen X. STAT proteins: Signal transducers and activators of transcription. Methods Enzymol. 2001;333:138–151. doi: 10.1016/s0076-6879(01)33052-5. [DOI] [PubMed] [Google Scholar]
- Buck M.J., Lieb J.D., Lieb J.D. ChIP-chip: Considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics. 2004;83:349–360. doi: 10.1016/j.ygeno.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Cawley S., Bekiranov S., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Bekiranov S., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Sementchenko V., Cheng J., Williams A.J., Cheng J., Williams A.J., Williams A.J., et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- Chen J., Sadowski I., Sadowski I. Identification of the mismatch repair genes PMS2 and MLH1 as p53 target genes by using serial analysis of binding elements. Proc. Natl. Acad. Sci. 2005;102:4813–4818. doi: 10.1073/pnas.0407069102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi J., Penland L., Brown P.O., Bittner M.L., Meltzer P.S., Ray M., Chen Y., Su Y.A., Trent J.M., Penland L., Brown P.O., Bittner M.L., Meltzer P.S., Ray M., Chen Y., Su Y.A., Trent J.M., Brown P.O., Bittner M.L., Meltzer P.S., Ray M., Chen Y., Su Y.A., Trent J.M., Bittner M.L., Meltzer P.S., Ray M., Chen Y., Su Y.A., Trent J.M., Meltzer P.S., Ray M., Chen Y., Su Y.A., Trent J.M., Ray M., Chen Y., Su Y.A., Trent J.M., Chen Y., Su Y.A., Trent J.M., Su Y.A., Trent J.M., Trent J.M. Use of a cDNA microarray to analyse expression patterns in cancer. Nat. Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, Indentification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007 doi: 10.1038/nature05874. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euskirchen G., Royce T.E., Bertone P., Martone R., Rinn J.L., Nelson F.K., Sayward F., Luscombe N.M., Miller P., Gerstein M., Royce T.E., Bertone P., Martone R., Rinn J.L., Nelson F.K., Sayward F., Luscombe N.M., Miller P., Gerstein M., Bertone P., Martone R., Rinn J.L., Nelson F.K., Sayward F., Luscombe N.M., Miller P., Gerstein M., Martone R., Rinn J.L., Nelson F.K., Sayward F., Luscombe N.M., Miller P., Gerstein M., Rinn J.L., Nelson F.K., Sayward F., Luscombe N.M., Miller P., Gerstein M., Nelson F.K., Sayward F., Luscombe N.M., Miller P., Gerstein M., Sayward F., Luscombe N.M., Miller P., Gerstein M., Luscombe N.M., Miller P., Gerstein M., Miller P., Gerstein M., Gerstein M., et al. CREB binds to multiple loci on human chromosome 22. Mol. Cell. Biol. 2004;24:3804–3814. doi: 10.1128/MCB.24.9.3804-3814.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman S.E., Bertone P., Nath A.K., Royce T.E., Gerstein M., Weissman S., Snyder M., Bertone P., Nath A.K., Royce T.E., Gerstein M., Weissman S., Snyder M., Nath A.K., Royce T.E., Gerstein M., Weissman S., Snyder M., Royce T.E., Gerstein M., Weissman S., Snyder M., Gerstein M., Weissman S., Snyder M., Weissman S., Snyder M., Snyder M. Global changes in STAT target selection and transcription regulation upon interferon treatments. Genes & Dev. 2005;19:2953–2968. doi: 10.1101/gad.1371305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde P., Qi R., Abernathy K., Gay C., Dharap S., Gaspard R., Earle-Hughes J., Snesrud E., Lee N., Quackenbush J., Qi R., Abernathy K., Gay C., Dharap S., Gaspard R., Earle-Hughes J., Snesrud E., Lee N., Quackenbush J., Abernathy K., Gay C., Dharap S., Gaspard R., Earle-Hughes J., Snesrud E., Lee N., Quackenbush J., Gay C., Dharap S., Gaspard R., Earle-Hughes J., Snesrud E., Lee N., Quackenbush J., Dharap S., Gaspard R., Earle-Hughes J., Snesrud E., Lee N., Quackenbush J., Gaspard R., Earle-Hughes J., Snesrud E., Lee N., Quackenbush J., Earle-Hughes J., Snesrud E., Lee N., Quackenbush J., Snesrud E., Lee N., Quackenbush J., Lee N., Quackenbush J., Quackenbush J. A concise guide to cDNA microarray analysis. Biotechniques. 2000;29:548–562. doi: 10.2144/00293bi01. [DOI] [PubMed] [Google Scholar]
- Horak C.E., Mahajan M.C., Luscombe N.M., Gerstein M., Weissman S.M., Snyder M., Mahajan M.C., Luscombe N.M., Gerstein M., Weissman S.M., Snyder M., Luscombe N.M., Gerstein M., Weissman S.M., Snyder M., Gerstein M., Weissman S.M., Snyder M., Weissman S.M., Snyder M., Snyder M. GATA-1 binding sites mapped in the β-globin locus by using mammalian ChIP-chip analysis. Proc. Natl. Acad. Sci. 2002;99:2924–2929. doi: 10.1073/pnas.052706999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug B.A., Ahmed N., Robbins J.A., Lazar M.A., Ahmed N., Robbins J.A., Lazar M.A., Robbins J.A., Lazar M.A., Lazar M.A. A chromatin immunoprecipitation screen reveals protein kinase cβ as a direct RUNX1 target gene. J. Biol. Chem. 2004;279:825–830. doi: 10.1074/jbc.M309524200. [DOI] [PubMed] [Google Scholar]
- Impey S., McCorkle S.R., Cha-Molstad H., Dwyer J.M., Yochum G.S., Boss J.M., McWeeney S., Dunn J.J., Mandel G., Goodman R.H., McCorkle S.R., Cha-Molstad H., Dwyer J.M., Yochum G.S., Boss J.M., McWeeney S., Dunn J.J., Mandel G., Goodman R.H., Cha-Molstad H., Dwyer J.M., Yochum G.S., Boss J.M., McWeeney S., Dunn J.J., Mandel G., Goodman R.H., Dwyer J.M., Yochum G.S., Boss J.M., McWeeney S., Dunn J.J., Mandel G., Goodman R.H., Yochum G.S., Boss J.M., McWeeney S., Dunn J.J., Mandel G., Goodman R.H., Boss J.M., McWeeney S., Dunn J.J., Mandel G., Goodman R.H., McWeeney S., Dunn J.J., Mandel G., Goodman R.H., Dunn J.J., Mandel G., Goodman R.H., Mandel G., Goodman R.H., Goodman R.H. Defining the CREB regulon: A genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- Kim J., Bhinge A.A., Morgan X.C., Iyer V.R., Bhinge A.A., Morgan X.C., Iyer V.R., Morgan X.C., Iyer V.R., Iyer V.R. Mapping DNA–protein interactions in large genomes by sequence tag analysis of genomic enrichment. Nat. Methods. 2005a;2:47–53. doi: 10.1038/nmeth726. [DOI] [PubMed] [Google Scholar]
- Kim T.H., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Richmond T.A., Wu Y., Green R.D., Ren B., Wu Y., Green R.D., Ren B., Green R.D., Ren B., Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005b;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.I., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K.I., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K.I., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K.I., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K.I., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K.I., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K.I., Chevalier B., Johnstone S.E., Cole M.F., Isono K.I., Johnstone S.E., Cole M.F., Isono K.I., Cole M.F., Isono K.I., Isono K.I., et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D.E., Darnell J.E., Jr., Darnell J.E., Jr. Stats: Transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- Liu X., Brutlag D.L., Liu J.S., Brutlag D.L., Liu J.S., Liu J.S. BioProspector: Discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac. Symp. Biocomput. 2001:127–138. [PubMed] [Google Scholar]
- Liu X.S., Brutlag D.L., Liu J.S., Brutlag D.L., Liu J.S., Liu J.S. An algorithm for finding protein–DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat. Biotechnol. 2002;20:835–839. doi: 10.1038/nbt717. [DOI] [PubMed] [Google Scholar]
- Loh Y.H., Wu Q., Chew J.L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Wu Q., Chew J.L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Chew J.L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Chen X., Bourque G., George J., Leong B., Liu J., Bourque G., George J., Leong B., Liu J., George J., Leong B., Liu J., Leong B., Liu J., Liu J., et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Berka J., Braverman M.S., Chen Y.J., Chen Z., Braverman M.S., Chen Y.J., Chen Z., Chen Y.J., Chen Z., Chen Z., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martone R., Euskirchen G., Bertone P., Hartman S., Royce T.E., Luscombe N.M., Rinn J.L., Nelson F.K., Miller P., Gerstein M., Euskirchen G., Bertone P., Hartman S., Royce T.E., Luscombe N.M., Rinn J.L., Nelson F.K., Miller P., Gerstein M., Bertone P., Hartman S., Royce T.E., Luscombe N.M., Rinn J.L., Nelson F.K., Miller P., Gerstein M., Hartman S., Royce T.E., Luscombe N.M., Rinn J.L., Nelson F.K., Miller P., Gerstein M., Royce T.E., Luscombe N.M., Rinn J.L., Nelson F.K., Miller P., Gerstein M., Luscombe N.M., Rinn J.L., Nelson F.K., Miller P., Gerstein M., Rinn J.L., Nelson F.K., Miller P., Gerstein M., Nelson F.K., Miller P., Gerstein M., Miller P., Gerstein M., Gerstein M., et al. Distribution of NF-κB binding sites across human chromosome 22. Proc. Natl. Acad. Sci. 2003;100:12247–12252. doi: 10.1073/pnas.2135255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P., Wei C.L., Sung W.K., Chiu K.P., Lipovich L., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., Wei C.L., Sung W.K., Chiu K.P., Lipovich L., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., Sung W.K., Chiu K.P., Lipovich L., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., Chiu K.P., Lipovich L., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., Lipovich L., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., Gupta S., Shahab A., Ridwan A., Wong C.H., Shahab A., Ridwan A., Wong C.H., Ridwan A., Wong C.H., Wong C.H., et al. Gene identification signature (GIS) analysis for transcriptome characterization and genome annotation. Nat. Methods. 2005;2:105–111. doi: 10.1038/nmeth733. [DOI] [PubMed] [Google Scholar]
- Nuwaysir E.F., Huang W., Albert T.J., Singh J., Nuwaysir K., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., Huang W., Albert T.J., Singh J., Nuwaysir K., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., Albert T.J., Singh J., Nuwaysir K., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., Singh J., Nuwaysir K., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., Nuwaysir K., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., Richmond T., Gorski T., Berg J.P., Ballin J., Gorski T., Berg J.P., Ballin J., Berg J.P., Ballin J., Ballin J., et al. Gene expression analysis using oligonucleotide arrays produced by maskless photolithography. Genome Res. 2002;12:1749–1755. doi: 10.1101/gr.362402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberley M.J., Farnham P.J., Farnham P.J. Probing chromatin immunoprecipitates with CpG-island microarrays to identify genomic sites occupied by DNA-binding proteins. Methods Enzymol. 2003;371:577–596. doi: 10.1016/S0076-6879(03)71043-X. [DOI] [PubMed] [Google Scholar]
- Odom D.T., Zizlsperger N., Gordon D.B., Bell G.W., Rinaldi N.J., Murray H.L., Volkert T.L., Schreiber J., Rolfe P.A., Gifford D.K., Zizlsperger N., Gordon D.B., Bell G.W., Rinaldi N.J., Murray H.L., Volkert T.L., Schreiber J., Rolfe P.A., Gifford D.K., Gordon D.B., Bell G.W., Rinaldi N.J., Murray H.L., Volkert T.L., Schreiber J., Rolfe P.A., Gifford D.K., Bell G.W., Rinaldi N.J., Murray H.L., Volkert T.L., Schreiber J., Rolfe P.A., Gifford D.K., Rinaldi N.J., Murray H.L., Volkert T.L., Schreiber J., Rolfe P.A., Gifford D.K., Murray H.L., Volkert T.L., Schreiber J., Rolfe P.A., Gifford D.K., Volkert T.L., Schreiber J., Rolfe P.A., Gifford D.K., Schreiber J., Rolfe P.A., Gifford D.K., Rolfe P.A., Gifford D.K., Gifford D.K., et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B., Cam H., Takahashi Y., Volkert T., Terragni J., Young R.A., Dynlacht B.D., Cam H., Takahashi Y., Volkert T., Terragni J., Young R.A., Dynlacht B.D., Takahashi Y., Volkert T., Terragni J., Young R.A., Dynlacht B.D., Volkert T., Terragni J., Young R.A., Dynlacht B.D., Terragni J., Young R.A., Dynlacht B.D., Young R.A., Dynlacht B.D., Dynlacht B.D. E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints. Genes & Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J.L., Euskirchen G., Bertone P., Martone R., Luscombe N.M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Euskirchen G., Bertone P., Martone R., Luscombe N.M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Bertone P., Martone R., Luscombe N.M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Martone R., Luscombe N.M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Luscombe N.M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Nelson F.K., Miller P., Gerstein M., Miller P., Gerstein M., Gerstein M., et al. The transcriptional activity of human chromosome 22. Genes & Dev. 2003;17:529–540. doi: 10.1101/gad.1055203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh T.Y., Cuddapah S., Zhao K., Cuddapah S., Zhao K., Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes & Dev. 2005;19:542–552. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Service R.F. The race for the $1000 genome. Science. 2006;311:1544–1546. doi: 10.1126/science.311.5767.1544. [DOI] [PubMed] [Google Scholar]
- Shendure J., Porreca G.J., Reppas N.B., Lin X., McCutcheon J.P., Rosenbaum A.M., Wang M.D., Zhang K., Mitra R.D., Church G.M., Porreca G.J., Reppas N.B., Lin X., McCutcheon J.P., Rosenbaum A.M., Wang M.D., Zhang K., Mitra R.D., Church G.M., Reppas N.B., Lin X., McCutcheon J.P., Rosenbaum A.M., Wang M.D., Zhang K., Mitra R.D., Church G.M., Lin X., McCutcheon J.P., Rosenbaum A.M., Wang M.D., Zhang K., Mitra R.D., Church G.M., McCutcheon J.P., Rosenbaum A.M., Wang M.D., Zhang K., Mitra R.D., Church G.M., Rosenbaum A.M., Wang M.D., Zhang K., Mitra R.D., Church G.M., Wang M.D., Zhang K., Mitra R.D., Church G.M., Zhang K., Mitra R.D., Church G.M., Mitra R.D., Church G.M., Church G.M. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- Urban A.E., Korbel J.O., Selzer R., Richmond T., Hacker A., Popescu G.V., Cubells J.F., Green R., Emanuel B.S., Gerstein M.B., Korbel J.O., Selzer R., Richmond T., Hacker A., Popescu G.V., Cubells J.F., Green R., Emanuel B.S., Gerstein M.B., Selzer R., Richmond T., Hacker A., Popescu G.V., Cubells J.F., Green R., Emanuel B.S., Gerstein M.B., Richmond T., Hacker A., Popescu G.V., Cubells J.F., Green R., Emanuel B.S., Gerstein M.B., Hacker A., Popescu G.V., Cubells J.F., Green R., Emanuel B.S., Gerstein M.B., Popescu G.V., Cubells J.F., Green R., Emanuel B.S., Gerstein M.B., Cubells J.F., Green R., Emanuel B.S., Gerstein M.B., Green R., Emanuel B.S., Gerstein M.B., Emanuel B.S., Gerstein M.B., Gerstein M.B., et al. High-resolution mapping of DNA copy alterations in human chromosome 22 using high-density tiling oligonucleotide arrays. Proc. Natl. Acad. Sci. 2006;103:4534–4539. doi: 10.1073/pnas.0511340103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman W., Sandelin A., Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 2004;5:267–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- Wei C.L., Wu Q., Vega V.B., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., Wu Q., Vega V.B., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., Vega V.B., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., Shahab A., Yong H.C., Fu Y., Weng Z., Yong H.C., Fu Y., Weng Z., Fu Y., Weng Z., Weng Z., et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Weinmann A.S., Bartley S.M., Zhang T., Zhang M.Q., Farnham P.J., Bartley S.M., Zhang T., Zhang M.Q., Farnham P.J., Zhang T., Zhang M.Q., Farnham P.J., Zhang M.Q., Farnham P.J., Farnham P.J. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol. Cell. Biol. 2001;21:6820–6832. doi: 10.1128/MCB.21.20.6820-6832.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann A.S., Pearlly S.Y., Oberley M.J., Huang T.H.-M., Farnham P.J., Pearlly S.Y., Oberley M.J., Huang T.H.-M., Farnham P.J., Oberley M.J., Huang T.H.-M., Farnham P.J., Huang T.H.-M., Farnham P.J., Farnham P.J. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes & Dev. 2002;16:235–244. doi: 10.1101/gad.943102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Smith L.T., Plass C., Huang T.H.-M., Smith L.T., Plass C., Huang T.H.-M., Plass C., Huang T.H.-M., Huang T.H.-M. ChIP-chip comes of age for genome-wide functional analysis. Cancer Res. 2006;66:6899–6902. doi: 10.1158/0008-5472.CAN-06-0276. [DOI] [PubMed] [Google Scholar]