Abstract
The completion of the human genome project has fueled the search for regulatory elements by a variety of different approaches. Many successful analyses have focused on examining primary DNA sequence and/or chromatin structure. However, it has been difficult to detect common sequence motifs within the feature of chromatin structure most closely associated with regulatory elements, DNase I hypersensitive sites (DHSs). Considering just the nucleotide sequence and/or the chromatin structure of regulatory elements may neglect a critical feature of what is recognized by the regulatory machinery—DNA structure. We introduce a new computational method to detect common DNA structural motifs in a large collection of DHSs that are found in the ENCODE regions of the human genome. We show that DHSs have common DNA structural motifs that show no apparent sequence consensus. One such structural motif is much more highly enriched in experimentally identified DHSs that are in CpG islands and near transcription start sites (TSSs), compared to DHSs not in CpG islands and farther from TSSs, suggesting that DNA structural motifs may participate in the formation of functional regulatory elements. We propose that studies of the conservation of DNA structure, independent of sequence conservation, will provide new information about the link between the nucleotide sequence of a DNA molecule and its experimentally demonstrated function.
Since the completion of the sequence of the human genome (Lander et al. 2001; Venter et al. 2001), a major goal of genome research has been the identification of non-coding functional genomic elements (The ENCODE Project Consortium 2004). Searches for functional elements have focused on the experimental determination of chromatin structure and sites of protein binding and the computational detection of conserved non-coding DNA sequences (The ENCODE Project Consortium 2007). These approaches have been highly productive, and we now have a growing catalog of annotated functional elements in the human genome. However, the physical nature of functional sites in genomic DNA remains an important open question. The regulatory machinery that assembles on genomic DNA does so by recognizing in some way the presence of a functional element in the genomic DNA. While nucleotide sequence might be expected to be the key determinant of a functional element, local DNA structure is, in fact, what the regulatory machinery “senses” when scanning the genome for functional elements.
Regions of the genome that are hypersensitive to digestion by deoxyribonuclease I (called DNase I hypersensitive sites, DHSs) have been shown to be associated with a wide variety of functional genomic elements, including promoters, enhancers, origins of replication, and centromeres (Gross and Garrard 1988; Felsenfeld 1992; Felsenfeld and Groudine 2003). High-throughput methods recently have been developed to locate DHSs throughout large stretches of a genome (Crawford et al. 2004, 2006a, b; Dorschner et al. 2004; Sabo et al. 2004a, b, 2006), including the entire set of ENCODE regions (The ENCODE Project Consortium 2004) that encompass 1% of the human genome.
Although DHSs occur nonrandomly in the genome, it has been difficult to detect specific DNA sequence motifs that are held in common by DHSs (Noble et al. 2005). Here we ask whether, instead of a common nucleotide sequence, a particular local structure of genomic DNA is associated with genomic loci that are hypersensitive to DNase I. To address this question, we introduce a new method to identify short regions of shared local DNA structure in genomic DNA. Our measure of local structure is the hydroxyl radical cleavage pattern (Price and Tullius 1993). We call this new measure of conserved local DNA structure the Conserved OH Radical Cleavage Signature (CORCS). We show here that there is a DNA structural element that is highly enriched in DHSs that are associated with CpG islands and near transcription start sites (TSSs), and that this structural element is not predictable on the basis of DNA sequence information alone. Our results suggest that consideration of local DNA structure, as well as nucleotide sequence, will be important to understanding the mechanistic underpinnings of functional genomic elements.
Results
Hydroxyl radical cleavage as a measure of local DNA structure
While there are many algorithms that can find regions in a genome that are similar in nucleotide sequence, locating regions that have similar three-dimensional shape or structure is not as straightforward. In order to identify these regions, some measure of structure must be obtained. Chemical probes are capable of providing such structural information for long stretches of DNA (Nielsen 1990). A nearly ideal chemical probe for mapping genomic DNA structure is the hydroxyl radical, a small and highly reactive free radical that cleaves DNA nonspecifically by abstracting a hydrogen atom from a deoxyribose residue in the DNA backbone (Pogozelski and Tullius 1998). The cleavage pattern (the extent of cleavage at each nucleotide) is revealed by high resolution, quantitative denaturing gel electrophoresis (Shadle et al. 1997).
We have shown previously that the extent of hydroxyl radical cleavage of a given nucleotide in duplex DNA is governed by its exposure to solvent (Balasubramanian et al. 1998). The hydroxyl radical cleavage pattern of a particular DNA sequence therefore provides a map of the local variation in the shape of the DNA backbone.
For the purpose of the work presented here, a key feature of the hydroxyl radical cleavage experiment is that different DNA sequences can produce similar cleavage patterns (Price and Tullius 1993; Greenbaum et al. 2007). Since the same local DNA structure can be adopted by different sequences of nucleotides, it is possible that a region of the genome that functions by virtue of its structure may not be conserved in nucleotide sequence.
We have determined the hydroxyl radical cleavage patterns of a substantial collection of DNA sequences, constructed a database of these patterns, and then used this database to develop an algorithm to predict the cleavage pattern of any DNA sequence (Greenbaum et al. 2007; see Methods). We used this algorithm to predict the cleavage pattern of the 30 Mb of DNA within the ENCODE regions. These predicted cleavage patterns are available for display and analysis in the UCSC Genome Browser (Karolchik et al. 2003). The browser track for our data can be accessed through the following link: http://genome.ucsc.edu/cgi-bin/hgTrackUi?hgsid=73232656&g=encodeBu_ ORChID1.
Detection of segments of common local DNA structurein DNase hypersensitive sites
To find hydroxyl radical cleavage patterns that are shared by a set of DHSs, we developed a computer program that implements the Gibbs sampling algorithm (Lawrence et al. 1993). Gibbs sampling is a statistical technique that can facilitate rapid searches of large data sets for similar patterns. Our program, which we call the CORCS Screening Utility (CORCSScrU), is specifically tailored to work with predicted hydroxyl radical cleavage intensity data, in two modes. In one mode, CORCSScrU “discretizes”—i.e., bins into discrete elements—the continuous-value predicted hydroxyl radical cleavage pattern based on percentile rank and then executes the Gibbs sampling algorithm. In the alternative mode, CORCSScrU operates directly on the continuous-value predicted hydroxyl radical cleavage pattern, without first discretizing the data. We implemented both modes so that we could compare the performance of the quicker, but presumably less accurate, discrete mode to the continuous-value mode.
We used the CORCSScrU program to align DHSs by their predicted hydroxyl radical cleavage patterns. The DHSs we studied were derived from several individual data sets and from the union of some of these data sets (3150 DHSs total). The DHS data sets are publicly available via the UCSC Genome Browser (see Methods).
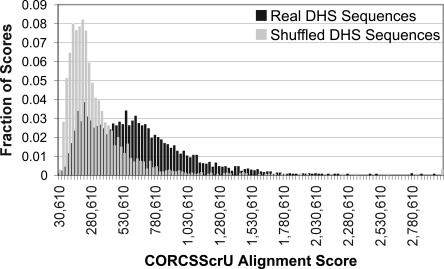
To assess the significance of the cleavage pattern alignment scores, we shuffled the DHS sequences, preserving sequence composition, and then ran the CORCSScrU program on the shuffled sequences. This process was repeated 5000 times. Histograms of alignment scores for the two data sets are shown in Figure 1. Visual inspection indicates that these two distributions are clearly different. A Kolmogorov-Smirnov (KS) test confirms this conclusion, with P < 10−17.
Figure 1.
Histogram of alignment scores of shuffled versus DHS sequences. The CORCSScrU program was run 3204 times until convergence, using either real or shuffled sequences from the MPSS DHS data set. The window size was preset to 12. The resulting alignment scores were binned and are represented here as two histograms. The alignment scores for the real sequences are generally higher than those from the shuffled sequences. A Kolmogorov-Smirnov test indicates that these two distributions are significantly different (p = 10−17).
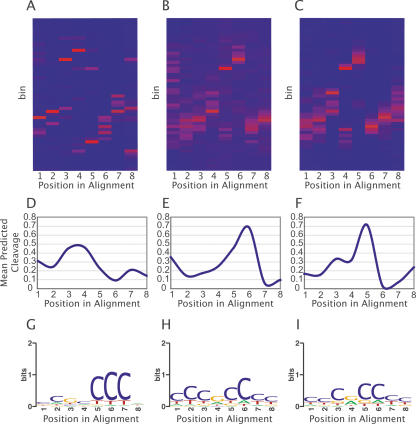
CORCSScrU identified several common hydroxyl radical cleavage motifs within DHSs. Three representative motifs, CORCS1, CORCS2, and CORCS3, are depicted as heat maps in Figure 2, A–C. Here, the X-axis represents each position in the identified motif and the Y-axis represents cleavage value bins. Dark blue cells in the heat map indicate no cleavage values for bin Y at position X are present, whereas red cells indicate a large proportion of the cleavage values for that column. If cleavage values were randomly distributed, each column would be uniformly colored. The motifs illustrated here were discovered in separate runs of the CORCSScrU program. CORCS1 was discovered using CORCSScrU in discretized mode with the smaller MPSS DHS data set, while CORCS2 and CORCS3 were discovered using CORCSScrU in discretized and continuous mode, respectively, with the Union DHS data set (see Methods section for more details about data sets). Close inspection reveals that the two CORCS found by aligning DHSs from the Union data set are similar, but offset by one nucleotide (Fig. 2, cf. B and C). To quantitatively assess the similarity of these two CORCS, we calculated the correlation between the mean predicted hydroxyl radical cleavage intensity for positions 2–8 from CORCS2 and for positions 1–7 from CORCS3. A highly significant correlation (Pearson’s r = 0.951; p < 0.0005) confirms that CORCS2 and CORCS3 are, indeed, similar. The stochastic nature of the Gibbs sampling algorithm makes it unlikely that exactly the same motif is converged on in every run. However, we found that a very similar, if slightly offset, signal emerged consistently in repeated runs.
Figure 2.
Analysis of representative high-scoring CORCS. (A–C) Heat maps of CORCS found in (A) the MPSS data set and (B) the Union data set using the discrete sampler, and in (C) the Union data set using the continuous sampler. The X-axis represents each position in the CORCS and the Y-axis represents cleavage value bins. Dark blue cells in the heat map indicate no cleavage values for bin Y at position X are present, whereas red cells indicate a large proportion of the cleavage values for that column. (D–F) Mean predicted hydroxyl radical cleavage patterns of CORCS found in (D) the MPSS data set and (E) the Union data set using the discrete sampler, and in (F) the Union data set using the continuous sampler. (G–I) Sequence logos of CORCS found in (G) the MPSS data set and in (H) the Union data set using the discrete sampler, and in (I) the Union data set using the continuous sampler.
The fact that similar CORCS were recovered from a data set when CORCSScrU was run either in discretized or continuous mode suggests that use of the quicker discretized mode does not result in a significant loss of information. To investigate this point further, we plotted the mean predicted hydroxyl radical cleavage pattern values for each position in each CORCS (Fig. 2D–F). The mean cleavage intensity at any given position mirrors closely what CORCSScrU finds in discretized mode (e.g., Fig. 2, cf. B and E).
To determine whether a CORCS arises simply as the result of finding similar DNA sequences in different DHSs, we examined the corresponding nucleotide alignments between human DHSs. We found little similarity between nucleotide patterns within CORCS, which can be summarized in the form of sequence logos (Fig. 2G–I; Schneider and Stephens 1990). Further examination of the sequence logos from CORCS2 and CORCS3 when they are shifted in a manner as to align the cleavage patterns (positions 2–8 from CORCS2 compared to positions 1–7 from CORCS3) reveals that the nucleotide composition at these aligned positions is not the same. This result shows that DNA motifs with different sequence composition can have similar cleavage patterns.
A CORCS represents a common DNA structural pattern having little sequence similarity
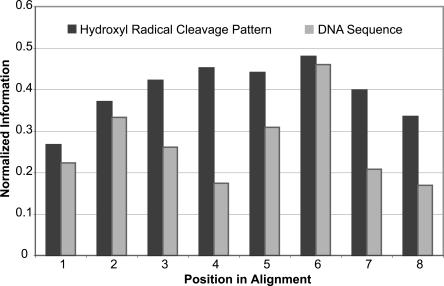
We calculated the similarity of both sequence and structure in the CORCS in terms of information content. More specifically, we computed the maximum entropy minus the observed entropy at each position (Schneider and Stephens 1990) of the alignments, normalized by the maximum entropy. Entropy is a measure of degeneracy or uncertainty. Information is a measure of the decrease of uncertainty. Therefore, an alignment with a higher information content is more conserved. This analysis revealed that in all cases, the hydroxyl radical cleavage pattern alignment contained more information than the corresponding nucleotide sequence alignment. Each position in CORCS2 has more information in the cleavage pattern alignment than in the nucleotide sequence alignment (Fig. 3). For CORCS1, CORCS3, and other high-scoring CORCS, we found that a majority of positions have more information in the cleavage pattern alignment as compared to the nucleotide sequence alignment. Although certain positions of a CORCS may be more similar in sequence than others, the total information content of the cleavage pattern alignment for each CORCS motif is greater than the corresponding information content of the nucleotide sequence alignment.
Figure 3.
Conservation of nucleotide sequence versus structure in CORCS2. Plotted here is the normalized information present at each nucleotide position in CORCS2 for the hydroxyl radical cleavage pattern alignment (dark gray) and the nucleotide sequence alignment (light gray).
Enrichment analysis of CORCS1
The three CORCS we show in Figure 2 were found by running CORCSScrU on a set of annotated DHSs found in the ENCODE regions. We next asked what the distribution of one of these CORCS was in the entire set of ENCODE regions, to see how specific the CORCS is for DHSs. To do this, we used a MatInspector-like algorithm (Quandt et al. 1995), modified to use hydroxyl radical cleavage data, to locate all sequences within the ENCODE regions that have a similar structural profile to CORCS1. We examined the ENCODE-wide distribution of CORCS1 because it was discovered using the MPSS DHS data set, which is considerably smaller than, and disjoint from, the Union DHS data set (which is based on data from only the GM06990 cell line) (The ENCODE Project Consortium 2007; see Methods). This approach allowed us to test whether CORCS1 is found in annotated DHSs identified by other experimental means in a different cell line.
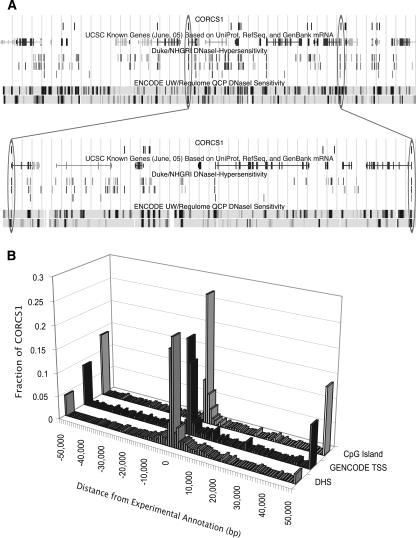
We scanned CORCS1 across the predicted hydroxyl radical cleavage patterns of the ENCODE regions and scored each overlapping segment. We found 588 cleavage patterns with a similarity score to CORCS1 above the 99.999-th percentile threshold. These sequences were extracted and their coordinates recorded into a browser extensible data (BED) format file, for viewing in the UCSC Genome Browser (Fig. 4A) and for enrichment analysis.
Figure 4.
Location of CORCS1 sites relative to experimental annotations. (A) UCSC Genome Browser shot of CORCS1 in ENCODE region ENm002. Data types are indicated by labels above each track. For the NHGRI DHSs, the top, middle, and bottom tracks correspond to GM06990 (DNase-Chip method), CD4+ T cells (DNase-chip method), and CD4+ T cells (MPSS method), respectively. The latter data set was the training set for discovering CORCS1. For the UW/Regulome DHSs, the upper and lower tracks contain data from the GM06990 cell line and the SKNSH cell line, respectively. (Below) A segment of the browser shot is expanded to highlight a few examples. The oval on the right indicates a CORCS1 site that aligns with a DHS that was discovered by three different methods in three different cell lines. The oval on the left indicates a CORCS1 site that aligns with a DHS that is not in the training set. (B) Clustering of CORCS1 near experimental annotations. The distance (in base pairs) of each of the 588 CORCS1 sites to the nearest experimental annotation was measured. The three histograms show that CORCS1 clusters near annotated DHSs, TSSs, and CpG islands.
We show in Figure 4A that although not every example of CORCS1 found in the ENCODE regions aligns with an annotated DHS, the majority do. Most striking is the overlap of several of the CORCS with DHSs in data sets other than the training data set. A portion of this figure is enlarged to highlight a few examples. The oval on the right shows a CORCS1 site that aligns with a DHS discovered by three different methods across three different cell lines. The oval on the left shows a CORCS1 site that aligns with a DHS that is not in the training set.
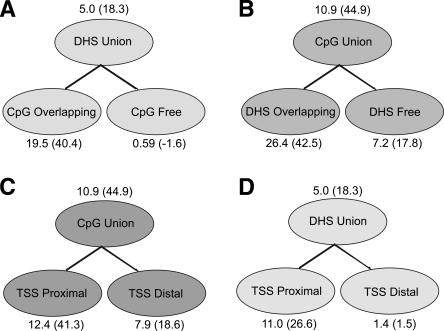
We found that CORCS1 is 5.0-fold (Z-score = 18.3) enriched for experimentally identified DNase hypersensitive sites. This enrichment is reinforced by the histogram in Figure 4B, which shows a tighter clustering of CORCS1 sites near annotated DHSs compared to either TSSs or CpG islands.
CORCS1 is found preferentially in CpG islands that harbor a DHS
In addition to finding that CORCS1 is highly enriched in DHSs (5.0-fold; Z-score = 18.3), we found that it is even more enriched in DHSs that overlap with CpG islands (19.5-fold; Z-score = 40.4) (Fig. 5A). To further investigate this observation, we divided the set of all CpG islands in ENCODE into DHS-rich and DHS-poor, and found that the occurrence of CORCS1 was significantly biased for DHS-rich CpG islands (26.4-fold enrichment; Z-score = 42.5) compared to DHS-poor CpG islands (7.2-fold enrichment; Z-score = 17.8) (Fig. 5B). That is, although CORCS1 is enriched for CpG islands in general, this CORCS is much more specific for CpG islands that contain DNase hypersensitive sites.
Figure 5.
Enrichment of CORCS1 in DHSs, CpG islands, and TSSs. Numbers above or below ovals represent fold enrichment; the corresponding Z-score is appended in parentheses.
Although similar in sequence composition, by definition, to their TSS-distal counterparts, TSS-proximal CpG islands have distinctly different functional roles (Takai and Jones 2002), which include gene silencing (Bird 2002), imprinting (Feil and Khosla 1999), X-chromosome inactivation (Panning and Jaenisch 1998), and carcinogenesis (Baylin et al. 1998; Jones and Laird 1999). Because we found that CORCS1 tends to be found in DHS-containing CpG islands and DHSs are known to be involved in gene regulation, we sought to address whether CORCS1 occurs preferentially in TSS-proximal CpG islands. We segregated annotated CpG islands into TSS-proximal (within 2.5 kb of the nearest annotated TSS) and TSS-distal (>2.5 kb) and performed an enrichment analysis for CORCS1. Figure 5C shows that CORCS1 is enriched 12.4-fold (Z-score = 41.3) for TSS-proximal compared to 7.9-fold (Z-score = 18.6) for TSS-distal CpG islands. It is interesting to speculate that a local DNA structural motif that is more common among TSS-proximal CpG islands may have a role in ascribing unique functional properties to these CpG islands compared to their TSS-distal relatives.
CORCS1 is found preferentially in DHSs near TSSs
Given the above results, along with the presumption that CpG islands are involved in regulation of transcription (Takai and Jones 2002; Saxonov et al. 2006), we sought to determine whether or not CORCS1 occurs preferentially in DHSs near TSSs. Clustering analysis reveals that CORCS1 tends to be located in close proximity to annotated DHSs and TSSs (Fig. 4B), which is suggestive but not demonstrative. To address this question directly, we divided all DHSs into TSS-proximal (within 2.5 kb of the nearest annotated TSS) and TSS-distal (>2.5 kb) and performed an enrichment analysis for CORCS1. We found that CORCS1 is much more enriched in TSS-proximal DHSs (11.0-fold; Z-score = 26.6) compared to TSS-distal DHSs (1.4-fold; Z-score = 1.5) (Fig. 5D).
CORCS2 and CORCS3 are also found preferentially in DHSs near TSSs and DHSs overlapping CpG islands
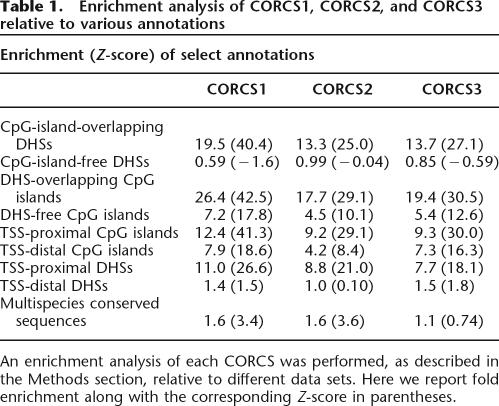
Because CORCS1 showed enrichment for DHSs near TSSs and DHSs overlapping CpG islands, we considered the possibility that CORCS2 and CORCS3 may show similar trends. Table 1 shows enrichment values for CORCS1, CORCS2, and CORCS3 relative to various annotations. Interestingly, the enrichment results for CORCS2 and CORCS3 are consistent with what we observed for CORCS1; that is, CORCS2 and CORCS3 are also preferentially found in DHSs near TSSs and DHSs overlapping CpG islands.
Table 1.
Enrichment analysis of CORCS1, CORCS2, and CORCS3 relative to various annotations
An enrichment analysis of each CORCS was performed, as described in the Methods section, relative to different data sets. Here we report fold enrichment along with the corresponding Z-score in parentheses.
The observation that all CORCS reported here are more enriched for TSS-proximal CpG islands compared to TSS-distal CpG islands (Table 1) and the minor enrichment for the cytosine nucleotide in the CORCS motifs (Fig. 2G–I) prompted us to investigate G+C% in TSS-proximal and TSS-distal CpG islands. TSS-proximal CpG islands have higher G+C% than TSS-distal CpG islands (mean = 63.9%, 60.7%, respectively; p < 1 × 10−24), which may explain the slight enrichment in cytosine among the identified CORCS motifs.
CORCS are moderately enriched for multispecies conserved sequences
We conducted an enrichment analysis for the three CORCS presented here against multispecies conserved sequences. CORCS1 and CORCS2 are slightly enriched for conserved sequences (Table 1). We found that CORCS3 is not enriched for conserved sequences (Table 1). One interesting point to consider is that although the CORCS show slight enrichment for conserved sequences, these motifs are based on conserved structures, which suggests that searching for orthologous DNA structural conservation may reveal a higher level of enrichment.
Discussion
When using computational techniques to search for functional non-coding sequences, considering sequence information alone has the potential to overlook important functional elements that are manifested at the level of DNA structure. This raises the tantalizing possibility that some non-coding functional elements may be under evolutionary selection at the level of structure rather than sequence. This concept accords well with the finding by the ENCODE Consortium that Regulatory Factor Binding Regions (RFBRs) often are only weakly enriched in identifiable transcription factor-binding motifs, and that there is a surprisingly low level of sequence constraint in experimentally identified non-coding elements (The ENCODE Project Consortium 2007).
Here we have used the hydroxyl radical cleavage pattern to identify regions in DNase hypersensitive sites that are more similar in structure than in nucleotide sequence. We identified CORCS (Conserved OH Radical Cleavage Signatures) in a large collection of annotated DHSs from the ENCODE regions of the human genome. The striking correlation of the location of CORCS1 with annotated DHSs (Fig. 4B), combined with the results of enrichment analysis (Fig. 5) makes a compelling argument for the existence of common structures within or near DNase hypersensitive sites. The question of whether such common structural features are responsible for conferring nuclease hypersensitivity on these regions remains open. Observing the effect on DNase I sensitivity of deletion or mutation of a CORCS could test this hypothesis directly.
A striking result of our analysis is the finding that CORCS1 is much more highly enriched in CpG islands that harbor DNase hypersensitive sites (Fig. 5B), suggesting that there may be a structural difference between CpG islands that are in regions of open chromatin compared to those that are not. Further speculation raises the possibility that these differences in structure are the underlying determinant of the functional differences between these elements. The broad implications of verifying this hypothesis, along with the compelling evidence presented in this study, warrant its further investigation.
The methods we describe here can be applied to other types of functional genomic elements and are particularly suited to the analysis of elements that show no apparent sequence consensus. The work presented here suggests that the identification of common DNA structural motifs to distinguish among functional elements may be a plausible and cost-effective initiative.
Methods
Data sets used for this work
All data sets we used are freely available for download from the UCSC Genome Browser (http://genome.ucsc.edu/encode/). To discover CORCS1, we used DHS sequences from the NHGRI CD4+ T cell MPSS data set (229 sequences in total) (Crawford et al. 2006b). To discover CORCS2 and CORCS3, we iteratively selected 300 sequences at random from a Union DHS data set (3150 sequences in total) derived from GM06990 lymphoblastoid cells (The ENCODE Project Consortium 2007). Multispecies conserved sequences used for enrichment analysis were based on the “Moderate” (encodeMsaEIModerate) track available from the ENCODE Comparative Genomics section of the UCSC Genome Browser and were provided by the ENCODE Multispecies Sequence Analysis group (Margulies et al. 2007).
Gibbs sampling of cleavage patterns in DNase hypersensitive sites: CORCSScrU
We collected experimentally determined hydroxyl radical cleavage patterns for 56 DNA sequences (mean length = 41 bp) and assembled them into a database that we named the OH Radical Cleavage Intensity Database (ORChID). Using the experimental cleavage patterns in this database, we developed an algorithm to predict the cleavage pattern of any DNA sequence with a high degree of accuracy (mean Pearson correlation for leave-one-out cross-validation = 0.88) (Greenbaum et al. 2007).
The computer program we developed for Gibbs sampling is based on an algorithm previously reported for the identification of protein and DNA sequence motifs (Lawrence et al. 1993). The first step was to predict the cleavage patterns of a set of 3150 annotated DNase hypersensitive sites. Next, in order to make the hydroxyl radical cleavage data amenable to analysis, the patterns were “discretized.” Discretization was performed on a percentile basis, and 50 bins were created. Other binning schemes also were tested. A pre-determined window size of 8 nt was sampled. We chose this size because it represents a reasonable size for the footprint of a DNA-binding factor (Maston et al. 2006). The sampler was run until convergence, which was defined as M iterations with no improvement in the alignment score, where M is equal to five times the size of the data set. The phase was shifted by one-fourth of the window size after every 1000 iterations in order to escape local minima.
Alternatively, we skipped the discretization step and applied the Gibbs sampling algorithm to continuous-value predicted hydroxyl radical cleavage data. Owing to the nature of these data, we made one modification during sampling: We scored each window of length L in the chosen sequence by calculating the probability that the predicted cleavage intensity at position i, where 1 ≤ i ≤ L, occurs in the observed alignment distribution at position i versus the probability that it occurs in the distribution of background predicted cleavage intensities.
Heat maps
We constructed position weight matrices based on bin counts at each position of a CORCS and used the program matrix2png (Pavlidis and Noble 2003) with the −z option to generate heat maps. For CORCS discovered in continuous mode, we subsequently discretized the continuous cleavage values, as described above, and then generated heat maps.
Calculation of hydroxyl radical cleavage conservation
Conservation of cleavage was calculated via Equation 1 (Schneider and Stephens 1990):
Rcleavage(l) represents the amount of information present at position l, 5.64 is the maximum amount of uncertainty possible for 50 bins (in bits), and H(l) is the uncertainty at position l (Schneider and Stephens 1990). Sequence conservation was calculated similarly. In order to make a direct comparison between conservation of sequence and cleavage, values were divided by their respective maximum possible entropies: 5.64 for cleavage intensity, and 2 for DNA sequence. This calculation gave us the normalized information for each position in the alignment for both the cleavage pattern and the sequence. We use this as a measure of conservation.
Identification of regions in ENCODE having cleavage patterns similar to CORCS
Several converged alignments were made into position weight matrices. All overlapping windows were scored for their similarity to these matrices (Quandt et al. 1995). As speed was not an issue, the pre-calculation of core score was omitted. The positions of windows that scored within the top 0.001% were recorded to a BED file for further analysis.
Enrichment analysis
Enrichment of CORCS in a particular type of experimentally annotated genomic segment was evaluated using a simple and elegant algorithm provided by Yutao Fu. First, two BED files were compared against one another, and the number of nucleotides that overlap were counted (O). Next, the elements of one of the data sets were shuffled and the calculation was performed again to obtain R, the number of nucleotides that overlap in the random data set. Shuffling was repeated 1000 times, and the mean overlap of the shuffled data sets was calculated. The enrichment (E) is then defined as:
By this scheme, data sets with no relation will have an enrichment of 1. Data sets that contain elements that colocalize will have a value >1, and those that show significant anticorrelation with respect to position will have a value <1. The standard deviation for R was recorded so that a Z-score for E could be calculated.
Acknowledgments
We thank The ENCODE Project Consortium for making their data publicly available, and the ENCODE Chromatin and Replication analysis group for providing the DHS data and for helpful suggestions. We are grateful to John Stamatoyannopoulos and Scott Kuehn for providing us with access to their analysis pipeline for determining the proximity of CORCS to genes, transcripts, and other experimentally annotated features within the ENCODE regions. This work was funded by a grant from the National Human Genome Institute of the National Institutes of Health (R01 HG003541).
Footnotes
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.5602807
References
- Balasubramanian B., Pogozelski W.K., Tullius T.D., Pogozelski W.K., Tullius T.D., Tullius T.D. DNA strand breaking by the hydroxyl radical is governed by the accessible surface areas of the hydrogen atoms of the DNA backbone. Proc. Natl. Acad. Sci. 1998;95:9738–9743. doi: 10.1073/pnas.95.17.9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin S.B., Herman J.G., Graff J.R., Vertino P.M., Issa J.P., Herman J.G., Graff J.R., Vertino P.M., Issa J.P., Graff J.R., Vertino P.M., Issa J.P., Vertino P.M., Issa J.P., Issa J.P. Alterations in DNA methylation: A fundamental aspect of neoplasia. Adv. Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes & Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Crawford G., Holt I., Mullikin J., Tai D., Blakesley R., Bouffard G., Young A., Masiello C., Green E., Wolfsberg T., Holt I., Mullikin J., Tai D., Blakesley R., Bouffard G., Young A., Masiello C., Green E., Wolfsberg T., Mullikin J., Tai D., Blakesley R., Bouffard G., Young A., Masiello C., Green E., Wolfsberg T., Tai D., Blakesley R., Bouffard G., Young A., Masiello C., Green E., Wolfsberg T., Blakesley R., Bouffard G., Young A., Masiello C., Green E., Wolfsberg T., Bouffard G., Young A., Masiello C., Green E., Wolfsberg T., Young A., Masiello C., Green E., Wolfsberg T., Masiello C., Green E., Wolfsberg T., Green E., Wolfsberg T., Wolfsberg T., et al. Identifying 174 gene regulatory elements by genome-wide recovery of DNase hypersensitive sites. Proc. Natl. Acad. Sci. 2004;101:992–997. doi: 10.1073/pnas.0307540100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford G.E., Davis S., Scacheri P.C., Renaud G., Halawi M., Erdos M.R., Green R., Meltzer P.S., Wolfsberg T.G., Collins F.S., Davis S., Scacheri P.C., Renaud G., Halawi M., Erdos M.R., Green R., Meltzer P.S., Wolfsberg T.G., Collins F.S., Scacheri P.C., Renaud G., Halawi M., Erdos M.R., Green R., Meltzer P.S., Wolfsberg T.G., Collins F.S., Renaud G., Halawi M., Erdos M.R., Green R., Meltzer P.S., Wolfsberg T.G., Collins F.S., Halawi M., Erdos M.R., Green R., Meltzer P.S., Wolfsberg T.G., Collins F.S., Erdos M.R., Green R., Meltzer P.S., Wolfsberg T.G., Collins F.S., Green R., Meltzer P.S., Wolfsberg T.G., Collins F.S., Meltzer P.S., Wolfsberg T.G., Collins F.S., Wolfsberg T.G., Collins F.S., Collins F.S. DNase-chip: A high resolution method to identify DNaseI hypersensitive sites using tiled microarrays. Nat. Methods. 2006a;3:503–509. doi: 10.1038/NMETH888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford G.E., Holt I.E., Whittle J., Webb B.D., Tai D., Davis S., Margulies E.H., Chen Y., Bernat J.A., Ginsburg D., Holt I.E., Whittle J., Webb B.D., Tai D., Davis S., Margulies E.H., Chen Y., Bernat J.A., Ginsburg D., Whittle J., Webb B.D., Tai D., Davis S., Margulies E.H., Chen Y., Bernat J.A., Ginsburg D., Webb B.D., Tai D., Davis S., Margulies E.H., Chen Y., Bernat J.A., Ginsburg D., Tai D., Davis S., Margulies E.H., Chen Y., Bernat J.A., Ginsburg D., Davis S., Margulies E.H., Chen Y., Bernat J.A., Ginsburg D., Margulies E.H., Chen Y., Bernat J.A., Ginsburg D., Chen Y., Bernat J.A., Ginsburg D., Bernat J.A., Ginsburg D., Ginsburg D., et al. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS) Genome Res. 2006b;16:123–131. doi: 10.1101/gr.4074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorschner M.O., Hawrylycz M., Humbert R., Wallace J.C., Shafer A., Kawamoto J., Mack J., Hall R., Goldy J., Sabo P.J., Hawrylycz M., Humbert R., Wallace J.C., Shafer A., Kawamoto J., Mack J., Hall R., Goldy J., Sabo P.J., Humbert R., Wallace J.C., Shafer A., Kawamoto J., Mack J., Hall R., Goldy J., Sabo P.J., Wallace J.C., Shafer A., Kawamoto J., Mack J., Hall R., Goldy J., Sabo P.J., Shafer A., Kawamoto J., Mack J., Hall R., Goldy J., Sabo P.J., Kawamoto J., Mack J., Hall R., Goldy J., Sabo P.J., Mack J., Hall R., Goldy J., Sabo P.J., Hall R., Goldy J., Sabo P.J., Goldy J., Sabo P.J., Sabo P.J., et al. High-throughput localization of functional elements by quantitative chromatin profiling. Nat. Methods. 2004;1:219–225. doi: 10.1038/nmeth721. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, The ENCODE (ENCyclopedia of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007 doi: 10.1038/nature05874. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R., Khosla S., Khosla S. Genomic imprinting in mammals: An interplay between chromatin and DNA methylation? Trends Genet. 1999;15:431–435. doi: 10.1016/s0168-9525(99)01822-3. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992;355:219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G., Groudine M., Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- Greenbaum J.A., Pang B., Tullius T.D., Pang B., Tullius T.D., Tullius T.D. Construction of a genome-scale structural map at single-nucleotide resolution. Genome Res. 2007 doi: 10.1101/gr.6073107. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross D., Garrard W., Garrard W. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- Jones P.A., Laird P.W., Laird P.W. Cancer epigenetics comes of age. Nat. Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Schwartz M., Sugnet C.W., Thomas D.J., Sugnet C.W., Thomas D.J., Thomas D.J., et al. The UCSC Genome Browser database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E., Linton L., Birren B., Nusbaum C., Zody M., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Linton L., Birren B., Nusbaum C., Zody M., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Birren B., Nusbaum C., Zody M., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Nusbaum C., Zody M., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Zody M., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Devon K., Dewar K., Doyle M., FitzHugh W., Dewar K., Doyle M., FitzHugh W., Doyle M., FitzHugh W., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lawrence C., Altschul S., Boguski M., Liu J., Neuwald A., Wootton J., Altschul S., Boguski M., Liu J., Neuwald A., Wootton J., Boguski M., Liu J., Neuwald A., Wootton J., Liu J., Neuwald A., Wootton J., Neuwald A., Wootton J., Wootton J. Detecting subtle sequence signals: A Gibbs sampling strategy for multiple alignment. Science. 1993;262:208–214. doi: 10.1126/science.8211139. [DOI] [PubMed] [Google Scholar]
- Margulies E.H., Cooper G.M., Asimenos G., Thomas D.J., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Cooper G.M., Asimenos G., Thomas D.J., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Asimenos G., Thomas D.J., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Thomas D.J., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Birney E., Keefe D., Schwartz A.S., Hou M., Keefe D., Schwartz A.S., Hou M., Schwartz A.S., Hou M., Hou M., et al. Analyses of deep mammalian sequence alignments and constraint predictions for 1% of the human genome. Genome Res. 2007 doi: 10.1101/gr.6034307. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maston G.A., Evans S.K., Green M.R., Evans S.K., Green M.R., Green M.R. Transcriptional regulatory elements in the human genome. Annu. Rev. Genomics Hum. Genet. 2006;7:29–59. doi: 10.1146/annurev.genom.7.080505.115623. [DOI] [PubMed] [Google Scholar]
- Nielsen P.E. Chemical and photochemical probing of DNA complexes. J. Mol. Recognit. 1990;3:1–25. doi: 10.1002/jmr.300030102. [DOI] [PubMed] [Google Scholar]
- Noble W.S., Kuehn S., Thurman R., Stamatoyannopoulos J., Kuehn S., Thurman R., Stamatoyannopoulos J., Thurman R., Stamatoyannopoulos J., Stamatoyannopoulos J. Predicting the in vivo signature of human gene regulatory sequences. Bioinformatics. 2005;(Suppl. 1) 21:i338–i343. doi: 10.1093/bioinformatics/bti1047. [DOI] [PubMed] [Google Scholar]
- Panning B., Jaenisch R., Jaenisch R. RNA and the epigenetic regulation of X chromosome inactivation. Cell. 1998;93:305–308. doi: 10.1016/s0092-8674(00)81155-1. [DOI] [PubMed] [Google Scholar]
- Pavlidis P., Noble W.S., Noble W.S. Matrix2png: A utility for visualizing matrix data. Bioinformatics. 2003;19:295–296. doi: 10.1093/bioinformatics/19.2.295. [DOI] [PubMed] [Google Scholar]
- Pogozelski W.K., Tullius T.D., Tullius T.D. Oxidative strand scission of nucleic acids: Routes initiated by hydrogen abstraction from the sugar moiety. Chem. Rev. 1998;98:1089–1107. doi: 10.1021/cr960437i. [DOI] [PubMed] [Google Scholar]
- Price M.A., Tullius T.D., Tullius T.D. How the structure of an adenine tract depends on sequence context. A new model for the structure of TnAn DNA sequences. Biochemistry. 1993;32:127–136. doi: 10.1021/bi00052a018. [DOI] [PubMed] [Google Scholar]
- Quandt K., Frech K., Karas H., Wingender E., Werner T., Frech K., Karas H., Wingender E., Werner T., Karas H., Wingender E., Werner T., Wingender E., Werner T., Werner T. MatInd and MatInspector: New fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo P.J., Hawrylycz M., Wallace J.C., Humbert R., Yu M., Shafer A., Kawamoto J., Hall R., Mack J., Dorschner M.O., Hawrylycz M., Wallace J.C., Humbert R., Yu M., Shafer A., Kawamoto J., Hall R., Mack J., Dorschner M.O., Wallace J.C., Humbert R., Yu M., Shafer A., Kawamoto J., Hall R., Mack J., Dorschner M.O., Humbert R., Yu M., Shafer A., Kawamoto J., Hall R., Mack J., Dorschner M.O., Yu M., Shafer A., Kawamoto J., Hall R., Mack J., Dorschner M.O., Shafer A., Kawamoto J., Hall R., Mack J., Dorschner M.O., Kawamoto J., Hall R., Mack J., Dorschner M.O., Hall R., Mack J., Dorschner M.O., Mack J., Dorschner M.O., Dorschner M.O., et al. Discovery of functional noncoding elements by digital analysis of chromatin structure. Proc. Natl. Acad. Sci. 2004a;101:16837–16842. doi: 10.1073/pnas.0407387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo P.J., Humbert R., Hawrylycz M., Wallace J.C., Dorschner M.O., McArthur M., Stamatoyannopoulos J.A., Humbert R., Hawrylycz M., Wallace J.C., Dorschner M.O., McArthur M., Stamatoyannopoulos J.A., Hawrylycz M., Wallace J.C., Dorschner M.O., McArthur M., Stamatoyannopoulos J.A., Wallace J.C., Dorschner M.O., McArthur M., Stamatoyannopoulos J.A., Dorschner M.O., McArthur M., Stamatoyannopoulos J.A., McArthur M., Stamatoyannopoulos J.A., Stamatoyannopoulos J.A. Genome-wide identification of DNaseI hypersensitive sites using active chromatin sequence libraries. Proc. Natl. Acad. Sci. 2004b;101:4537–4542. doi: 10.1073/pnas.0400678101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo P.J., Kuehn M.S., Thurman R., Grant C., Johnson B., Johnson S., Kao H., Yu M., Goldy J., Weaver M., Kuehn M.S., Thurman R., Grant C., Johnson B., Johnson S., Kao H., Yu M., Goldy J., Weaver M., Thurman R., Grant C., Johnson B., Johnson S., Kao H., Yu M., Goldy J., Weaver M., Grant C., Johnson B., Johnson S., Kao H., Yu M., Goldy J., Weaver M., Johnson B., Johnson S., Kao H., Yu M., Goldy J., Weaver M., Johnson S., Kao H., Yu M., Goldy J., Weaver M., Kao H., Yu M., Goldy J., Weaver M., Yu M., Goldy J., Weaver M., Goldy J., Weaver M., Weaver M., et al. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat. Methods. 2006;3:511–518. doi: 10.1038/nmeth890. [DOI] [PubMed] [Google Scholar]
- Saxonov S., Berg P., Brutlag D.L., Berg P., Brutlag D.L., Brutlag D.L. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T., Stephens R., Stephens R. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadle S.E., Allen D.F., Guo H., Pogozelski W.K., Bashkin J.S., Tullius T.D., Allen D.F., Guo H., Pogozelski W.K., Bashkin J.S., Tullius T.D., Guo H., Pogozelski W.K., Bashkin J.S., Tullius T.D., Pogozelski W.K., Bashkin J.S., Tullius T.D., Bashkin J.S., Tullius T.D., Tullius T.D. Quantitative analysis of electrophoresis data: Novel curve fitting methodology and its application to the determination of a protein–DNA binding constant. Nucleic Acids Res. 1997;25:850–861. doi: 10.1093/nar/25.4.850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai D., Jones P.A., Jones P.A. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl. Acad. Sci. 2002;19:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Smith H.O., Yandell M., Evans C.A., Holt R.A., Yandell M., Evans C.A., Holt R.A., Evans C.A., Holt R.A., Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]