Abstract
The Encyclopedia of DNA Elements (ENCODE) project aims to identify and characterize all functional elements in a representative chromosomal sample comprising 1% of the human genome. Data generated by members of The ENCODE Project Consortium are housed in a number of public databases, such as the UCSC Genome Browser, NCBI’s Gene Expression Omnibus (GEO), and EBI’s ArrayExpress. As such, it is often difficult for biologists to gather all of the ENCODE data from a particular genomic region of interest and integrate them with relevant information found in other public databases. The ENCODEdb portal was developed to address this problem. ENCODEdb provides a unified, single point-of-access to data generated by the ENCODE Consortium, as well as to data from other source databases that lie within ENCODE regions; this provides the user a complete view of all known data in a particular region of interest. ENCODEdb Genomic Context searches allow for the retrieval of information on functional elements annotated within ENCODE regions, including mRNA, EST, and STS sequences; single nucleotide polymorphisms, and UniGene clusters. Information is also retrieved from GEO, OMIM, and major genome sequence browsers. ENCODEdb Consortium Data searches allow users to perform compound queries on array-based ENCODE data available both from GEO and from the UCSC Genome Browser. Results are retrieved from a specific genomic area of interest and can be further manipulated in a variety of contexts, including the UCSC Genome Browser and the Galaxy large-scale genome analysis platform. The ENCODEdb portal is freely accessible at http://research.nhgri.nih.gov/ENCODEdb.
With the completion of human genome sequencing, one of the major challenges of genomic biology is to comprehensively identify the structural and functional components encoded in the human genome. To this end, the Encyclopedia of DNA Elements (ENCODE) project aims to identify and characterize all functional elements in a representative chromosomal sample comprising 1% of the human genome (The ENCODE Project Consortium 2004). Members of The ENCODE Project Consortium are employing a variety of laboratory-based and computationally based methods in a highly collaborative fashion, with the goal of providing a complete catalog of protein-coding genes, nonprotein-coding genes, transcriptional regulatory elements, DNase hypersensitive sites, epigenetic modifications, and other sequence elements that mediate chromatin structure and function. The selected 30 Mb of the human genome is divided among 44 regions, each of which was selected to assure that a wide range of genomic features were included. Having such a comprehensive catalog of structural and functional elements in hand, even for just a representative 1% of the human genome, will be critical for understanding human biology well enough to decipher the biology of human health and disease.
The data management challenges of any such consortium-based effort are substantial. Early on in the planning of the ENCODE project, it became apparent that much of the data generated by members of the ENCODE Consortium should be stored in already-existing public repositories in order to maximize the visibility and availability of these data within the biological community. Data that can be directly linked to specific genomic coordinates are available through the UCSC Genome Browser (Hinrichs et al. 2006), while array-based data (e.g., expression and chromatin immunoprecipitation [ChIP-chip] data) are deposited in NCBI’s Gene Expression Omnibus (GEO) (Barrett et al. 2005) or EBI’s ArrayExpress (Parkinson et al. 2005).
Concurrent with the generation and deposition of experimental data into these allied databases, substantial progress has been made in the development of new tools intended to facilitate the analysis of genomic data sets. The assembly of genome-wide data sets has been facilitated by the introduction of tools such as the UCSC Table Browser, which is intended to retrieve specific subsets of coordinate-based data, and the Genome Alignment and Annotation Databases (GALA), which merge genome annotations with multi-species alignment data (Giardine et al. 2003). Once a genome-wide data set has been generated, high-throughput analysis can be performed using tools such as Galaxy, a simple Web portal that enables users to combine data from independent queries; visualize results; perform operations such as intersections, unions, and subtractions; and submit data to numerous computational tools that are useful in genomic analysis (Giardine et al. 2005).
While the ENCODE Consortium is generating large amounts of data related to the targeted 1% of the genome, it is important to remember that there are other significant sources of biological information, accumulated over many years, that contain data that lie within ENCODE regions; data from these very rich sources of information should also be considered in order to have a complete view of all known information regarding a particular region of interest. These data include mRNA, EST, and STS sequences from GenBank (Benson et al. 2006); single nucleotide polymorphisms stored in NCBI’s Database of Single Nucleotide Polymorphisms (dbSNP) (Wheeler et al. 2006); sequence clusters within the NCBI UniGene database (Wheeler et al. 2006); disease-centric information in the Online Mendelian Inheritance in Man (OMIM) database (Hamosh et al. 2005); array-based data from NCBI GEO (Barrett et al. 2005); and genomic data from the UCSC Genome Browser (Hinrichs et al. 2006). While it would be possible to query each of these databases individually, it became obvious that a “portal” was necessary to enable and facilitate the process of gathering all of these disparate types of data from all of these source databases for users interested in either a genome-wide or gene-centric perspective of these data. It also became obvious that, once these data are gathered, it is still very difficult for the average user to apply basic analysis or visualization tools such as the ones described above to these data.
The ENCODEdb portal was developed to provide a unified, single point-of-access to data generated by the ENCODE Consortium, as well as to data from other source databases that lie within ENCODE regions, regardless of which public database the primary data are housed in. This provides the user a complete view of all known data in a particular region of interest. ENCODEdb users can both browse data in genomic regions of interest, as well as easily assemble custom data sets that can be visualized with the UCSC Genome Browser or used for downstream analysis with tools such as Galaxy. This report focuses on the functional aspects of ENCODEdb, illustrating the kinds of biological insights that can be made using the data generated by the ENCODE Consortium and other sources. The ENCODEdb portal is freely accessible at http://research.nhgri.nih.gov/ENCODEdb.
Results and Discussion
Genomic Context searches
Genomic Context searches allow the user to retrieve information on functional elements that have been annotated within ENCODE regions, and the data returned by these searches are not limited to data that have been generated by the ENCODE Consortium. As such, the Genomic Context searches are intended to provide the user a compendium of all relevant genomic information that is known about each individual ENCODE region, without requiring the user to query each individual source database separately. Using data provided by UCSC, NCBI, or NCBI GEO, the ENCODEdb database stores the genomic position of the relevant functional elements. Query terms, whether they be gene based (e.g., gene symbol, GenBank accession number) or region based (e.g., ENCODE region, cytological band), are all translated into their genomic coordinates before the search is issued, and all elements that overlap these coordinates are reported.
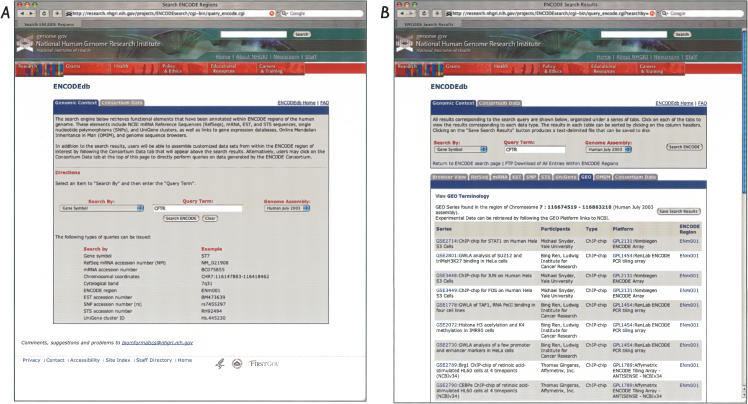
To illustrate some of the types of data that are returned by a Genomic Context query, the gene CFTR will be used as an example. CFTR is the cystic fibrosis conductance regulator gene (OMIM:602421) and lies within ENCODE region ENm001. A user would initiate the search by entering the term “CFTR” as the query term and selecting Gene Symbol from the pull-down menu, as shown in Figure 1A. Note that searches can be done on a variety of terms, including RefSeq mRNA accession number, chromosomal coordinates, cytological band, and UniGene cluster ID; the user does not need to know in advance whether the genomic region of interest actually lies in an ENCODE region, or which ENCODE region it lies in. The user may also select which genome assembly to search (Human July 2003 [hg16] or May 2004 [hg17]). Once the query is submitted, the user is provided a summary of detailed annotations on the CFTR locus, organized as a series of tabs in the results window (Fig. 1B). Each one of the tabs corresponds to one of the queried source databases, providing links back to the source database so that the user can examine the primary database entries directly. A tab is shown only when data from that source database are available in the region of interest.
Figure 1.
Genomic Context searches using ENCODEdb. (A) Queries can be issued on any genomic region of interest, without knowing in advance whether the region actually lies in an ENCODE region or which ENCODE region it lies in. Searches may be issued using a gene symbol, RefSeq mRNA accession number (NM number), mRNA accession number, chromosomal coordinates, cytological band, ENCODE region, EST accession number, SNP accession number, STS accession number, or UniGene cluster ID; examples of proper syntax for each search is shown in the figure. At present, users may select either the Human July 2003 (hg16) or May 2004 (hg17) assemblies for their searches. (B) The result of a gene symbol-based search, using CFTR as the query. The GEO tab has been highlighted. All GEO Series (GSE) in the CFTR region are shown. Clicking on any of the GSE numbers will spawn a new window, showing the GEO series entry page within GEO. Clicking on any of the GEO platform (GPL) numbers takes the user to the appropriate GEO platform page, giving full information on the array platform used in this experiment. Clicking on ENCODE region numbers takes the user to the appropriate view within the UCSC Genome Browser. A “GEO Terminology” link is provided for users who are unfamiliar with the structure or content of the GEO database.
Many users are probably familiar with the UCSC Genome Browser, the NCBI MapViewer, and Ensembl for browsing and retrieving genomic annotations. At the time of this writing, the NCBI MapViewer provides no access to ENCODE-specific data, and Ensembl provides only limited views of these data. The GEO and OMIM links that are provided as part of the results of ENCODEdb Genomic Context searches are not currently available through the UCSC Genome Browser. The results provided in the RefSeq, mRNA, EST, SNP, and STS tabs are also available from the UCSC Genome Browser, albeit as a graphic not as text. Although UCSC does also provide data in text format through the Table Browser, we believe that the user interface available through ENCODEdb will be more straightforward for many bench biologists.
Another important feature of the ENCODEdb Genomic Context searches is that array-based data housed in GEO for a region of interest can be retrieved by simply clicking on the GEO tab, producing the view shown in Figure 1B; this “one-click” method is easier for the user since a region-based search cannot easily be done at the GEO Web site. The tabular view used here provides the user a quick overview of the data in a more readable, compact, and informative format than can be obtained using GEO directly, allowing users to quickly focus in on particular array-based data of interest. The results are organized by GEO series (GSE numbers), which are defined as related samples that make up an experiment. There are also links to the appropriate GEO platform (GPL numbers), which describe the list of elements (e.g., oligonucleotide probe sets, cDNAs, or SAGE tags) being assayed or that may be detected and quantified in that experiment.
In addition to the summary data stored under each individual tab, users can select the Browser View tab to obtain a graphical view of all annotations in the region of interest. The user can select to view either a UCSC Genome Browser “default view”, with track selection preset, or a view that they can configure based on their own needs. Two Ensembl-based views are also available under the Browser View tab: Ensembl CytoView, which provides a genomic overview of sequence-based features in this region, and Ensembl MultiSpecies view, which provides an alignment of the human region of interest with corresponding regions in other selected organisms. We anticipate providing similar access to the NCBI Map Viewer in the future.
Consortium Data searches
Consortium Data searches allow users to perform compound queries on data generated by the ENCODE Consortium. Query options include array-based data stored in GEO, as well as data available through the UCSC Genome Browser. As with Genomic Context searches, Consortium Data search terms are also translated into their respective genomic coordinates, and data are retrieved from UCSC and GEO in a region-specific manner.
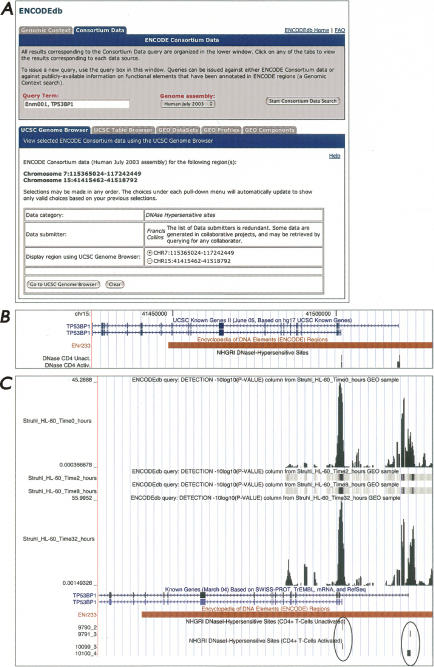
There are two ways in which users can perform a Consortium Data search. If a user has already performed a Genomic Context search and then clicks on the Consortium Data tab, the query term will be “passed through” to the Consortium Data search. Alternatively, a user can perform a Consortium Data search directly by clicking on the Consortium Data link on the ENCODEdb home page (Fig. 1A); this would take the user to the view shown in Figure 2A. Unlike the Genomic Context search, Consortium Data searches can be performed on multiple regions at the same time. Multiple search terms are separated by commas, which are an implicit Boolean OR, so the search term “ENm001, TP53BP1” (as shown in Fig. 2A) would return ENCODE data that fall into either of these two regions. As before, users can issue their query against either the Human July 2003 (hg16) or May 2004 (hg17) assemblies. Queries are conducted against five different data sources.
Figure 2.
The UCSC Genome Browser query form. (A) A query using “ENm001, TP53BP1” as the search term, searching against the Human July 2003 sequence assembly. Users can select desired data categories and data submitters from the pull-down menus provided on the form; here, DNase hypersensitive sites has been chosen as the data category, and Francis Collins has been chosen as the data submitter. The choices under each pull-down menu will dynamically update to show only valid choices based on any previous selections; this feature was built into ENCODEdb to assist the user, since there is no similar functionality available through the GEO Web site itself. Because the query was performed using two search terms, users have the option to go to the region of ENm001 (on chromosome 7) or TP53BP1 (on chromosome 15). Once the Go To UCSC Genome Browser button is pressed, a new window is spawned, showing the selected experiments within the context of the UCSC Genome Browser (B). Here, the image is centered on two forms of TP53BP1 in the UCSC Known Genes tracks. The Known Genes track was opened manually, and the window was shifted to the right for better viewing of the 5′ ends of the transcripts. Through this type of view, the overlap between known genes and the DNase I hypersensitivity data requested through ENCODEdb becomes obvious. (C) ENCODEdb can be used to test specific hypotheses. This panel illustrates the results of a query issued to determine whether an alternative promoter corresponds to a downstream hypersensitive site. The view was generated by clicking the GEO Components tab shown in Figure 2A and then changing the selections to the following: ChIP-on-chip assay, experimental group “Pol2 ChIP-chip of RA-stimulated HL60 cells at four timepoints” in the HL60 cell line, for the region Chromosome 15:41486698–41590028. Wiggle was chosen as the output format, and the data column to display in this format is Detection. The resulting UCSC Genome Browser view shows the annotation tracks seen in Figure 2B, along with the newly requested ChIP-chip data. The 0- and 32-h timepoint tracks have been manually expanded to illustrate the correlation between hypersensitive sites, the 5′ ends of the genes, and the presence of PolII.
UCSC Genome Browser
If the user selects UCSC Genome Browser as the target for their query, the user will be taken to a query form similar to that shown in Figure 2A. As with the Genomic Context search, results are organized under a series of tabs, so that the user can easily switch between results from the five different data sources. The chromosomal regions specified by the query are shown above the table with the pull-down menus. The user can now filter what data they wish to view by making selections in the Data Category and Data Submitter pull-downs; the choices under each pull-down dynamically update, showing only valid choices based on any previous selections. The ability to prefilter data is important, since the ENCODE Consortium has generated huge amounts of data; to date, there are 81,563,202 individual data points across 10 cell lines, 17 data categories, and 31 data providers. Simply displaying all of these data at the same time may prove overwhelming to the user, so users can narrow down what data are displayed to those data that are relevant to their own research interests.
As an example, requesting data on DNase I-hypersensitive (DNase HS) sites generated by the Collins laboratory (Crawford et al. 2004) would produce a view similar to that shown in Figure 2B, which shows each identified hypersensitive site as a tick mark in the DNase HS track. Using the pull-down menus in the UCSC Genome Browser window, the view has been expanded to show the two transcripts of the TP53BP1 gene that are displayed in the Known Genes annotation track (for details, see figure legend). The view shown in Figure 2B illustrates DNase HS sites upstream of the two transcripts, which, as depicted, have different transcription start sites. The upper transcript is for the known TP53BP1 gene, while the lower transcript, with the downstream (internal) transcription start site, is from an uncharacterized cDNA. This view shows the DNase HS sites immediately 5′ of the transcription start sites that correspond to putative regulatory elements of the TP53BP1 gene.
UCSC Table Browser
Similarly, the form under the UCSC Table Browser tab can be used to limit a search by both Data Category and Data Submitter. This form is intended as a preliminary step to using the UCSC Table Browser, a powerful tool for downloading data stored at UCSC. When a user preselects ENCODE-specific choices using the ENCODEdb portal, the task of selecting the desired tables within the Table Browser becomes decidedly simpler. For instance, pull-down menus specific to an experimental group, track (e.g., DNase hypersensitive sites), and data table (e.g., microarray chip or massively parallel signature sequencing) can easily be adjusted to suit the user’s particular needs. The choices under each pull-down menu dynamically update, based on any previous selections made by the user. Since there are more than 700 tables to choose from through UCSC’s Table Browser, the simplified preselection menu offered through ENCODEdb helps reduce this number to one that is more manageable for the novice user.
GEO DataSets and GEO Profiles
GEO DataSets are curated sets of GEO records that have been assembled at NCBI into biologically meaningful and statistically comparable groupings; these DataSets provide a coherent synopsis about an experiment and serve as the basis for downstream data mining. The GEO DataSets tab allows for the retrieval of these data in a genome coordinate–based way that is not easily done through the GEO Web site itself. ENCODEdb can perform searches on experimental assay type, experiment group (GEO series), and data submitter; as before, the choices under each pull-down dynamically update, showing only valid choices based on any previous selections. The resulting GEO DataSets entries include a summary of the experiment, an indication of what type of array-based technique was used, information on the individual samples used in the study, and links to GEO Series data used to create the GEO data set. The GEO Profiles tab is similar to the GEO DataSets tab in function but differs in the type of data returned. As implied by its name, submitting a query through this part of ENCODEdb returns genome coordinate–specific information on GEO Profiles, which contain individual gene expression profiles. GEO Profiles become available through ENCODEdb as they are posted on the GEO Web site.
GEO Components
The final tab, GEO Components, is the most versatile of the array-based query tools in ENCODEdb since it provides the user the ability to both create a data set and either visualize or perform downstream analyses on these data using tools not currently available through GEO. The term “component” is used to encompass all cross-linked Platform, Series, and Sample entries within GEO, and the form under the GEO Components tab can be used to retrieve array-based data using parameter-specific query options. Once the initial query is issued, users can again filter down what data they wish to view by making selections in a series of pull-down menus; these menus allow selection on experimental assay type, experimental groups (GEO Series), data submitter, and cell line. As before, the choices under each pull-down dynamically update, showing only valid choices based on any previous selections. The resulting data sets can be obtained in either browser-extensible data (BED) format, in UCSC Custom Track format, or in UCSC “wiggle track” format, where data are converted into a smoothed-curve format for visualization. The results of a GEO Components search viewed as a UCSC Custom Wiggle Track are shown in Figure 2C. Finally, the user can have their data set sent directly to Galaxy, a powerful platform for interactive large-scale genome analysis (Giardine et al. 2005). Neither the data formatting options nor the link to Galaxy is provided by NCBI’s GEO interface.
Regardless of which of these output options is selected, users are taken to a new page, in which they are asked to select which specific data fields they are interested in. The ability to choose specific data fields depends on the output option selected in the previous step. For example, if Download BED File or either of the UCSC display options was selected, the user can only select a single field, due to the limitations of the file formats themselves. If Download Selected Columns or Send Query to Galaxy was selected, any or all of the data fields can be selected. Choosing Download Selected Columns allows users to identify any differences between the GEO data, which is often provided as replicate assays, and the array-based data provided through the UCSC Genome or Table Browser, which is displayed as an average of the replicate values. Furthermore, the UCSC Table Browser presents transformed data, usually in the form of averages of replicate P-values or binding sites. Data obtained directly from GEO are raw data (i.e., user-normalized for missing or aberrant spots) and are available as mean and median intensities, intensities normalized against background, the log ratio of intensities, flagged data (to identify bad spots), and P-value data. The ability to select specific data fields of interest was built into ENCODEdb since these data are not easily obtained on a per-column basis from the GEO Web site, even though these data are all provided within the source GEO records. The ability to access these types of data is useful for anyone who wishes to evaluate ENCODE (or genome-wide data, for that matter) as it becomes available. The ability to easily create data sets for downstream analysis, as described above, is one of the key features of ENCODEdb, making these data more accessible to the average user.
The utility of the GEO Components feature of ENCODEdb can be best illustrated by returning to the example considering DNase I–hypersensitive sites discussed above. Using ENCODEdb, it is simple to ask whether the previously identified DNase HS sites overlap with other experimentally determined promoter elements. Figure 2C shows the results of retrieving, from GEO, the PolII binding sites reported in ChIP-chip analyses, then displaying the resulting data as a UCSC Genome Browser custom track. Four time points are shown (0, 2, 8, and 32 h), with the tracks for 0 and 32 h expanded; the heights of the peaks represent the P-values of the ChIP-chip results. From this view, it is apparent that RNA PolII binds to both positions exhibiting DNase I hypersensitivity and, therefore, that the promoters of both TP53BP1 transcripts, characterized and uncharacterized, have been experimentally verified. The most important aspect of this example is that a user is able to export data out of GEO and visualize them alongside other, disparate types of data found within the UCSC Genome Browser. Without a resource such as ENCODEdb, this would be virtually impossible for anyone but the most expert of users to do.
Concluding remarks
In summary, the ENCODEdb portal provides a unified, single point-of-access to data generated by the ENCODE Consortium, as well as to data from other source databases that lie within ENCODE regions. ENCODEdb vastly simplifies the process of amassing data from numerous public sources, using a front end that should be intuitive to most biologists with little to no familiarity with the query tools provided by most public data repositories. Because all the data within ENCODEdb can be searched by its genomic coordinates, the portal is a powerful tool for users interested in both gene-centric and genome-scale analyses. In particular, ENCODE-specific GEO data can be retrieved by gene-based or coordinate-based queries, a functionality not available through GEO itself. ENCODEdb’s integration with the UCSC Genome Browser makes it ideal for visualizing data of interest, while its integration with the Galaxy system vastly simplifies the process of analyzing large amounts of genomic data. Continued development of ENCODEdb will focus on the integration of new types of data being generated by the ENCODE Consortium, and it is hoped that investigators studying a variety of biological systems will use tools such as ENCODEdb to fulfill the goal of deciphering the biology underlying human health and disease.
Methods
ENCODEdb currently provides unified access to data from the following sources through a publicly available Web front end: the NCBI mRNA Reference Sequences (RefSeq) database, GenBank (for mRNA, EST, and STS sequences), NCBI’s dbSNP, the NCBI UniGene database, OMIM, the NCBI GEO, and the UCSC Genome Browser. Queries of Consortium data through ENCODEdb allow display of the data at the UCSC Browser. Results can also be transferred to the Galaxy server for further manipulation and comparison. ENCODEdb is updated at least monthly with new data from NCBI and UCSC.
This database is implemented in Perl and uses an Oracle database back end, with cookies and JavaScript enabled. Metadata from various GEO experiments are stored in database tables, with the actual raw data stored as text flatfiles in GEO SOFT format, allowing the data structure flexibility of this format to be maintained. The ENCODEdb portal is freely accessible at http://research.nhgri.nih.gov/ENCODEdb, using any up-to-date Web browser such as Safari or Firefox.
Additional information regarding the implementation of ENCODEdb can be found on the ENCODEdb Web site.
Acknowledgments
We thank Webb Miller, Ross Hardison, Gretchen Gibney, Elise Feingold, Peter Good, and Laura Liefer for their thoughtful insights and feedback during the development of ENCODEdb. This research was supported by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health.
Footnotes
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.5582207
References
- Barrett T., Suzek T.O., Troup D.B., Wilhite S.E., Ngau W.C., Ledoux P., Rudnev D., Lash A.E., Fujibuchi W., Edgar R., Suzek T.O., Troup D.B., Wilhite S.E., Ngau W.C., Ledoux P., Rudnev D., Lash A.E., Fujibuchi W., Edgar R., Troup D.B., Wilhite S.E., Ngau W.C., Ledoux P., Rudnev D., Lash A.E., Fujibuchi W., Edgar R., Wilhite S.E., Ngau W.C., Ledoux P., Rudnev D., Lash A.E., Fujibuchi W., Edgar R., Ngau W.C., Ledoux P., Rudnev D., Lash A.E., Fujibuchi W., Edgar R., Ledoux P., Rudnev D., Lash A.E., Fujibuchi W., Edgar R., Rudnev D., Lash A.E., Fujibuchi W., Edgar R., Lash A.E., Fujibuchi W., Edgar R., Fujibuchi W., Edgar R., Edgar R. NCBI GEO: Mining millions of expression profiles–database and tools. Nucleic Acids Res. 2005;33:D562–D566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L., Lipman D.J., Ostell J., Wheeler D.L., Ostell J., Wheeler D.L., Wheeler D.L. GenBank. Nucleic Acids Res. 2006;34:D16–D20. doi: 10.1093/nar/gkj157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford G.E., Holt I.E., Mullikin J.C., Tai D., Blakesley R., Bouffard G., Young A., Masiello C., Green E.D., Wolfsberg T.G., Holt I.E., Mullikin J.C., Tai D., Blakesley R., Bouffard G., Young A., Masiello C., Green E.D., Wolfsberg T.G., Mullikin J.C., Tai D., Blakesley R., Bouffard G., Young A., Masiello C., Green E.D., Wolfsberg T.G., Tai D., Blakesley R., Bouffard G., Young A., Masiello C., Green E.D., Wolfsberg T.G., Blakesley R., Bouffard G., Young A., Masiello C., Green E.D., Wolfsberg T.G., Bouffard G., Young A., Masiello C., Green E.D., Wolfsberg T.G., Young A., Masiello C., Green E.D., Wolfsberg T.G., Masiello C., Green E.D., Wolfsberg T.G., Green E.D., Wolfsberg T.G., Wolfsberg T.G., et al. Identifying gene regulatory elements by genome-wide recovery of DNase hypersensitive sites. Proc. Natl. Acad. Sci. 2004;101:992–997. doi: 10.1073/pnas.0307540100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, The ENCODE (ENCyclopedia of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- Giardine B., Elnitski L., Riemer C., Makalowska I., Schwartz S., Miller W., Hardison R.C., Elnitski L., Riemer C., Makalowska I., Schwartz S., Miller W., Hardison R.C., Riemer C., Makalowska I., Schwartz S., Miller W., Hardison R.C., Makalowska I., Schwartz S., Miller W., Hardison R.C., Schwartz S., Miller W., Hardison R.C., Miller W., Hardison R.C., Hardison R.C. GALA, a database for genomic sequence alignments and annotations. Genome Res. 2003;13:732–741. doi: 10.1101/gr.603103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B., Riemer C., Hardison R.C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., Riemer C., Hardison R.C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., Hardison R.C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., Zhang Y., Blankenberg D., Albert I., Taylor J., Blankenberg D., Albert I., Taylor J., Albert I., Taylor J., Taylor J., et al. Galaxy: A platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamosh A., Scott A.F., Amberger J.S., Bocchini C.A., McKusick V.A., Scott A.F., Amberger J.S., Bocchini C.A., McKusick V.A., Amberger J.S., Bocchini C.A., McKusick V.A., Bocchini C.A., McKusick V.A., McKusick V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs A.S., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., Diekhans M., Furey T.S., Harte R.A., Hsu F., Furey T.S., Harte R.A., Hsu F., Harte R.A., Hsu F., Hsu F., et al. The UCSC Genome Browser Database: Update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson H., Sarkans U., Shojatalab M., Abeygunawardena N., Contrino S., Coulson R., Farne A., Lara G.G., Holloway E., Kapushesky M., Sarkans U., Shojatalab M., Abeygunawardena N., Contrino S., Coulson R., Farne A., Lara G.G., Holloway E., Kapushesky M., Shojatalab M., Abeygunawardena N., Contrino S., Coulson R., Farne A., Lara G.G., Holloway E., Kapushesky M., Abeygunawardena N., Contrino S., Coulson R., Farne A., Lara G.G., Holloway E., Kapushesky M., Contrino S., Coulson R., Farne A., Lara G.G., Holloway E., Kapushesky M., Coulson R., Farne A., Lara G.G., Holloway E., Kapushesky M., Farne A., Lara G.G., Holloway E., Kapushesky M., Lara G.G., Holloway E., Kapushesky M., Holloway E., Kapushesky M., Kapushesky M., et al. ArrayExpress—a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2005;33:D553–D555. doi: 10.1093/nar/gki056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio R., Edgar S., Federhen L.Y., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio R., Edgar S., Federhen L.Y., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio R., Edgar S., Federhen L.Y., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio R., Edgar S., Federhen L.Y., Canese K., Chetvernin V., Church D.M., DiCuccio R., Edgar S., Federhen L.Y., Chetvernin V., Church D.M., DiCuccio R., Edgar S., Federhen L.Y., Church D.M., DiCuccio R., Edgar S., Federhen L.Y., DiCuccio R., Edgar S., Federhen L.Y., Edgar S., Federhen L.Y., Federhen L.Y., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2006;34:D173–D180. doi: 10.1093/nar/gkj158. [DOI] [PMC free article] [PubMed] [Google Scholar]