Abstract
Identifying the genome-wide binding sites of transcription factors is important in deciphering transcriptional regulatory networks. ChIP-chip (Chromatin immunoprecipitation combined with microarrays) has been widely used to map transcription factor binding sites in the human genome. However, whole genome ChIP-chip analysis is still technically challenging in vertebrates. We recently developed STAGE as an unbiased method for identifying transcription factor binding sites in the genome. STAGE is conceptually based on SAGE, except that the input is ChIP-enriched DNA. In this study, we implemented an improved sequencing strategy and analysis methods and applied STAGE to map the genomic binding profile of the transcription factor STAT1 after interferon treatment. STAT1 is mainly responsible for mediating the cellular responses to interferons, such as cell proliferation, apoptosis, immune surveillance, and immune responses. We present novel algorithms for STAGE tag analysis to identify enriched loci with high specificity, as verified by quantitative ChIP. STAGE identified several previously unknown STAT1 target genes, many of which are involved in mediating the response to interferon-γ signaling. STAGE is thus a viable method for identifying the chromosomal targets of transcription factors and generating meaningful biological hypotheses that further our understanding of transcriptional regulatory networks.
The ENCODE project has suggested that a larger fraction of the human genome than previously suspected may be transcriptionally active (The ENCODE Project Consortium 2006). Correspondingly, a significant fraction of the genome is likely to be involved in regulating gene expression and other aspects of human biology. Much of the regulatory potential of cis-acting sequences in the genome involves interactions of proteins with DNA. Identifying the genomic binding sites of regulatory proteins such as transcription factors is important for cataloging the regulatory potential encoded in the human genome and reconstructing transcriptional regulatory networks. Chromatin immunoprecipitation (ChIP) combined with microarray hybridization (ChIP-chip) has enabled global mapping of transcription factor binding sites in the human genome (Kim et al. 2005a; Lee et al. 2006). Although whole-genome oligonucleotide tiling arrays are becoming available for ChIP-chip analyses, they remain expensive and entail specialized resources. Another limitation with the use of tiling arrays is that they typically do not cover repetitive sequences, which account for a significant fraction of the genome. For example, recent “whole-genome” tiling arrays included only ∼50% of the genome that was nonrepetitive (Kim et al. 2005a; Lee et al. 2006). Binding sites and functional elements that lie near repetitive sequences are therefore likely to be undetected through the use of such arrays. Many tiling array platforms currently need seven to a few dozen arrays to cover the genome, requiring significant scale up of antibody, cell culture material, and effort, especially if replicate experiments are performed.
We have developed an unbiased genomic method to map transcription factor binding sites called STAGE (Sequence Tag Analysis of Genomic Enrichment), based on sequencing “tags” or short oligonucleotide signatures from ChIP-enriched DNA (Kim et al. 2005b). Since it is not constrained by the availability of tiling microarrays for any particular organism, STAGE makes it possible to experimentally determine whether the target genes of a transcription factor in one species are also targets in a related species. Similar sequencing-based approaches for identifying transcription factor targets have recently also been independently developed in other labs (Impey et al. 2004; Roh et al. 2004, 2005; Chen and Sadowski 2005; Loh et al. 2006; Wei et al. 2006).
In order to make STAGE more competitive with genome-wide tiling arrays, we have now developed modifications that exploit new developments in sequencing technology. Here we use STAGE for analysis of the targets of the transcription factor, STAT1. We used bead-based pyrosequencing (454) technology to improve the throughput and cost-effectiveness of sequencing and significantly reduce the time and effort needed to perform STAGE (Margulies et al. 2005). STAT (Signal Transducer and Activator of Transcription) proteins are transcription factors that mediate cytokine and growth factor signaling. Interferons modulate cell proliferation, apoptosis, immune surveillance, and immune responses primarily via the JAK-STAT pathway (Platanias 2005). Interferon (IFNG) specifically activates STAT1 which forms homodimers, translocates to the nucleus, and binds to promoters bearing the gamma-activation sequence (GAS) motif and activates (IFNG) inducible genes (Ramana et al. 2000). ChIP-chip analysis of STAT1 targets on chromosome 22 revealed that STAT1 regulates several genes involved in cell growth, apoptosis, immune responses, and lipid metabolism (Hartman et al. 2005). We used STAGE to identify genome-wide STAT1 binding targets after interferon (IFNG) treatment. We also developed improved analysis algorithms to identify target sites with high specificity. Our results indicate that IFNG-induced STAT1 binds to a large number of sites genome-wide and that many of these sites lie proximal to genes that are involved in biological processes modulated by IFNG.
Results
Identifying STAGE tags for STAT1 by bead-based pyrosequencing
DNA bound by STAT1 in IFNG-treated HeLa cells was isolated by ChIP. We generated ditags as described before (Kim et al. 2005b) and amplified ditags by PCR. Amplified ditags were sequenced by 454 Inc., but without the initial nebulization step normally used in their procedure to shear the DNA. Thus, each read typically contained a complete STAGE ditag, flanked by primer sequences. We sequenced a total of 179,954 reads from the STAT1 STAGE tag library, representing about 17 Mb of sequence from one run. After removing duplicate reads, we were able to extract 162,577 tags; 31,353 tags (19%) could not be matched to any location in the genome and were considered orphans. The remaining 131,224 tags were used for further analysis.
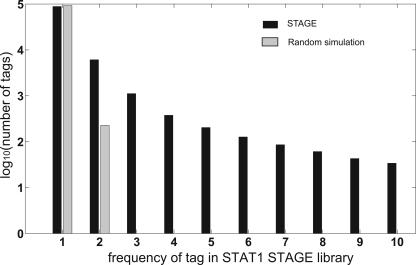
If STAGE tags are derived from ChIP-enriched DNA, then the distribution of tags in the STAGE library should deviate from a randomly selected population of tags. We simulated background tag libraries in silico by randomly selecting the same number of tags (131,224 for STAT1) from the entire genome multiple times. Tags that had more than one hit, i.e., a perfect match, on the genome were ignored. The average frequency distribution of single-hit tags in the random library was compared with the experimental STAT1 STAGE library. For a frequency of occurrence of 1, the numbers of tags in the random and real data were similar. However, for a frequency of occurrence of 2 and more, there was strong enrichment in the STAGE library over background (Fig. 1). Thus, the STAGE tags generated by 454 sequencing represented DNA that was distinct from simulated random genomic DNA.
Figure 1.
Comparison of the STAT1 STAGE tag library with a simulated randomly generated background library. A background library was generated to simulate STAGE tag libraries by randomly selecting the same number of tags from the genome as the experimental STAGE library. This procedure was repeated 20 times and the values were averaged. Only tags with a single, unique hit on the genome were used in this analysis. The numbers of single-hit tags (Y-axis) were plotted against the frequencies of those tags in the random (gray bars) and experimental (black bars) tag library (X-axis). For frequencies of 2 and above, the STAGE tag library for STAT1 shows a clear enrichment over a randomly generated tag library.
STAGE targets for STAT1
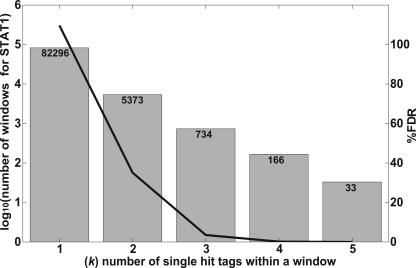
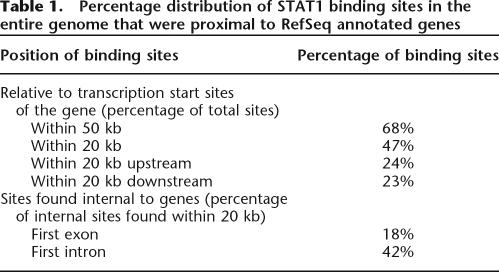
Since ∼50% of the human genome consists of repeat sequences, a given tag in the STAGE library may map to multiple locations in the genome. A tag that is represented in the genome at multiple locations would be more likely to be found in the STAGE library by random chance. Hence, a higher frequency of occurrence of a tag in the STAGE library does not necessarily reflect the enrichment of the tag in the ChIP-enriched DNA. To exclude such ambiguous tags in our analysis, we calculated the probability that a given tag was truly enriched over background by ChIP. Each tag was first assigned a probability of enrichment by assuming that the selection of tags from the genome follows a binomial distribution. Details of the calculations and the algorithm we developed to identify significant targets are included in the Methods. Since STAGE tags are derived from ChIP-enriched DNA, multiple tags can be expected to cluster within short regions in the genome similar in size to the fragments isolated by ChIP, as compared to a random library representing no enrichment, where the tags would be expected to be sampled uniformly across wide regions in the genome. We used this rationale to define binding targets. We performed a simulation where we scanned windows of different sizes across each chromosome and counted the frequencies of windows containing different numbers of single-hit tags. For each window size, we determined whether there were a larger number of windows containing a given number of single-hit tags in the real STAGE library as compared to a simulated random library of STAGE tags. A window of 500 bp gave a false discovery rate (FDR) based on simulations of <5% for STAT1 while the number of targets detected was 734 (Fig. 2). The complete set of data for all window sizes used is given in Supplemental Table 1. We used a window of 500 bp for all further analysis. To improve the specificity of target detection, a window was considered a target only if at least one tag within that window was deemed to be enriched. Thus, for each window we calculated two probabilities, namely, the probability of finding a given number of single-hit tags and the probability that at least one of those tags was statistically likely to be enriched. To avoid assigning high probabilities to windows that contained only a single enriched tag, we gave greater weight to the probability of finding a given number of single-hit tags within a window than to the probability of simply finding any enriched tags in that window. This combined probability calculation gave us a false discovery rate of <1% at a probability threshold of 0.95. It should be noted, however, that this false discovery rate is based on in silico analysis under the assumption that selection of STAGE tags follows a binomial distribution. It is possible that experimental manipulations introduce biases that were not modeled in the simulation. STAGE detected 381 binding sites for STAT1 in the entire genome at this threshold. Based on annotations in the RefSeq gene database (Pruitt et al. 2005), 68% of the STAT1 binding sites found by STAGE were within 50 kb of the transcription start site (TSS) of a gene, 70% of which were found within 20 kb (Table 1).
Figure 2.
Determination of optimal window size used for target identification. Windows of different sizes (300, 500, 1000, and 2000 bp) were scanned across the entire genome. For each window, we defined k as the number of single-hit tags found within the window. The number of windows observed for a given k in the STAGE tag data was compared with the number observed in random simulated data. A window size of 500 bp gave an optimal separation between random and real data. Data shown is for a window size of 500 bp. The gray bars indicate log10 of the number of windows detected based on STAT1 tags, with actual numbers of windows at each k listed at the top of the column. The black line shows the decline in the false discovery rate (FDR) with increasing k. The FDR was calculated as the ratio of the number of windows found in the random simulated library to the number of windows detected in the experimental STAT1 library. The raw data for other window sizes is included in Supplemental Table 1.
Table 1.
Percentage distribution of STAT1 binding sites in the entire genome that were proximal to RefSeq annotated genes
Verification of STAT1 targets by ChIP-chip and quantitative ChIP
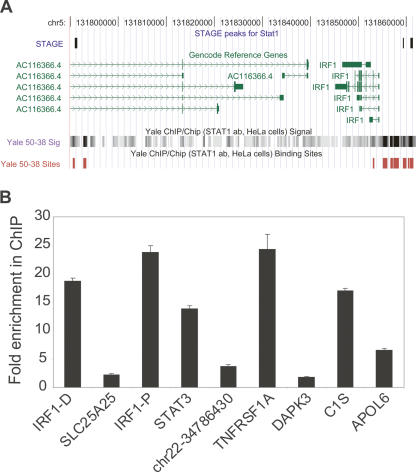
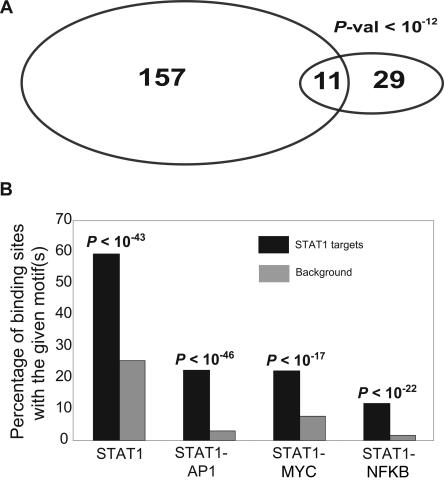
Seven of the 381 STAT1 binding sites identified in the genome by STAGE were within the ENCODE regions. Three of these seven targets overlapped with a ChIP-chip peak where the STAT1 ChIP-chip was performed on ENCODE region tiling oligonucleotide arrays (Fig. 3A; The ENCODE Project Consortium 2007). To obtain a quantitative estimate of the false positive rate of our STAGE analysis, we selected 10 target sites identified by STAGE that had probabilities ranging from 0.95 to 1.0 and assayed their enrichment in a biologically independent STAT1 ChIP sample. Nine out of these 10 sites showed a quantitative enrichment in the ChIP sample relative to the input, with eight of them showing an enrichment of more than twofold (Fig. 3B). Thus, we estimate our true positive rate to be ∼90% giving a false positive rate of 0.1. We also compared STAT1 target genes identified by STAGE to STAT1 target promoters that we identified by ChIP-chip using a global core-promoter microarray. The core-promoter microarray included 9764 different promoters where a promoter was defined as 1 kb upstream of and 200 bp downstream from the TSS of a gene. ChIP-chip revealed 157 promoters to be bound by STAT1 at an enrichment ratio greater than threefold. Twenty-nine out of these 9764 promoters had a high-confidence STAT1 binding site, as identified by STAGE, between 1 kb upstream of and 200 bp downstream from the TSS, and 11 out of these 29 were in common with the targets identified by ChIP-chip (Fig. 4A). Under a hypergeometric distribution, this overlap was significant at a P-value <10−12.
Figure 3.
(A) STAT1 binding sites in the ENCODE regions. A portion of the ENCODE region ENm002 is shown as displayed in the UCSC Human Genome Browser. Three out of the seven STAT1 binding sites identified by STAGE matched STAT1 binding sites identified by ChIP-chip analysis performed on NimbleGen ENCODE region tiling arrays. Transcripts identified in this region by the GENCODE project are shown in green. The bottom shows raw ratio data as well as peak calls for STAT1 binding sites from NimbleGen ChIP-chip data. (B) Quantitative ChIP analysis of binding sites identified by STAGE. Nine out of 10 binding sites detected by STAGE were validated as true binding loci by quantitative PCR. Columns show fold enrichment of each locus in the ChIP sample relative to input DNA, normalized to an unrelated control locus. STAGE detected two binding sites separated by >1500 bp in the IRF1 promoter which are indicated in the figure. IRF-D indicates the distal (IRF1-distal) and IRF1-P indicates the proximal site (IRF1-proximal). No genes were found in the proximity of the site indicated as chr22-34786430.
Figure 4.
(A) Overlap of STAT1 target genes identified by STAGE with STAT1 target genes identified by ChIP-chip using a core promoter array. STAGE identified 29 promoters out of the ∼9000 promoters present on the core promoter array as STAT1 target promoters. Eleven out of these 29 overlapped with the 157 promoters identified as STAT1 targets by ChIP-chip analysis at an enrichment ratio greater than threefold. The enrichment ratio refers to the ratio of the fluorescence intensity of ChIP DNA to that of reference DNA at each spot on the core promoter microarray. (B) Motif analysis. The Y-axis shows the percentage counts of the number of sites bearing the given motif(s) out of the 381 STAT1 binding sites detected by STAGE. Almost 60% of the 381 binding sites had the STAT1 motif TTCNNNGAA as compared to 27% in the background. We also detected an enrichment for the co-occurrence of the binding motifs for STAT1 and AP1 (TGAG/CTCA), STAT1 and MYC (CACA/GTG), and STAT1 and NFKB (GGGA/GNNC/TC/TCC) in accordance with the fact that STAT1 exhibits cooperative binding with these factors to regulate downstream promoters.
Enrichment of motifs in STAT1 targets
If a STAT1 binding site detected by STAGE occurred within 1 kb upstream of and 200 bp downstream from the TSS of a gene, we considered that gene to be a STAT1 target. STAGE detected 59 genes in RefSeq as STAT1 targets by the above criteria (Supplemental Table 2). Sixty-two percent of these target genes (37/59) had the GAS STAT1 motif TTCNNNGAA within 1 kb upstream of and 200 bp downstream from the TSS of the gene. This represented a motif enrichment among target promoters of more than twofold compared to background. The background in this case was considered as 1 kb upstream of and 200 bp downstream from the TSS of all genes in RefSeq. This enrichment was statistically significant (P-value <10−8) assuming a hypergeometric distribution.
We applied the same analysis for all STAT1 binding sites in the entire genome. For each window detected as a STAT1 binding site, we searched for the STAT1 GAS motif in that window extending our search to 250 bp on either side of the window. Out of 381 binding sites detected by STAGE, 226 (59.32%) had the GAS consensus sequence. This represents an enrichment of more than twofold over background level of occurrence of the GAS motif in randomly selected windows from the entire genome (P-value <10−43) (Fig. 4B). Additionally, in accordance with the fact that STAT1 is known to exhibit cooperative binding with other transcription factors like AP1, MYC, and NFKB, we found an enrichment for the STAT1 motif along with motifs for AP1 (Eferl and Wagner 2003), MYC (Adhikary and Eilers 2005), and NFKB (Martone et al. 2003) (Fig. 4B).
Genes proximal to STAT1 binding sites
STAGE identified several previously unknown STAT1 target genes (Supplemental Table 2), many of which are involved in IFNG signaling. One of these was DAPK3 (death-associated protein kinase 3), a positive regulator of programmed cell death. DAPK3 induces apoptosis by associating with the pro-apoptotic protein DAXX. IFNG is known to increase DAPK3–DAXX complex formation and this complex is necessary for induction of caspases and IFNG-mediated apoptosis (Kawai et al. 2003). STAT1 modulation of DAPK3 could thus represent one mechanism by which IFNG can induce apoptosis. DAPK3 phosphorylates MDM2 and (CDKN1A), components of the TP53 pathway (Burch et al. 2004), and its identification as a STAT1 target suggests a novel collaboration between the IFNG/STAT1 apoptotic pathway and the TP53 tumor suppressor pathway. Another possible mechanism for IFNG-mediated apoptosis was suggested by the observation that APOL6, which induces mitochondria-mediated apoptosis characterized by the release of cytochrome-c and activation of caspase-9 (Liu et al. 2005), was also identified as a STAT1 target by STAGE.
STAT3 is anti-apoptotic and induces cell proliferation while STAT1 promotes growth arrest and apoptosis (Stephanou et al. 2000; Stephanou and Latchman 2005). In mouse embryonic fibroblasts, it was shown that IFNG induces high levels of expression of STAT1 while STAT3 levels remain low. However, in the absence of STAT1, i.e., in STAT1−/− cells, IFNG stimulation induces high levels of STAT3 gene expression (Ramana et al. 2005). Our data implicating STAT3 as a direct transcriptional target of STAT1 suggest that STAT1 represses STAT3 during IFNG signaling, further promoting its own apoptotic function.
Tumor necrosis factor (TNF) is cytokine that is involved in a plethora of cellular responses including cell differentiation, survival, and apoptosis. TNF binds to its receptor TNFRSF1A (Tumor Necrosis Factor Receptor Super Family 1A) and causes NFKB activation, which is crucial for the expression of many proinflammatory cytokines, chemokines, and multiple regulators of apoptosis and cell differentiation. In the absence of IFNG stimulation, cytoplasmic STAT1 binds to TNFRSF1A and maintains a tight control over TNF-mediated NFKB activation. However, IFNG stimulation was shown to increase sensitivity of cells to further TNF stimulation (Wesemann and Benveniste 2003). STAGE identified a STAT1 binding site in the first intron of TNFRSF1A, suggesting the possibility that IFNG dependent increased sensitivity to TNF could be a direct result of activation of TNFRSF1A by IFNG-stimulated STAT1. All the target sites and genes described above were verified by quantitative ChIP from an independent ChIP sample (Fig. 3B). We also identified other previously known STAT1 targets such as IRF1, HLA-E, ICAM1, as well as STAT1 itself, whose expression is known to be induced by IFNG. The complete list of STAT1 targets identified by STAGE is provided in Supplemental Table 2.
Identification of MYC targets within the ENCODE regions by STAGE
We also used STAGE to identify the targets of MYC, an important oncogenic transcription factor. We carried out ChIP using an antibody against MYC in HeLa cells followed by the STAGE procedure. We sequenced ∼4500 clones using standard sequencing methodology for generating the MYC STAGE library. Each clone contained on average ∼20–30 STAGE tags. Out of a total of 127,351 tags extracted for MYC, 19,867 (15%) were orphans that could not be mapped to the human genome. We used the remaining 107,484 tags for further analysis. Based on extrapolations from our ChIP-chip data (below) and previous observations (Cawley et al. 2004), MYC is likely to have between 17,000 and 25,000 binding sites on the genome. Because our depth of sequencing of STAGE tags for MYC was slightly lower than for STAT1, and the possibility that MYC may have a larger number of binding targets on the genome, the high specificity algorithm we developed for identifying STAT1 targets did not yield a significant number of binding targets for MYC. We therefore used a more relaxed algorithm as described in Methods to identify 2218 binding sites for MYC in the entire genome at a probability threshold of 0.8. We calculated the false discovery rate based on simulations at this threshold to be 5%. Twenty-six of the MYC binding sites identified by STAGE occurred within the ENCODE region. We also identified MYC binding sites within the ENCODE regions by ChIP-chip using NimbleGen oligonucleotide tiling arrays (The ENCODE Project Consortium 2007). The ChIP-chip analysis included three biological replicates, and we defined MYC binding peaks in the ENCODE regions using NimbleGen SignalMap software. Fourteen out of the 26 MYC binding sites identified by STAGE within the ENCODE regions were within 500 bp of a ChIP-chip peak in at least one of the three biological replicate experiments.
Discussion
Bead-based pyrosequencing technology has several advantages for STAGE over standard sequencing approaches (Margulies et al. 2005). First, there is no requirement for cloning and isolation of independent recombinant clones. Rather, tags generated by the STAGE procedure can be directly sequenced. Potential biases introduced by cloning in bacteria can thus be avoided. Second, the water-in-oil emulsion that is generated in making the library can be stored, and only a portion of this sample is used to generate on the order of 200,000 sequence reads in a single run of the instrument. Thus from a single chromatin immunoprecipitation reaction performed from a normally grown culture of mammalian cells, it is possible to sequence many samples and together generate more than one million sequence reads amounting to >100 Mb of sequence using STAGE, greatly improving the depth of sequencing and coverage of targets enriched in the ChIP sample. Third, bead-based pyrosequencing is more cost-effective. In our experience, the price of sequencing a STAGE tag using 454 Inc.’s service was about one-fifth that of standard clone-based sequencing (2.5 cents per tag for 454 vs. 14 cents per tag for clone-based sequencing). It is possible to modify the STAGE procedure such that each pyrosequencing read covers four tags, improving the cost-effectiveness and coverage by twofold.
We have developed analysis algorithms to detect genomic binding loci with high specificity. A recently developed algorithm, START, is also aimed at detecting transcription factor targets using ChIP-derived tag libraries (Marinescu et al. 2006). START uses a gene-centric approach where a user-defined window upstream of or downstream from a gene is searched to map tags and genes are denoted as targets using a z-score. START is thus limited to detecting binding sites near the 5′ end of a gene. Our approach defines enriched loci in the whole genome and then identifies genes that lie proximal to these binding sites, enabling identification of binding sites that may have long-range effects on the regulated gene. START does not assign statistical significance to clusters of tags that are not centered on a gene. Finally, START does not make any attempt to distinguish tags that are enriched from tags that might simply be noise, while we assign each tag a probability of enrichment to better distinguish noise from signal.
The currently implemented algorithm is an improvement over the previously employed algorithm (Kim et al. 2005b) to assign probabilities to STAGE-detected binding sites. Though the older algorithm assigned probabilities based on tag enrichment and rewarded clustering of tags, it did not make any attempt to differentiate if a given cluster is significant or not. The current algorithm assigns each cluster a probability of significance and employs individual tag enrichment as an additional criterion to compute a combined probability. This enables a more stringent assessment of whether a given window is a binding site or not. Overall, our results indicate that in depth sequencing using STAGE can identify biologically relevant direct binding targets of transcription factors throughout the genome.
Methods
ChIP for STAT1 and MYC
STAT1 ChIP was performed in HeLa S3 cells that were induced with 5 ng/mL human recombinant IFNG (R&D Systems) for 30 min and then fixed with 1% formaldehyde at room temperature for 10 min. Fixation was quenched with 125 mM glycine and cells were lysed in hypotonic lysis buffer (20 mM HEPES, pH 7.9, 10 mM KCl, 1 mM EDTA, pH 8, 10% glycerol, 1 mM DTT, 0.5 mM PMSF, 0.1 mM sodium orthovanadate, and protease inhibitors). Cell lysates were homogenized and nuclear pellets were collected and lysed in RIPA buffer (10 mM Tris-Cl, pH 8.0, 140 mM NaCl, 1% Triton X-100, 0.1% SDS, 1% deoxycholic acid, 0.5 mM PMSF, 1 mM DTT, 0.1 mM sodium orthovanadate, and protease inhibitors). Nuclear lysates were sonicated with a Branson 250 Sonifier (output 20%, 100% duty cycle) to shear the chromatin to ∼1 kb in size. Clarified lysates were incubated overnight at 4°C with anti-STAT1 alpha p91 (C-24) rabbit polyclonal antibody (sc-345 from Santa Cruz Biotechnology). Protein–DNA complexes were precipitated by protein A agarose and immunoprecipitates were washed three times in 1× RIPA, once in PBS, and then eluted. Crosslinks were reversed overnight at 65°C, and ChIP DNA was purified by Proteinase K treatment followed by extraction with phenol:chloroform:isoamyl alcohol extraction and precipitation with ethanol. Chromatin immunoprecipitation was performed for MYC in HeLa cells using anti-myc antibody (SC-764x from Santa Cruz Biotechnology) using the same protocol as described previously for E2F4 (Kim et al. 2005b).
STAT1 and MYC tag libraries
The STAGE procedure was modified for generating the STAT1 tag library. All steps leading to the generation of ditags from ChIP-enriched DNA were performed exactly as for MYC below. Gel-purified ditags were amplified by PCR using linker specific primers and sequenced by 454 Inc. Duplicate reads were removed by a Perl script. For MYC, the STAGE procedure was carried out as described previously (Kim et al. 2005b). Purified clones were sequenced by Agencourt Inc. Twenty-one-base-pair tags were extracted from each read using Perl scripts.
Generating hits for STAGE tags on the genome
We used the May 2004 Build 35 Human Genome assembly available at http://genome.ucsc.edu for all analyses. Twenty-one-base-pair tags were matched to the genome as described previously (Kim et al. 2005b). Briefly, an indexed, custom database of all CATG(N)17 sequences in the genome was first created. This represents a database of all possible STAGE tags using NlaIII, where each tag sequence was keyed to its chromosome and nucleotide coordinates. Each STAGE library tag was now mapped to the indexed genome-wide tag database by a simple binary search algorithm (Cormen et al. 2001).
Assigning probabilities for tag enrichment
We defined the number of distinct positions in the genome containing a perfect match to a given tag in the STAGE library as nhit. Thus, a tag with a nhit of 1 meant that this tag mapped to a single unique location in the human genome. We defined the number of occurrences of the tag in the sequenced STAGE library, that is, the number of times a given tag was observed in the STAGE library, as nocc. The selection of N tags at random from the entire genome could be modeled as a binomial distribution where the success of an event is defined as selecting a tag with a given nhit. The background probability of selection of a tag with a given nhit was calculated as p = nhit/total number of tags in the genome. If an observed tag with a given nhit has a nocc = f, we calculated the probability of selecting a tag with the observed nhit and nocc ≥ f under a random model. This probability was calculated as 1 minus the cumulative binomial probability of selecting that particular tag with a frequency ≤f − 1, which was calculated as
where p is the background probability of selection of the tag and x iterates from 0 to f − 1.
Multiplying this probability by the total number of tags found in the genome having the given nhit yields the expected frequency of selecting tags with the given nhit and nocc ≥ f
Thus, the expected frequency of a tag with a given nhit and nocc = f when N tags are selected at random was calculated as
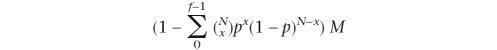 |
where p = nhit/T, and T is the total number of 21 bp CATG(N17) tags found in the entire genome (27,429,149). M = number of tags with a given nhit.
If the expected frequency was greater than the observed frequency, the tag was assigned a low enrichment probability of 0.001.
STAGE target calls for STAT1
A window size of 500 bp was used as described above. For each window, we defined k = number of tags assigned to the window with a single hit on the genome.
Probability that the window is a target = wt_nhit* Phit+ wt_nocc *Pnocc, where
The expected frequency of a window with a given k was obtained from random simulations. It is also possible to calculate this expected frequency and avoid time-consuming random simulations.
Pnocc was calculated as the probability that at least one tag assigned to the window was not random:
where p(tagi) is the probability that tagi was enriched. wt_nhit and wt_nocc were empirically derived weights and were set to 0.9 and 0.1, respectively.
STAGE target calls for MYC
A window of size 500 bp was scanned across each chromosome, and tags mapping within the window were assigned to the window. For the MYC analysis, we discarded tags that had more than 10 hits on the genome.
Quantitative ChIP PCR for binding sites identified by STAGE
We performed quantitative PCR on an independent IFN-γ-stimulated STAT1 ChIP DNA sample. We selected 10 sites to test, spanning a range of final STAGE probability scores. For each of the 10 selected binding sites, we extended the site by 100 bp on either side. Primers were designed to amplify 60–100 bp fragments within the extended window. Quantitative PCR reactions were performed in triplicate in a 96-well optical reaction plate (ABI PRISM) using SYBR Green PCR Master Mix (Applied Biosytems) on an ABI 7900 instrument. The –ΔΔCt values for each locus were calculated with respect to the ChIP input DNA, normalized to a reference locus (GAPDH gene promoter) as described (Livak and Schmittgen 2001). Data for the nine sites that could be confirmed are shown in Figure 3B. Primer sequences are provided in Supplemental Table 3.
Acknowledgments
We thank R. Green, M. Singer, and N. Jiang at NimbleGen Inc. for facilitating the MYC ChIP-chip analysis; K. Rosenbloom and UCSC browser staff for data upload; and 454 Inc. and Agencourt Inc. for sequencing. This work was supported by an NIH/NHGRI ENCODE Technology Development grant (HG003532) and a grant from the US Army Breast Cancer Research Program to V.R.I.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.5574907
References
- Adhikary S., Eilers M., Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat. Rev. Mol. Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- Burch L.R., Scott M., Pohler E., Meek D., Hupp T., Scott M., Pohler E., Meek D., Hupp T., Pohler E., Meek D., Hupp T., Meek D., Hupp T., Hupp T. Phage-peptide display identifies the interferon-responsive, death-activated protein kinase family as a novel modifier of MDM2 and p21WAF1. J. Mol. Biol. 2004;337:115–128. doi: 10.1016/j.jmb.2003.10.081. [DOI] [PubMed] [Google Scholar]
- Cawley S., Bekiranov S., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Bekiranov S., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Sementchenko V., Cheng J., Williams A.J., Cheng J., Williams A.J., Williams A.J., et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- Chen J., Sadowski I., Sadowski I. Identification of the mismatch repair genes PMS2 and MLH1 as p53 target genes by using serial analysis of binding elements. Proc. Natl. Acad. Sci. 2005;102:4813–4818. doi: 10.1073/pnas.0407069102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormen T., Leiserson C., Rivest R., Leiserson C., Rivest R., Rivest R. Introduction to algorithms. MIT Press; Cambridge, MA: 2001. [Google Scholar]
- Eferl R., Wagner E.F., Wagner E.F. AP-1: A double-edged sword in tumorigenesis. Nat. Rev. Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007 doi: 10.1038/nature05874. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman S.E., Bertone P., Nath A.K., Royce T.E., Gerstein M., Weissman S., Snyder M., Bertone P., Nath A.K., Royce T.E., Gerstein M., Weissman S., Snyder M., Nath A.K., Royce T.E., Gerstein M., Weissman S., Snyder M., Royce T.E., Gerstein M., Weissman S., Snyder M., Gerstein M., Weissman S., Snyder M., Weissman S., Snyder M., Snyder M. Global changes in STAT target selection and transcription regulation upon interferon treatments. Genes & Dev. 2005;19:2953–2968. doi: 10.1101/gad.1371305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S., McCorkle S.R., Cha-Molstad H., Dwyer J.M., Yochum G.S., Boss J.M., McWeeney S., Dunn J.J., Mandel G., Goodman R.H., McCorkle S.R., Cha-Molstad H., Dwyer J.M., Yochum G.S., Boss J.M., McWeeney S., Dunn J.J., Mandel G., Goodman R.H., Cha-Molstad H., Dwyer J.M., Yochum G.S., Boss J.M., McWeeney S., Dunn J.J., Mandel G., Goodman R.H., Dwyer J.M., Yochum G.S., Boss J.M., McWeeney S., Dunn J.J., Mandel G., Goodman R.H., Yochum G.S., Boss J.M., McWeeney S., Dunn J.J., Mandel G., Goodman R.H., Boss J.M., McWeeney S., Dunn J.J., Mandel G., Goodman R.H., McWeeney S., Dunn J.J., Mandel G., Goodman R.H., Dunn J.J., Mandel G., Goodman R.H., Mandel G., Goodman R.H., Goodman R.H. Defining the CREB regulon: A genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- Kawai T., Akira S., Reed J.C., Akira S., Reed J.C., Reed J.C. ZIP kinase triggers apoptosis from nuclear PML oncogenic domains. Mol. Cell. Biol. 2003;23:6174–6186. doi: 10.1128/MCB.23.17.6174-6186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.H., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Richmond T.A., Wu Y., Green R.D., Ren B., Wu Y., Green R.D., Ren B., Green R.D., Ren B., Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005a;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Bhinge A.A., Morgan X.C., Iyer V.R., Bhinge A.A., Morgan X.C., Iyer V.R., Morgan X.C., Iyer V.R., Iyer V.R. Mapping DNA–protein interactions in large genomes by sequence tag analysis of genomic enrichment. Nat. Methods. 2005b;2:47–53. doi: 10.1038/nmeth726. [DOI] [PubMed] [Google Scholar]
- Lee T.I., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Johnstone S.E., Cole M.F., Isono K., Cole M.F., Isono K., Isono K., et al. Control of developmental regulators by polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Lu H., Jiang Z., Pastuszyn A., Hu C.A., Lu H., Jiang Z., Pastuszyn A., Hu C.A., Jiang Z., Pastuszyn A., Hu C.A., Pastuszyn A., Hu C.A., Hu C.A. Apolipoprotein l6, a novel proapoptotic Bcl-2 homology 3-only protein, induces mitochondria-mediated apoptosis in cancer cells. Mol. Cancer Res. 2005;3:21–31. [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Loh Y.H., Wu Q., Chew J.L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Wu Q., Chew J.L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Chew J.L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Chen X., Bourque G., George J., Leong B., Liu J., Bourque G., George J., Leong B., Liu J., George J., Leong B., Liu J., Leong B., Liu J., Liu J., et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Berka J., Braverman M.S., Chen Y.J., Chen Z., Braverman M.S., Chen Y.J., Chen Z., Chen Y.J., Chen Z., Chen Z., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinescu V.D., Kohane I.S., Kim T.K., Harmin D.A., Greenberg M.E., Riva A., Kohane I.S., Kim T.K., Harmin D.A., Greenberg M.E., Riva A., Kim T.K., Harmin D.A., Greenberg M.E., Riva A., Harmin D.A., Greenberg M.E., Riva A., Greenberg M.E., Riva A., Riva A. START: An automated tool for serial analysis of chromatin occupancy data. Bioinformatics. 2006;22:999–1001. doi: 10.1093/bioinformatics/btl062. [DOI] [PubMed] [Google Scholar]
- Martone R., Euskirchen G., Bertone P., Hartman S., Royce T.E., Luscombe N.M., Rinn J.L., Nelson F.K., Miller P., Gerstein M., Euskirchen G., Bertone P., Hartman S., Royce T.E., Luscombe N.M., Rinn J.L., Nelson F.K., Miller P., Gerstein M., Bertone P., Hartman S., Royce T.E., Luscombe N.M., Rinn J.L., Nelson F.K., Miller P., Gerstein M., Hartman S., Royce T.E., Luscombe N.M., Rinn J.L., Nelson F.K., Miller P., Gerstein M., Royce T.E., Luscombe N.M., Rinn J.L., Nelson F.K., Miller P., Gerstein M., Luscombe N.M., Rinn J.L., Nelson F.K., Miller P., Gerstein M., Rinn J.L., Nelson F.K., Miller P., Gerstein M., Nelson F.K., Miller P., Gerstein M., Miller P., Gerstein M., Gerstein M., et al. Distribution of NFKB-binding sites across human chromosome 22. Proc. Natl. Acad. Sci. 2003;100:12247–12252. doi: 10.1073/pnas.2135255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platanias L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- Pruitt K.D., Tatusova T., Maglott D.R., Tatusova T., Maglott D.R., Maglott D.R. NCBI Reference Sequence (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramana C.V., Chatterjee-Kishore M., Nguyen H., Stark G.R., Chatterjee-Kishore M., Nguyen H., Stark G.R., Nguyen H., Stark G.R., Stark G.R. Complex roles of STAT1 in regulating gene expression. Oncogene. 2000;19:2619–2627. doi: 10.1038/sj.onc.1203525. [DOI] [PubMed] [Google Scholar]
- Ramana C.V., Kumar A., Enelow R., Kumar A., Enelow R., Enelow R. STAT1-independent induction of SOCS-3 by interferon-gamma is mediated by sustained activation of Stat3 in mouse embryonic fibroblasts. Biochem. Biophys. Res. Commun. 2005;327:727–733. doi: 10.1016/j.bbrc.2004.12.074. [DOI] [PubMed] [Google Scholar]
- Roh T.Y., Ngau W.C., Cui K., Landsman D., Zhao K., Ngau W.C., Cui K., Landsman D., Zhao K., Cui K., Landsman D., Zhao K., Landsman D., Zhao K., Zhao K. High-resolution genome-wide mapping of histone modifications. Nat. Biotechnol. 2004;22:1013–1016. doi: 10.1038/nbt990. [DOI] [PubMed] [Google Scholar]
- Roh T.Y., Cuddapah S., Zhao K., Cuddapah S., Zhao K., Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes & Dev. 2005;19:542–552. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephanou A., Latchman D.S., Latchman D.S. Opposing actions of STAT-1 and STAT-3. Growth Factors. 2005;23:177–182. doi: 10.1080/08977190500178745. [DOI] [PubMed] [Google Scholar]
- Stephanou A., Brar B.K., Knight R.A., Latchman D.S., Brar B.K., Knight R.A., Latchman D.S., Knight R.A., Latchman D.S., Latchman D.S. Opposing actions of STAT-1 and STAT-3 on the Bcl-2 and Bcl-x promoters. Cell Death Differ. 2000;7:329–330. doi: 10.1038/sj.cdd.4400656. [DOI] [PubMed] [Google Scholar]
- Wei C.L., Wu Q., Vega V.B., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., Wu Q., Vega V.B., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., Vega V.B., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., Shahab A., Yong H.C., Fu Y., Weng Z., Yong H.C., Fu Y., Weng Z., Fu Y., Weng Z., Weng Z., et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Wesemann D.R., Benveniste E.N., Benveniste E.N. STAT-1 alpha and IFN-gamma as modulators of TNF-alpha signaling in macrophages: Regulation and functional implications of the TNF receptor 1:STAT-1 alpha complex. J. Immunol. 2003;171:5313–5319. doi: 10.4049/jimmunol.171.10.5313. [DOI] [PubMed] [Google Scholar]