Abstract
A previous study documented the presence of mutations in mprF that accompanied the loss of daptomycin susceptibility among Staphylococcus aureus isolates following exposure to the drug. An association between the development of glycopeptide-intermediate S. aureus and daptomycin nonsusceptibility has also been recently described. We report that among three clinical S. aureus isolates which developed vancomycin heteroresistance, as well as daptomycin nonsusceptibility despite a lack of exposure to this drug, there were no mutations resulting in amino acid substitutions in MprF.
Daptomycin is a cyclic lipopeptide antibiotic which has bactericidal activity toward many gram-positive bacteria (4). With increasing resistance among Staphylococcus aureus strains to other classes of antibiotics, daptomycin has become an attractive therapeutic option. However, there have been recent reports of daptomycin treatment failures associated with the emergence of daptomycin-nonsusceptible S. aureus strains (5, 6).
Friedman et al. (3) recently demonstrated that for S. aureus isolates with reduced daptomycin susceptibilities selected by in vitro exposure to serially increasing concentrations of daptomycin, the initial increase in daptomycin MICs was temporally associated with mutations in mprF. Similarly, they showed that mutations resulting in amino acid substitutions in MprF were present in all three daptomycin-nonsusceptible clinical isolates of methicillin-resistant S. aureus (MRSA) that they studied (3).
Several groups have now reported an association between the development of glycopeptide-intermediate S. aureus (GISA) strains and daptomycin nonsusceptibility (2, 8, 11). Sakoulas et al. (11) described three sets of clinical MRSA isolates (each set of isolates being collected from a separate patient) in which GISA or hetero-GISA (hGISA) phenotypes arose during vancomycin therapy and which also demonstrated increases in daptomycin MICs and daptomycin heteroresistance. While the emergence of daptomycin resistance associated with amino acid substitutions in MprF as reported by Friedman and colleagues (3) occurred in the context of daptomycin exposure, none of the GISA or hGISA isolates associated with daptomycin nonsusceptibility as reported by Sakoulas et al. (11) had been collected from patients treated with daptomycin.
Thus, we determined the DNA sequences of the mprF genes of these daptomycin-nonsusceptible GISA isolates, which had never been exposed to daptomycin. As a control, we sequenced the mprF gene of the daptomycin-nonsusceptible MRSA isolate from a previously reported pair of MRSA isolates, one daptomycin susceptible and the other nonsusceptible, that did not demonstrate vancomycin heteroresistance and had been recovered from a patient who had been treated with daptomycin (6).
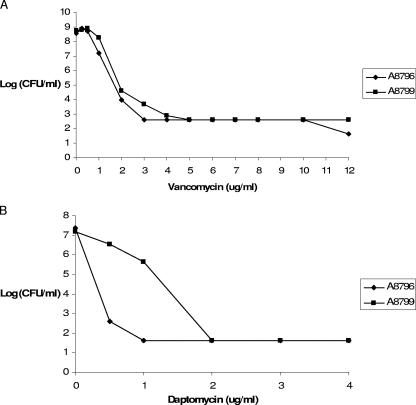
Table 1 lists the S. aureus isolates described in this report and the corresponding MICs. Vancomycin MICs for S. aureus A8796 and A8799, which were not provided in the published reference, were determined by agar dilution by using CLSI methods (1). Vancomycin and daptomycin population analyses for both isolates were performed using methods described previously (12). Brain heart infusion agar was used on plates containing vancomycin. Mueller-Hinton II broth containing 1.5% agar and supplemented to contain a 50- to 55-μg/ml concentration of ionized calcium was used on plates containing daptomycin. Compared to S. aureus A8796, S. aureus A8799 demonstrated heteroresistance to daptomycin but not to vancomycin (Fig. 1). S. aureus A8796 and A8799 had previously been sequenced through Cubist Pharmaceuticals, and the nonsusceptible member of the pair showed a single-base-pair mutation within the mprF-2 amplicon leading to a serine-to-leucine change at amino acid 337 (L. Friedman, personal communication). We resequenced mprF from the resistant isolate in this study.
TABLE 1.
MRSA isolate characteristics
| Strain designation | MIC (μg/ml)a of:
|
MprF amino acid substitution | |
|---|---|---|---|
| Daptomycin | Vancomycin | ||
| A8090 | 0.5 | 1 | |
| A8094 | 2 | 8 | None |
| A6224 | 0.5 | 2 | |
| A6226 | 2 | 8 | None |
| A6300 | 1 | 2 | |
| A6298 | 2 | 4 | None |
| A8796 | 0.5-1 | 1 | |
| A8799 | 2-4 | 2 | S337L |
Daptomycin MICs determined by using cation-adjusted Mueller-Hinton II broth supplemented with calcium to a concentration of 50 μg/ml are from reference 11, except those for S. aureus A8796 and S. aureus A8799, which are reported in reference 6. Vancomycin MICs determined by agar dilution are from reference 11, except those for S. aureus A8796 and S. aureus A8799, which were determined in this study.
FIG. 1.
Vancomycin (A) and daptomycin (B) population analyses for S. aureus A8796 and A8799.
Genomic DNA from all isolates was obtained using the GenElute bacterial genomic DNA kit (Sigma). PCR using the mprF-2 forward and reverse primers (3) was first performed by using genomic DNA from S. aureus A8799. PCR conditions consisted of 95°C for 5 min; followed by 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min; and then 1 cycle of 72°C for 10 min for a final extension step. Following the purification of the PCR product with ExoSAP-IT (USB), sequencing was performed at the Molecular Biology Core Facilities, Dana-Farber Cancer Institute, Boston, MA, by using the mprF-2 sequencing primers (3). Nucleotide sequence data were translated into amino acid sequences by using Editseq (DNASTAR Inc., Madison, WI), and sequences were aligned with the MprF protein sequence of S. aureus MW2 by using Megalign (DNASTAR). These results confirmed the presence of the S337L amino acid substitution in S. aureus A8799.
Having confirmed the presence of a mutation in mprF from S. aureus A8799, we amplified, sequenced, and analyzed the entire mprF open reading frames from the MRSA-GISA pairs shown in Table 1 by using the mprF1 to mprF4 amplification and sequencing primers described by Friedman et al. (3). Notably, compared with their daptomycin-susceptible, vancomycin-susceptible progenitors, the daptomycin-nonsusceptible GISA isolates (A8094, A6226, and A6298) exhibited no mutations conferring amino acid substitutions in MprF throughout the entire sequenced region.
MprF is involved in the lysylation of cell membrane phosphotidylglycerol and has been postulated to facilitate bacterial resistance to cationic antimicrobial peptides by reducing the negative charge of the cell membrane (9). Jung and colleagues have shown that daptomycin, in the presence of calcium, behaves similarly to cationic peptides in its interaction with phospholipid membranes (4). Therefore, it is not unreasonable to expect mutations in MprF to affect the activity of daptomycin.
However, there have been conflicting reports concerning the role of MprF in S. aureus susceptibility to vancomycin. Ruzin et al. (10) reported that the inactivation of mprF allows for more binding of vancomycin to S. aureus cell membranes and results in increased susceptibility to vancomycin among GISA strains. While Nishi et al. (7) confirmed these findings for GISA, they also noted that the inactivation of mprF in vancomycin-susceptible S. aureus results in decreased vancomycin susceptibility and a rightward shift in the population analysis profile for vancomycin-treated mprF mutants compared to that for the wild type. To explain these discrepant findings, Nishi and colleagues hypothesized that in the context of the GISA phenotype, mprF inactivation influences cell wall synthesis and results in increased vancomycin susceptibility whereas in a glycopeptide-susceptible background, mprF inactivation alters the cell membrane charge, causing changes in the pattern of vancomycin binding to the membrane, resulting in decreased susceptibility to glycopeptides (7). In contrast to these findings with laboratory-generated mutants, Wootton et al. (13) demonstrated no differences in the expression of mprF among multiple clinically related glycopeptide-susceptible S. aureus and GISA (or hGISA) isolates and suggested that the expression of this gene is not directly related to the GISA phenotype.
In support of the results of the recent study by Friedman and colleagues (3), we confirm the presence of a mutation affecting the amino acid sequence of MprF in another MRSA isolate in which daptomycin nonsusceptibility developed during daptomycin therapy. However, we also demonstrate that among clinical GISA or hGISA strains arising from susceptible progenitors exposed to vancomycin, but not daptomycin, reduced susceptibility to daptomycin was not associated with amino acid alterations in MprF.
Other investigators have observed the thickening of cell walls of GISA isolates and have noted a correlation between cell wall thickening and reduced daptomycin susceptibility (2). Whether the reduced susceptibility to daptomycin which emerges in some clinical isolates of MRSA after treatment with vancomycin in parallel with reductions in vancomycin susceptibility can be attributed to this or to other mechanisms remains to be established. However, our results point to the fact that mechanisms other than those affecting MprF are involved as the first step toward reduced susceptibility to daptomycin under some conditions.
Footnotes
Published ahead of print on 2 April 2007.
REFERENCES
- 1.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 7th ed. Document M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 2.Cui, L., E. Tominaga, H. M. Neoh, and K. Hiramatsu. 2006. Correlation between reduced daptomycin susceptibility and vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 50:1079-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedman, L., J. D. Alder, and J. A. Silverman. 2006. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 50:2137-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jung, D., A. Rozek, M. Okon, and R. E. Hancock. 2004. Structural transitions as determinants of the action of the calcium-dependent antibiotic daptomycin. Chem. Biol. 11:949-957. [DOI] [PubMed] [Google Scholar]
- 5.Mangili, A., I. Bica, D. R. Snydman, and D. H. Hamer. 2005. Daptomycin-resistant, methicillin-resistant Staphylococcus aureus bacteremia. Clin. Infect. Dis. 40:1058-1060. [DOI] [PubMed] [Google Scholar]
- 6.Marty, F. M., W. W. Yeh, C. B. Wennersten, L. Venkataraman, E. Albano, E. P. Alyea, H. S. Gold, L. R. Baden, and S. K. Pillai. 2006. Emergence of a clinical daptomycin-resistant Staphylococcus aureus isolate during treatment of methicillin-resistant Staphylococcus aureus bacteremia and osteomyelitis. J. Clin. Microbiol. 44:595-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishi, H., H. Komatsuzawa, T. Fujiwara, N. McCallum, and M. Sugai. 2004. Reduced content of lysyl-phosphatidylglycerol in the cytoplasmic membrane affects susceptibility to moenomycin, as well as vancomycin, gentamicin, and antimicrobial peptides, in Staphylococcus aureus. Antimicrob. Agents Chemother. 48:4800-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel, J. B., L. A. Jevitt, J. Hageman, L. C. McDonald, and F. C. Tenover. 2006. An association between reduced susceptibility to daptomycin and reduced susceptibility to vancomycin in Staphylococcus aureus. Clin. Infect. Dis. 42:1652-1653. [DOI] [PubMed] [Google Scholar]
- 9.Peschel, A., R. W. Jack, M. Otto, L. V. Collins, P. Staubitz, G. Nicholson, H. Kalbacher, W. F. Nieuwenhuizen, G. Jung, A. Tarkowski, K. P. van Kessel, and J. A. van Strijp. 2001. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J. Exp. Med. 193:1067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruzin, A., A. Severin, S. L. Moghazeh, J. Etienne, P. A. Bradford, S. J. Projan, and D. M. Shlaes. 2003. Inactivation of mprF affects vancomycin susceptibility in Staphylococcus aureus. Biochim. Biophys. Acta 1621:117-121. [DOI] [PubMed] [Google Scholar]
- 11.Sakoulas, G., J. Alder, C. Thauvin-Eliopoulos, R. C. Moellering, Jr., and G. M. Eliopoulos. 2006. Induction of daptomycin heterogeneous susceptibility in Staphylococcus aureus by exposure to vancomycin. Antimicrob. Agents Chemother. 50:1581-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakoulas, G., G. M. Eliopoulos, R. C. Moellering, Jr., C. Wennersten, L. Venkataraman, R. P. Novick, and H. S. Gold. 2002. Accessory gene regulator (agr) locus in geographically diverse Staphylococcus aureus isolates with reduced susceptibility to vancomycin. Antimicrob. Agents Chemother. 46:1492-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wootton, M., A. P. Macgowan, and T. R. Walsh. 2005. Expression of tcaA and mprF and glycopeptide resistance in clinical glycopeptide-intermediate Staphylococcus aureus (GISA) and heteroGISA strains. Biochim. Biophys. Acta 1726:326-327. [DOI] [PubMed] [Google Scholar]