Abstract
During 2005, 66 carbapenem-resistant isolates of Acinetobacter baumannii were collected from seven tertiary-care hospitals participating in a nationwide surveillance network in Colombia. The isolates were multidrug resistant and produced the carbapenemases OXA-23 and OXA-51. Forty-five belonged to four clones while 21 were unique pulsotypes. One clone was present in two hospitals within one city, while another had spread between two hospitals in different cities. Blood, secretions, and abdominal fluids were the most frequent sites of isolation. This is the first description of widespread dissemination of OXA-23 in South America.
Acinetobacter baumannii is an important nosocomial pathogen which appears to be increasing in frequency (8). Carbapenems have been the drugs of choice for treatment of severe Acinetobacter infections, but their efficacy is increasingly compromised by resistance (19).
According to the SENTRY reports, resistance rates for nosocomial gram-negative pathogens, including A. baumannii, are higher in Latin American countries than in the United States or Europe. The prevalence of carbapenem resistance in A. baumannii isolates across Latin America in the SENTRY database in 2001 was estimated at 25% (13, 24). During 2005, carbapenem resistance rates for A. baumannii were around 40% in 12 Colombian tertiary-care hospitals (18).
Carbapenem-hydrolyzing OXA enzymes are the most important cause of carbapenem resistance in A. baumannii worldwide (23). These began to be described over a decade ago, in 1993, with the description of ARI-1, later renamed OXA-23, in an imipenem-resistant A. baumannii strain from a patient in the Edinburgh Royal Infirmary (22). The strain was isolated in 1985, before the use of imipenem in the hospital. Imipenem resistance was subsequently demonstrated to be transferable (25). Since then, carbapenem-resistant isolates of A. baumannii carrying oxacillinases have been reported worldwide (4, 14, 29). It has been recognized that most A. baumannii strains have a chromosomal carbapenemase gene (a blaOXA-51-like gene) (10), though this is expressed at a high level only if an insertion sequence, such as ISAba1, is inserted upstream (30). In addition, a minority of A. baumannii strains have further OXA carbapenemase genes that are not part of the normal genomic repertoire of the species; these include the blaOXA-23-like gene, the blaOXA-24-like gene, and blaOXA-58. Although they are less-efficient hydrolyzers of carbapenems in vitro than are the metallo-β-lactamases (MβLs), these oxacillinases can inactivate carbapenems and their presence or activation by ISAba1 is demonstrably correlated with resistance (4, 30).
Based on the high rates of resistance to carbapenems in A. baumannii strains from 10 tertiary-care hospitals in the Colombian network, an investigation into the underlying mechanisms and strain structure was undertaken.
(This report was presented in part at the 46th Annual Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco, CA, 2006 [13a].)
MATERIALS AND METHODS
During 2005, the research facility Centro Internacional de Entrenamiento e Investigaciones Medicas (CIDEIM) conducted a study of nosocomial multidrug-resistant A. baumannii with the participation of the Colombian Nosocomial Bacterial Resistance Study Group, which included 10 tertiary-care institutions in six cities. Centers were selected if they provided tertiary care, had microbiologists and infectious-disease physicians on site, and agreed to participate.
Epidemiological and susceptibility data for all isolates from patients in general wards and intensive care units were sent to CIDEIM. This information was analyzed with WHONET 5.3 software (26). Initial susceptibilities were determined by the automated systems used in nine participating institutions (Microscan, Dade Behring Inc, Deerfield, IL, or Vitek, bioMérieux, Lyons, France) or, at one site, by the CLSI standard disk susceptibility method (20).
Seventy-one A. baumannii isolates that had been reported as carbapenem resistant based on an imipenem or meropenem MIC of ≥16 μg/ml (21) were available and sent to CIDEIM for further analysis. Seven of the 10 institutions sent isolates with this phenotype (Table 1).
TABLE 1.
A. baumannii isolates from 10 tertiary-care hospitals in Colombia
| City | Hospital | No. of carbapenem-resistant A. baumannii isolates sent to CIDEIM | Clone no. (no. of isolates) | No. of unique pulsotypes | No. of isolates positive by PCR for gene:
|
Isolates found by WHONET to be carbapenem resistant
|
||
|---|---|---|---|---|---|---|---|---|
| blaOXA-51-like | blaOXA-23-like | % of total | No. resistant/total no. | |||||
| Cali | A | 31 | 1 (21) | 10 | 31 | 31 | 29 | 78/268 |
| Bogotá | B | 12 | 2 (8) | 4 | 12 | 12 | 62 | 43/69 |
| C | 2 | 2 (2) | 2 | 2 | 33 | 3/9 | ||
| D | 0 | 11 | 3/27 | |||||
| Medellín | E | 10 | 3 (10) | 10 | 10 | 34 | 17/50 | |
| F | 0 | 50 | 3/6 | |||||
| Pereira | G | 7 | 1 (2); 4 (2) | 3 | 7 | 7 | 29 | 15/52 |
| Bucaramanga | H | 1 | 1 | 1 | 0 | 18 | 2/11 | |
| I | 0 | 40 | 10/25 | |||||
| Barranquilla | J | 3 | 3 | 3 | 3 | 32 | 8/25 | |
| Total | 66 | Four clones (45) | 21 | 66 | 65 | 34 (avg) | 182/542 | |
Bacterial identification and susceptibility testing.
Bacterial identification was confirmed by Vitek (bioMérieux, Lyons, France) with the GNI+ card, used according to the manufacturer's instructions. MICs were determined for imipenem (Merck Sharp & Dohme, Rahway, NJ) and meropenem (AstraZeneca, Alderley Park, United Kingdom) by the CLSI broth microdilution method (21).
Strain typing.
Pulsed-field gel electrophoresis (PFGE) was performed on genomic DNA of all A. baumannii isolates as described previously (28). A CHEF Mapper system (Bio-Rad Laboratories, Fremont, CA) was used to electrophorese SmaI-digested DNA (Promega, Madison, WI) at a voltage of 6 V/cm at 14°C, with pulse times of 1 s and 30 s for 19 h. The results were analyzed with Diversity software (Bio-Rad), and band-based dendrograms were produced using Dice coefficients (7). Indistinguishable and closely related (85% to 99% related) pulsotypes were considered clonal, and the major clones from each hospital were then compared with other major clones from other hospitals and cities.
Isoelectric focusing.
Isoelectric focusing of crude sonicates was done following the method described by Mathew et al. (17).
PCR amplification and sequencing.
Screening of carbapenem-resistant A. baumannii isolates was performed with a multiplex PCR assay using the primers described by Woodford et al. (30). To confirm the presence of the ISAba1 insertion upstream of OXA-23 and OXA-51, PCR was performed using the protocol described by Turton et al. (27).
To identify fully the carbapenemase genes in major clones, primers for blaOXA-23-like gene oxacillinases (11) were used to amplify the entire blaOXA gene from a genomic DNA template. The amplification products were purified using the QIAquick PCR purification kit (QIAGEN, Valencia, CA), cloned into plasmid pCR-XL-TOPO, and transformed into chemically competent Escherichia coli TOP10 (Invitrogen, Carlsbad, CA) by heat shock, as detailed by the manufacturer of the TOPO XL PCR cloning kit (Invitrogen). Recombinant plasmid DNA was isolated using the QIAfilter Midi plasmid preparation kit (QIAGEN), and both strands of the insert were sequenced by ACGT, Inc. (Wheeling, IL), using M13R and M13F(−20) primers.
MβL screening.
Screening for MβLs in selected isolates was performed with an MβL Etest (AB Biodisk, Solna, Sweden), used according to the manufacturer's protocol. MIC ratios (MIC of imipenem alone/MIC of imipenem plus EDTA) of ≥8 were considered indicative of MβL production.
Hybridization studies.
To determine the locations of β-lactamase genes, genomic and plasmid DNA preparations were evaluated, as previously described (15), from representative isolates of each major clone type and tested by hybridization to probes specific for blaOXA-23 or 16S rRNA genes, under conditions of high stringency. The probes consisted of the entire 822-bp blaOXA-23 amplicon, generated with primers OXA-23A and OXA-23B (11), or the 16S rRNA amplicon, generated with universal primers A and B (16). These were amplified by PCR and labeled with digoxigenin (Roche, Mannheim, Germany). An imipenem-resistant, OXA-40-producing A. baumannii isolate (15) was included as a negative control for both experiments. In the plasmid experiment, uncut plasmid DNA was used, the positive control consisted of a recombinant pCR-XL-TOPO-OXA-23 plasmid as described above, and the plasmids in Escherichia coli V517 (56.7, 5.8, 4.09, 3.15, 2.83, and 2.2 kb) were used as size standards.
RESULTS
During 2005, the carbapenem resistance rates for A. baumannii varied greatly among the hospitals but averaged 33.6% (range, 11 to 62%) (Table 1). In total, 542 A. baumannii isolates were identified at the 10 participating institutions, 182 of them reportedly carbapenem resistant. Blood (30%), secretions (15%), abdominal fluid (14%), catheters (10%), and urine (8%) were the most frequent sites of isolation for these resistant isolates; 71 of these, from seven institutions, were sent to CIDEIM for further investigation. Two isolates were excluded because they were not A. baumannii and another three because they were susceptible to both carbapenems (MICs, 0.5 to 2 μg/ml). The majority of isolates were resistant to both carbapenems, though six exhibited a carbapenem MIC resulting in either a susceptible or an intermediate designation. Examples of this occurred for both meropenem (n = 2) and imipenem (n = 4). All subsequent analysis was limited to the 66 isolates confirmed as resistant to at least one carbapenem.
The isolates were subjected to PFGE analysis, which revealed that 45 isolates clustered into four clones (Table 1). Clonal outbreaks were present in hospitals A, B and C, E, and G, in Cali, Bogotá, Medellín, and Pereira, respectively. Clone 2 was shared by hospitals B and C in Bogotá, while clone 1 was shared by hospitals A and G, located in Cali and Pereira, respectively. Twenty-one isolates were unrelated to other strains and were categorized as unique pulsotypes. Isoelectric focusing revealed multiple β-lactamase bands for each isolate, but all had bands with pIs of 6.7 or 6.8, consistent with OXA-23 (19). Sixty-five of the 66 were positive for blaOXA-23-like genes by multiplex PCR, all 66 were positive for blaOXA-51-like genes, and all were negative for blaOXA-58 and blaOXA-24-like genes.
Single representative isolates of clones 1, 2, and 3 were chosen for sequencing of the OXA gene. These represented the three most-numerous clones (clone 1, 23 isolates; clone 2, 10 isolates; and clone 3, 10 isolates). All were associated with clusters or outbreaks within single facilities. The results showed that each clone was 100% homologous with the classical blaOXA-23 (ARI-1) gene (GenBank accession no. AJ132105) from nucleotides 22 to 891.
The presence of the insertion sequence ISAba1, upstream of a carbapenemase gene, reportedly can affect the gene expression and contingent resistance (27). To determine if this association applied among our isolates, one isolate per clone was screened by PCR for linkage of ISAba1 to blaOXA. ISAba1 was present upstream of the blaOXA-23-like gene in clones 1, 2, 3, and 4, and this linkage was also seen in 18/20 unique pulsotypes but not in the remaining two. The latter isolates were both from hospital A; each isolate exhibited carbapenem MICs of between 16 and 32 μg/ml.
The single isolate from hospital H was negative for the blaOXA-23-like gene and did not have ISAba1 upstream of the intrinsic blaOXA-51-like carbapenemase gene of A. baumannii, a configuration previously associated with carbapenem resistance (27). Carbapenem resistance in these instances might be explained by other mechanisms, such as impermeability and/or another β-lactamase. MβLs were not present.
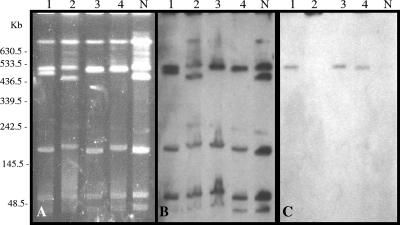
Genetic locations were investigated for the blaOXA-23-like genes in the representatives of the major A. baumannii clones. Total DNA was digested with the I-CeuI endonuclease and hybridized successively with 16S rRNA and OXA-23 probes. After digestion with the enzyme, the DNA was resolved into five to seven fragments, each of which hybridized with the 16S rRNA-specific probe (Fig. 1A and B), identifying them as chromosomal. The blaOXA-23-specific probe cohybridized with a single chromosomal fragment of isolates representing clones 1, 3, and 4 (Fig. 1C). No hybridizing chromosomal fragment was identified for clone 2.
FIG. 1.
Localization of the blaOXA-23 gene in I-CeuI-generated chromosome fragments of A. baumannii separated by PFGE. (A) Electrophoretic pattern after I-CeuI digestion; (B) hybridization with a probe specific for rRNA genes; (C) hybridization with a probe specific for the blaOXA-23-like gene. Lanes: 1 to 4, representatives of clones 1, 2, 3, and 4, respectively; N, imipenem-resistant OXA-40-producing A. baumannii.
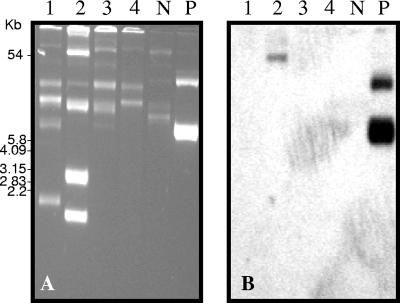
Plasmid DNA was extracted from the same A. baumannii isolates used in the I-CeuI experiments, and electrophoretic separation of uncut plasmid DNA detected multiple bands in each of the DNAs. Under conditions of high stringency, the OXA-23 probe hybridized with a single plasmid band in the representative of clone 2 (Fig. 2), whereas no hybridization was noted for the other clones. We conclude that blaOXA-23 was chromosomal in clones 1, 3, and 4 but plasmid mediated in clone 2.
FIG. 2.
Plasmid localization of the blaOXA-23-like gene in A. baumannii clones. (A) Undigested-plasmid profiles; (B) hybridization with a probe specific for blaOXA-23-like genes. Lanes: 1 to 4, representatives of clones 1, 2, 3, and 4, respectively; N and P, negative- and positive-control lanes, respectively.
DISCUSSION
During the past few years, OXA-23 enzymes have been reported in Acinetobacter strains from Brazil (6), China (GenBank accession number AY554200) (31), Ireland (3), Korea (12), the United Kingdom (27, 30), and Singapore (GenBank accession number AY795964). One clone with OXA-23 has spread to over 36 hospitals in southern England (5), while clones with OXA-40 (OXA-24 related) have spread widely in Spain and, more recently, in the United States (15).
We report here that OXA-23-like carbapenemases were present in A. baumannii isolates from multiple, widely separated cities in Colombia. The producers included both nonclonal and clonal isolates, with clonal spread having occurred between hospitals in the same city and between hospitals in different cities. In three of the four clones examined, blaOXA-23 was chromosomally encoded; in the fourth clone, it was plasmid encoded. Chromosomal mediation has been demonstrated or assumed for other OXA carbapenemases (4), though plasmid encoding of OXA-23 (19) and OXA-58 (1, 9) enzymes has also been reported. Chromosomal mediation of blaOXA-23 has been described previously for Proteus mirabilis (2).
Based on our data, there are two major contributors to the high prevalence of carbapenem-resistant A. baumannii isolates in Colombia. The first is the presence of OXA-23-like carbapenemases. A discrepancy in susceptibilities between imipenem and meropenem, noted for a subset of these isolates, raises the possibility of a more widespread dissemination of these OXA-23-like carbapenemases than that detected here. As clinical laboratories typically test only a single carbapenem in their automated panels, the presence of a potentially plasmid-mediated resistance mechanism in carbapenem-susceptible/intermediate isolates may go clinically unrecognized.
A second factor contributing to this high prevalence is the dissemination of resistant clones. We have previously published work on the clonal dissemination of carbapenem-resistant Pseudomonas aeruginosa in Colombian hospitals (28), where the MβL VIM-2 was detected in isolates from multiple cities. Some clones were local while others had spread between cities; in general, the prevalence of carbapenem resistance was related mostly to clonal spread. The present results illustrate similar patterns for a different species and enzyme class.
Our findings further illustrate the emerging global problems due to OXA-class carbapenemases in Acinetobacter spp. As carbapenems have been the drugs of choice for serious Acinetobacter infection, this is a major clinical problem. Given the proclivity of Acinetobacter for nosocomial spread and contamination of the environment, enhanced infection control measures will be of major importance. This is particularly true in light of the paucity of new agents active against this pathogen.
Acknowledgments
We thank the participating institutions from the Colombian Nosocomial Resistance Study Group, whose members are as follows: CIDEIM, Sandra Reyes; Cali, Ernesto Martínez, Lena Barrera, Luz Marina Gallardo, Alba Lucía Bohorquez, and Nancy Villamarín; Bogotá, Carlos Alquichire, Aura Lucía Leal, Martha Ruiz, Mariluz Páez, Pilar Hurtado, Andrés Torres, Adela Cubides, Henry Mendoza, Alba Lucia Sanín, Nancy Botía, Claudia Rodríguez, Sandra Reina, Martha Patricia Melendez, Sonia Cuervo, Jorge Cortés, Maria Cristina Paredes, and Patricia Arroyo; Medellín, Carlos Ignacio Gómez, Jaime López, Mónica Cuartas, Ana Lucia Correa, Jorge Donado, Julián Betancourth, Juan David Villa, Ana Cristina Quiroga, Luz Teresita Correa, Eugenia Loaiza, Luz Maria Melguiza, Martha Vallejo, Rubén Darío Trejos, Victoria García, and Dora Rivas; Barranquilla, Ezequiel Guijarro, Rubén Darío Camargo, Adriana Marín, and Ángela Mendoza; Pereira, Carmen Elisa Llano, Araceli Cano, Martha Lucía Gómez, and Liliana Villa; Bucaramanga, Claudia Bárcenas, Adriana Pinto, and Luis Ángel Villar; and Ibagué, Claudia Echeverry and Amparo Ovalle.
We are grateful to Bristol-Myers Squibb, Pfizer, and Merck Sharp & Dohme for their financial support.
Footnotes
Published ahead of print on 2 April 2007.
REFERENCES
- 1.Bertini, A., A. Giordano, P. Varesi, L. Villa, C. Mancini, and A. Carattoli. 2006. First report of the carbapenem-hydrolyzing oxacillinase OXA-58 in Acinetobacter baumannii isolates in Italy. Antimicrob. Agents Chemother. 50:2268-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonnet, R., H. Marchandin, C. Chanal, D. Sirot, R. Labia, C. De Champs, E. Jumas-Bilak, and J. Sirot. 2002. Chromosome-encoded class D β-lactamase OXA-23 in Proteus mirabilis. Antimicrob. Agents Chemother. 46:2004-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boo, T. W., F. Walsh, and B. Crowley. 2006. First report of OXA-23 carbapenemase in clinical isolates of Acinetobacter species in the Irish Republic. J. Antimicrob. Chemother. 58:1101-1102. [DOI] [PubMed] [Google Scholar]
- 4.Brown, S., and S. Amyes. 2006. OXA (beta)-lactamases in Acinetobacter: the story so far. J. Antimicrob. Chemother. 57:1-3. [DOI] [PubMed] [Google Scholar]
- 5.Coelho, J. M., J. F. Turton, M. E. Kaufmann, J. Glover, N. Woodford, M. Warner, M. F. Palepou, R. Pike, T. L. Pitt, B. C. Patel, and D. M. Livermore. 2006. Occurrence of carbapenem-resistant Acinetobacter baumannii clones at multiple hospitals in London and Southeast England. J. Clin. Microbiol. 44:3623-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalla-Costa, L. M., J. M. Coelho, H. A. Souza, M. E. Castro, C. J. Stier, K. L. Bragagnolo, A. Rea-Neto, S. R. Penteado-Filho, D. M. Livermore, and N. Woodford. 2003. Outbreak of carbapenem-resistant Acinetobacter baumannii producing the OXA-23 enzyme in Curitiba, Brazil. J. Clin. Microbiol. 41:3403-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dice, L. R. 1945. Measures of the amount of ecological association between species. Ecology 26:297-302. [Google Scholar]
- 8.Gaynes, R., and J. R. Edwards. 2005. Overview of nosocomial infections caused by gram-negative bacilli. Clin. Infect. Dis. 41:848-854. [DOI] [PubMed] [Google Scholar]
- 9.Heritier, C., A. Dubouix, L. Poirel, N. Marty, and P. Nordmann. 2005. A nosocomial outbreak of Acinetobacter baumannii isolates expressing the carbapenem-hydrolysing oxacillinase OXA-58. J. Antimicrob. Chemother. 55:115-118. [DOI] [PubMed] [Google Scholar]
- 10.Heritier, C., L. Poirel, P. E. Fournier, J. M. Claverie, D. Raoult, and P. Nordmann. 2005. Characterization of the naturally occurring oxacillinase of Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:4174-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heritier, C., L. Poirel, T. Lambert, and P. Nordmann. 2005. Contribution of acquired carbapenem-hydrolyzing oxacillinases to carbapenem resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:3198-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeong, S. H., I. K. Bae, K. O. Park, Y. J. An, S. G. Sohn, S. J. Jang, K. H. Sung, K. S. Yang, K. Lee, D. Young, and S. H. Lee. 2006. Outbreaks of imipenem-resistant Acinetobacter baumannii producing carbapenemases in Korea. J. Microbiol. 44:423-431. [PubMed] [Google Scholar]
- 13.Jones, R. N. 2001. Global aspects of antimicrobial resistance among key bacterial pathogens. Results from the 1997-1999 SENTRY Antimicrobial Program. Clin. Infect. Dis. 32:S81-S156. [DOI] [PubMed] [Google Scholar]
- 13a.Kattan, J. N., A. M. Guzman, A. Correa, S. Reyes, K. Lolans, N. Woodford, M. V. Villegas, J. P. Quinn, and D. M. Livermore. 2006. Abstr. 46th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C2-598.
- 14.Livermore, D. M. 2003. The threat from the pink corner. Ann. Med. 35:226-234. [DOI] [PubMed] [Google Scholar]
- 15.Lolans, K., T. W. Rice, L. S. Munoz-Price, and J. P. Quinn. 2006. Multicity outbreak of carbapenem-resistant Acinetobacter baumannii isolates producing the carbapenemase OXA-40. Antimicrob. Agents Chemother. 50:2941-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mammeri, H., L. Poirel, N. Mangeney, and P. Nordmann. 2003. Chromosomal integration of a cephalosporinase gene from Acinetobacter baumannii into Oligella urethralis as a source of acquired resistance to β-lactams. Antimicrob. Agents Chemother. 47:1536-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathew, A., A. M. Harris, M. J. Marshall, and G. W. Ross. 1975. The use of analytical isoelectric focusing for detection and identification of beta-lactamases. J. Gen. Microbiol. 88:169-178. [DOI] [PubMed] [Google Scholar]
- 18.Miranda, M. C., F. Perez, T. Zuluaga, M. R. Olivera, A. Correa, S. L. Reyes, M. V. Villegas, and Grupo de Resistencia Bacteriana Nosocomial de Colombia. 2006. Resistencia a antimicrobianos en bacilos Gram negativos aislados en unidades de cuidado intensivo en hospitales de Colombia, WHONET2003, 2004 y 2005. Biomedica 26:424-433. [PubMed] [Google Scholar]
- 19.Naas, T., M. Levy, C. Hirschauer, H. Marchandin, and P. Nordmann. 2005. Outbreak of carbapenem-resistant Acinetobacter baumannii producing the carbapenemase OXA-23 in a tertiary care hospital of Papeete, French Polynesia. J. Clin. Microbiol. 43:4826-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.NCCLS. 2003. Performance standards for antimicrobial disk susceptibility testing, 8th informational supplement. Approved standard M2-A7. NCCLS, Wayne, PA.
- 21.NCCLS. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A6. NCCLS, Wayne, PA.
- 22.Paton, R., R. S. Miles, J. Hood, and S. G. B. Amyes. 1993. ARI-1: β-lactamase-mediated imipenem resistance in Acinetobacter baumannii. Int. J. Antimicrob. Agents 2:81-88. [DOI] [PubMed] [Google Scholar]
- 23.Poirel, L., and P. Nordmann. 2006. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12:826-836. [DOI] [PubMed] [Google Scholar]
- 24.Sader, H. S., R. N. Jones, A. C. Gales, J. B. Silva, A. C. Pignatari, and the SENTRY Participants Group (Latin America). 2004. The SENTRY Participants Group (Latin America). SENTRY Antimicrobial Surveillance Program report: Latin American and Brazilian results for 1997 through 2001. Braz. J. Infect. Dis. 8:25-79. [DOI] [PubMed] [Google Scholar]
- 25.Scaife, W., H. K. Young, R. H. Paton, and S. G. Amyes. 1995. Transferable imipenem-resistance in Acinetobacter species from a clinical source. J. Antimicrob. Chemother. 36:585-586. [DOI] [PubMed] [Google Scholar]
- 26.Stelling, J. M., and T. F. O'Brien. 1997. Surveillance of antimicrobial resistance: the WHONET program. Clin. Infect. Dis. 24(Suppl. 1):S157-S168. [DOI] [PubMed] [Google Scholar]
- 27.Turton, J. F., M. E. Ward, N. Woodford, M. E. Kaufmann, R. Pike, D. M. Livermore, and T. L. Pitt. 2006. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 258:72-77. [DOI] [PubMed] [Google Scholar]
- 28.Villegas, M. V., K. Lolans, M. del Rosario Olivera, C. J. Suarez, A. Correa, A. M. Queenan, J. P. Quinn, and the Colombian Nosocomial Resistance Study Group. 2006. First detection of metallo-β-lactamase VIM-2 in Pseudomonas aeruginosa isolates from Colombia. Antimicrob. Agents Chemother. 50:226-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walsh, T. R., M. A. Toleman, L. Poirel, and P. Nordmann. 2005. Metallo-β-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18:306-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woodford, N., M. J. Ellington, J. M. Coelho, J. F. Turton, M. E. Ward, S. Brown, S. G. Amyes, and D. M. Livermore. 2006. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27:351-353. [DOI] [PubMed] [Google Scholar]
- 31.Yu, Y. S., Q. Yang, X. W. Xu, H. S. Kong, G. Y. Xu, and B. Y. Zhong. 2004. Typing and characterization of carbapenem-resistant Acinetobacter calcoaceticus-baumannii complex in a Chinese hospital. J. Med. Microbiol. 53:653-656. [DOI] [PubMed] [Google Scholar]