Abstract
β-Lactam resistance in Haemophilus parasuis is an emerging phenomenon that has not yet been characterized from a molecular perspective. Clinical high-level β-lactam-resistant isolates from Spain bore a novel plasmid, pB1000, expressing a functionally active ROB-1 β-lactamase. Pulsed-field gel electrophoresis was applied for the first time to H. parasuis and showed that β-lactam resistance is due to clonal spread of a resistant strain, BB1018, bearing pB1000.
Haemophilus parasuis is a gram-negative bacillus, responsible for Glässer's disease, a disease with worldwide distribution characterized by fibrinous polyserositis, polyarthritis, and meningitis in swine (2, 9, 14). The incidence and prevalence of H. parasuis infection are especially high in developed countries, where it is one of the main causes of lethality and economic loss (18). Since no definite vaccination is available, antimicrobial treatment is the sole weapon for fighting this pathogen once infection is established. Although tetracyclines are the major antimicrobials used against this bacterium, resistance has been found in many instances (4, 12, 20). In Spain, a recent report indicated that up to 40% of the clinical isolates are highly resistant to tetracyclines, showing that other, more effective molecules are needed to treat infected animals (3). For this reason, penicillins and aminopenicillins are being used as the alternative treatment of infections due to H. parasuis. Phenotypically, clinical isolates resistant to β-lactams have largely been found in Switzerland, the United Kingdom, and Spain (3, 20).
From 2002 to 2005, 90 H. parasuis clinical isolates from diseased pigs were obtained in our laboratory at the Veterinary School in Madrid in the course of routine diagnostics (5). Identification was performed using phenotypic characteristics in combination with a PCR based on species-specific amplification of the 16S rRNA gene with primers HPS-F and HPS-R (Table 1), essentially as described by Oliveira et al. (17). Bacteria were cultured on chocolate agar PolyViteX plates (BioMérieux) and in Haemophilus test medium broth (Wider; Francisco Soria Melguizo, SA, Madrid, Spain) at 37°C under microaerophilic conditions (5% CO2). To assess antimicrobial resistance of H. parasuis in Spain, a complete antimicrobial profile using disk diffusion and microdilution methods was performed as described by Aarestrup et al. (1) using Haemophilus test medium for fastidious bacteria. MIC determinations were performed using a commercially prepared, dehydrated panel (Sensititre). All plates were inoculated and interpreted following CLSI (formerly NCCLS) guidelines (16). Specific breakpoints for respiratory disease were used when available (1). Eight isolates (∼1%) were highly resistant to the β-lactams penicillin (MIC > 16) and amoxicillin (MIC > 256) (Table 2). All resistant isolates were susceptible to third-generation cephalosporins (Table 2) and to β-lactams in combination with clavulanate (data not shown). Further, all strains were positive in the nitrocefin test. Overall, these data implied that a non-inhibitor-resistant β-lactamase could be responsible for β-lactam resistance in H. parasuis.
TABLE 1.
Primers used in this study
| Primer name | Sequence (5′→3′) | Position | GenBank accession no. |
|---|---|---|---|
| rob-1F | TGTTGCAATCGCTGCC | 2952-2968 | DQ840517 |
| rob-1R | TTATCGTACACTTTCCA | 3392-3409 | DQ840517 |
| rob-1D | AATTGGTTGGACAATAACGCA | 3332-3353 | DQ840517 |
| rob-1U | ATCGTCATGCCTTTGCCAACG | 3043-3064 | DQ840517 |
| MAP-1 | GCTCTCTAATTCTTTCGATAA | 1617-1638 | DQ840517 |
| MAP-2 | TTTTGAAGAAAGCGACCTACC | 550-571 | DQ840517 |
| MAP-3 | TTCTGTGATGTCTGCTGAAAG | 1085-1106 | DQ840517 |
| MAP-4 | TAAAGCATTGGTATTAAAGGC | 3777-3798 | DQ840517 |
| MAP-5 | GATTTTATCAACTCAACGTGG | 1461-1482 | DQ840517 |
| HPS-F | GTGATGAGGAAGGGTGGTGT | 440-459 | M75065 |
| HPS-R | GGCTTCGTCACCCTCTGT | 1243-1260 | M75065 |
| univ1 | CTGGCTCAGGACGAACGCTG | 30-49 | EF221612 |
| univ2 | GTTGCGCTCGTTGCGGGACT | 1112-1131 | EF221612 |
TABLE 2.
Susceptibility to β-lactams of bacteria used in this study
| Strain | MIC (μg/ml)a
|
Source or reference | ||||
|---|---|---|---|---|---|---|
| PEN | AMX | OXA | CTX | CEC | ||
| ATCC 19417 | ≤0.03 | ≤2 | ≤0.25 | ≤0.03 | ≤1 | Collectionc |
| BB 1018 | >16 | >256 | ≤0.25 | 0.06 | >16 | This work |
| BB 1019 | >16 | >256 | ≤0.25 | 0.06 | >16 | This work |
| BB 1020 | >16 | >256 | ≤0.25 | ≤0.03 | >16 | This work |
| BB 1021 | >16 | >256 | ≤0.25 | 0.06 | >16 | This work |
| BB 1022 | >16 | >256 | ≤0.25 | 0.06 | >16 | This work |
| BB 1023 | >16 | >256 | ≤0.25 | 0.12 | >16 | This work |
| BB 1024 | >16 | >256 | ≤0.25 | 0.06 | >16 | This work |
| BB 1025 | >16 | >256 | ≤0.25 | 0.06 | >16 | This work |
| BB 1026b | 0.12 | ≤2 | ≤0.25 | ≤0.03 | ≤1 | This work |
| E. coli | >16 | 4 | >4 | 0.06 | 8 | Novagen |
| E. coli(pB1000) | >16 | >256 | >4 | 0.06 | 128 | This work |
AMX, amoxicillin; CEC, cefaclor; CTX, cefotaxime; OXA, oxacillin; PEN, penicillin.
β-Lactam-susceptible H. parasuis clinical isolate.
Collection of the Pasteur Institute.
In order to assess the type of β-lactamase responsible for the resistance phenotype in H. parasuis, a second-generation cephalosporin, cefaclor (Sigma Chemical Co., St. Louis, Mo), was used as a phenotypic marker. For Haemophilus influenzae, this molecule is used to specifically detect blaROB-1-expressing β-lactam-resistant clinical isolates (11). Analysis of H. parasuis showed that all β-lactam-resistant isolates also had high-level resistance to cefaclor (MIC, >16). In contrast, β-lactam-susceptible clinical isolates and type strain ATCC 19417, obtained from the Collection of the Pasteur Institute (Paris, France) (Table 2), were susceptible to cefaclor, indicating that resistance to this cephalosporin is not intrinsic to H. parasuis but is a characteristic associated with penicillin and amoxicillin resistance. To determine the β-lactamase responsible for this phenotype, a PCR was set up that specifically amplified the blaROB-1 gene (10) (Table 1). All resistant isolates were positive for blaROB-1, whereas all susceptible H. parasuis strains were negative (data not shown). The 821-bp DNA amplicon of all strains was purified and sequenced on both strands. Nucleotide sequences were 100% identical among all H. parasuis isolates. The predicted amino acid sequence was identical to that of ROB-1 of Actinobacillus porcitonsilarum (GenBank accession no. AJ830712.1), Actinobacillus pleuropneumoniae (GenBank accession no. S51028.1), Mannheimia haemolytica (GenBank accession no. Z21724.1), and H. influenzae (GenBank accession no. AF022114.1). Recently, blaROB-1 from H. influenzae could be altered in vitro into an enzyme that confers high-level resistance to clavulanate and cefotaxime, changing only two amino acids (R169W and A237T) (8). Thus, the presence of blaROB-1 in H. parasuis should not be underestimated, because this resistance determinant may elicit the spread of these bacteria among the animal population, and accumulation of mutations may lead to a novel extended-spectrum cephalosporinase resistant to β-lactamase inhibitors that may spread among animal and human pathogens (15).
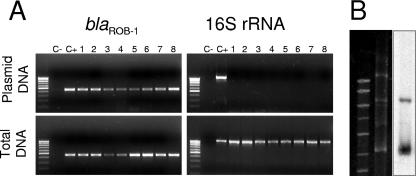
Plasmid extraction from the eight β-lactam-resistant H. parasuis isolates showed that all strains possessed plasmids. In order to determine whether blaROB-1 was located in one of these plasmids, a simple PCR-based technique that was named GPS-PCR (for Gene Positioning System) was developed. The technique is based on extraction and purification of plasmids and subsequent PCR with (i) primers of the gene to probe and (ii) universal primers of the chromosomally encoded 16S rRNA. A positive signal with the probe primers indicates that the gene is located in a plasmid, whereas a negative signal with the 16S rRNA primers shows that no chromosomal DNA is present in the preparations. Extraction and purification of plasmids from the β-lactam-resistant H. parasuis isolates was performed. GPS-PCR showed that all plasmid preparations were positive for the blaROB-1 gene, whereas PCRs of the 16S rRNA gene were negative (Fig. 1A). Hybridizations confirmed the plasmid location of the blaROB-1 gene (Fig. 1B). These data show that blaROB-1 of H. parasuis is encoded in an extra chromosomal plasmid and demonstrate that GPS-PCR is a valuable technique for assessing plasmid locations of genes in H. parasuis.
FIG. 1.
Location of blaROB-1 in H. parasuis. (A) GPS-PCR. Agarose gel electrophoresis of PCRs from plasmid extraction and total DNA. Note that plasmid extractions resulted in a positive PCR signal for blaROB-1 (upper left), whereas no amplification is observed using the chromosomally encoded 16S rRNA primers (upper right). Total DNA extraction serves as a PCR control for both reactions. The positive control is DNA from an A. pleuropneumoniae isolate bearing plasmid-encoded blaROB-1. Plasmid DNA extraction was carried out using the Plasmid Midi kit and QIAprep Spin Miniprep kit (QIAGEN, Inc., Chatworth, CA). PCR fragments were purified with QIAGEN PCR purification or gel extraction kits (QIAGEN, Inc., Chatworth, CA), following the manufacturer's instructions. (B) Southern blot of DNA extracted from a representative β-lactam-resistant H. parasuis isolate using blaROB-1 as a probe. The signal confirms that the blaROB-1 gene is located on a plasmid. Southern blotting was performed with DNA electrophoresed in 1% agarose gel and transferred onto Hybond N+ positively charged nylon membranes (Amersham Hybond; GE Healthcare). The blaROB-1 probe was obtained with primers MAP-4 and rob-1F (Table 1) prepared with the Nona primer kit (Q-BIO gene; MP Biomedicals).
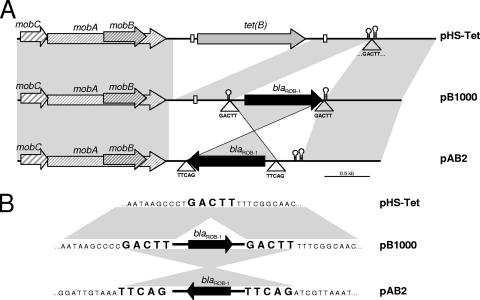
A pair of divergent primers was designed for blaROB-1, rob-1D and rob-1U, in order to amplify the complete replicon bearing the blaROB-1 gene through inverted PCR. All isolates gave a single amplicon of approximately 4.3 kb that was completely sequenced in one representative strain, BB1021, using the primers listed in Table 1. The complete plasmid had 4,613 bp and was designated pB1000. PCR mapping, using primer pairs MAP-1/MAP-2, MAP-3/rob-1U, MAP-4/rob-1F, and rob-1D/MAP-5 (Table 1), of all resistant strains together with restriction analysis with PstI showed that all H. parasuis isolates resistant to β-lactams bore plasmid pB1000 encoding blaROB-1. Sequence analysis of pB1000 showed that the coding sequence was preceded by putative −35 5′-TTGCTA and −10 5′-CGCCAAAAT boxes, together with a putative ribosome binding sequence, 5′-AAGGA, at an appropriate distance. The blaROB-1 gene was followed by a transcriptional terminator with a stem of 18 nucleotides (with two mismatches) and a loop formed by CTTGC. To ensure functionality of these signals and the blaROB-1 gene, pB1000 was transformed into Escherichia coli Novablue Singles competent cells (Novagen, Merck Chemicals Ltd., United Kingdom). The resulting transformants were highly resistant to penicillin (MIC > 16), amoxicillin (MIC > 256), and cefaclor (MIC > 16) (Table 2), demonstrating that the blaROB-1 gene was functionally active and responsible for the β-lactam resistance phenotype. Apart from this gene, pB1000 bore three genes, mobA, mobB, and mobC, encoding, respectively, three proteins of the relaxase family, MobA, MobB, and MobC (Fig. 2A). The genetic organization of pB1000 suggests that this replicon belongs to the recently described MOBHEN family (7). pB1000 was similar to plasmid pAB2 of a bovine M. haemolytica isolate containing blaROB-1 from Scotland (21). Interestingly, pB1000 was almost identical to an H. parasuis plasmid, pHS-Tet, recovered from a clinical strain in Australia, but with the blaROB-1 gene instead of the tet(B) gene (Fig. 2A) (13). Detailed analysis showed that the blaROB-1 gene was flanked by a perfect direct repeat, GACTT (Fig. 2B), in pB1000 and pAB2, indicating that these sequences can mediate insertion of the blaROB-1 gene. Supporting this notion, the pHS-Tet replicon contains a single copy of this sequence exactly in the insertion site of the blaROB-1 gene (Fig. 2B).
FIG. 2.
Genetic structure of pB1000. (A) Comparison of the genetic structures of pB1000, pHS-Tet, and pAB2. pB1000 contains the transcriptional terminator and a conserved direct repeat (white box) from pHS-Tet (13). Analogously, the transcriptional terminator of blaROB-1 is present in pHS-Tet. These data indicate that pB1000 and pHS-Tet might have evolved from a common ancestor bearing together the tet(B) and blaROB-1 genes. pAB2 has an inverted copy of blaROB-1. The empty triangle indicates the position of the GACTT sequence. (B) Potential recombination sites of blaROB-1. pHS-Tet has a single copy of the GACTT sequence. Further, in pB1000 and pAB2, blaROB-1 is embedded between the GACTT repeated sequences, indicating strongly that these sequences mediate mobilization of blaROB-1 via an intermediary hairpin structure and duplication of the insertion site.
All β-lactam-resistant H. parasuis isolates bore plasmid pB1000. In order to assess whether conjugation was implicated in the spread of pB1000, liquid and filter mating experiments were performed using E. coli as the recipient. None of the experiments gave transconjugants, showing that the conjugation machinery for pB1000 was absent in all isolates. To get insight into the diversity of the different isolates bearing pB1000, genetic characterization was performed. For this purpose, pulsed-field gel electrophoresis (PFGE) was applied for the first time to H. parasuis with a novel protocol performed as follows.
Preparation of chromosomal DNA.
H. parasuis colonies were resuspended in 2 ml TE buffer (1×) (10 mM Tris-HCl, 1 mM EDTA [pH 8]) and adjusted to an optical density at 600 nm of 2; 200 μl of this suspension was mixed with 10 μl of 20-mg/ml proteinase K. Agarose plugs were made from a 1:1 mixture of agarose D-1 (low electroendosmosis; Pronadisa SA) and the cell suspension. After solidification, plugs were incubated in 5 ml of lysis buffer (50 mM Tris-HCl, 50 mM EDTA, 1% lauroylsarcosine, 2 μg/ml proteinase K) for 2 h at 55°C with agitation. Cells were then washed two times with 10 ml of MiliQ water and four times with 10 ml buffer TE (1×) for 10 min each at 50°C with agitation.
Restriction endonuclease digestion.
For analysis, one-half of a DNA-agarose plug was digested for 16 h with 10 U of SmaI (Takara Bio, Inc.) at 30°C according to the manufacturer's instructions. Preliminary experiments using XbaI and BspI gave rise to small (<200 kb) or few (<4) bands/strain, respectively.
PFGE analysis.
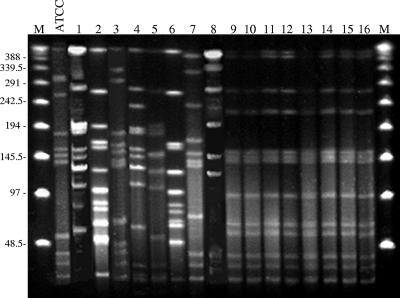
Plugs were loaded into a 1% agarose gel. PFGE was performed in 0.5× Tris-borate-EDTA buffer in a CHEF-DR III system (Bio-Rad). The following parameters were used: running time, 22 h; temperature, 14°C; field strength 6 V/cm; included angle, 120°; initial pulse time, 0.1 s; final pulse time, 25 s. The gels were stained with Sybr Safe (Invitrogen, Paisley, United Kingdom) for 20 min, distained in MiliQ water, and photographed under UV light. Lambda-ladder PFGE marker (New England Biolabs, Ipswich, MA) was used for molecular weight determinations. As a control, eight β-lactam-susceptible H. parasuis clinical isolates and type strain ATCC 19417 were included. All susceptible strains, including ATCC 19417, showed a different PFGE pattern, whereas the profiles of all β-lactam-resistant isolates were indistinguishable (Fig. 3). Thus, PFGE is a valuable technique for characterization of H. parasuis isolates that, in this study, has demonstrated that β-lactam resistance in H. parasuis is the consequence of clonal spread of a β-lactam-resistant strain. Analysis of the origin of the strains showed that they originated from different geographical regions. However, epidemiologic data strongly indicate that one of the farms could be the source of the strain, because it was the provider of piglets for the farms in which the rest of resistant strains were isolated.
FIG. 3.
PFGE fingerprint patterns of H. parasuis. Lanes 1 to 8, β-lactam-susceptible clinical isolates; lanes 9 to 16, β-lactam-resistant isolates. M stands for molecular marker and ATCC for β-lactam-susceptible H. parasuis strain ATCC 19417. Lane 1, BB1027; lane 2, BB1028; lane 3, BB1029; lane 4, BB1026; lane 5, BB1030; lane 6, BB1031; lane 7, BB1032; lane 8, BB1033; lane 9, BB1018; lane 10, BB1019; lane 11, BB1020; lane 12, BB1021; lane 13, BB1022; lane 14, BB1023; lane 15, BB1024; lane 16, BB1025.
Nucleotide sequence accession numbers.
Nucleotide sequences of this study have been deposited in GenBank under the following accession numbers: pB1000 from BB1021, DQ840517; blaROB-1 internal fragments, BB1018, DQ845801; BB1019, DQ845802; BB1020, DQ845803; BB1022, DQ845805; BB1023, DQ845806; BB1024, DQ845807; and BB1025, DQ845808.
This is the case for the isolates from Spain used in this study. We cannot discard the possibility that in other countries, resistance may be due to other factors. Such differences are very remarkable in the case of the geographic distribution of TEM-1- and ROB-1-mediated β-lactam resistance in H. influenzae. ROB-1-like β-lactamases are responsible for 0% of resistant isolates in Sweden, Argentina, or Israel, whereas the prevalence of ROB-1 is 31.6% in Mexico, 13.2% in the United States, and 9.2% in Canada (6). Such geographic differences may also be encountered with H. parasuis. However, to our knowledge, the present study is the first work characterizing β-lactam resistance in this species. Work in other regions will bring to light the implications of TEM-1 or ROB-1 enzymes in β-lactam resistance in H. parasuis.
(An initial report of this study was presented at the 16th European Congress for Clinical Microbiology and Infectious Diseases [19].)
Acknowledgments
We thank J. F. Fernandez-Garayzabal for critical reading of the manuscript. Patrice Courvalin and Bruno Perichon are acknowledged for helpful discussion and help with hybridizations and A. Casamayor for excellent technical support with PFGE. The VISAVET Group is acknowledged for the clinical isolates.
We thank the National Ramon y Cajal Program from the Spanish Ministry of Education and Science for support of B.G.-Z., the Spanish Ministry of Education and Science for supporting the Ph.D. scholarships of A.S.M. and A.C., and the Universidad Complutense de Madrid for the Ph.D. scholarship of J.A.E. This work was partially financed by project PR1-A/07-15397 from the Universidad Complutense de Madrid and S-0505/AGR/000265 (Vigilancia Sanitaria Program) from the Consejeria de Educacion, Comunidad de Madrid, Madrid, Spain.
Footnotes
Published ahead of print on 16 April 2007.
REFERENCES
- 1.Aarestrup, F. M., A. M. Seyfarth, and Ø. Angen. 2004. Antimicrobial susceptibility of Haemophilus parasuis and Histophilus somni from pigs and cattle in Denmark. Vet. Microbiol. 101:143-146. [DOI] [PubMed] [Google Scholar]
- 2.Biberstein, E. L., and D. C. White. 1969. A proposal for the establishment of two new Haemophilus species. J. Med. Microbiol. 2:75-77. [DOI] [PubMed] [Google Scholar]
- 3.de la Fuente, A. J., A. W. Tucker, J. Navas, M. Blanco, S. J. Morris, and C. B. Gutierrez-Martin. 2007. Antimicrobial susceptibility patterns of Haemophilus parasuis from pigs in the United Kingdom and Spain. Vet. Microbiol. 120:184-191. [DOI] [PubMed] [Google Scholar]
- 4.Eaves, L. E., P. J. Blackall, and M. Fegan. 1989. Characterization and antimicrobial sensitivity of haemophili isolated from pigs. Aust. Vet. J. 66:1-4. [DOI] [PubMed] [Google Scholar]
- 5.Escudero, J. A., A. San Millan, A. Catalan, A. G. de la Campa, E. Rivero, G. Lopez, L. Dominguez, M. A. Moreno, and B. Gonzalez-Zorn. 2007. First characterization of fluoroquinolone resistance in Streptococcus suis. Antimicrob. Agents Chemother. 51:777-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farrell, D. J., I. Morrissey, S. Bakker, S. Buckridge, and D. Felmingham. 2005. Global distribution of TEM-1 and ROB-1 beta-lactamases in Haemophilus influenzae. J. Antimicrob. Chemother. 56:773-776. [DOI] [PubMed] [Google Scholar]
- 7.Francia, M. V., A. Varsaki, M. P. Garcillan-Barcia, A. Latorre, A. Drainas, and F. de la Cruz. 2004. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol. Rev. 28:79-100. [DOI] [PubMed] [Google Scholar]
- 8.Galan, J. C., M. I. Morosini, M. R. Baquero, M. Reig, and F. Baquero. 2003. Haemophilus influenzae blaROB-1 mutations in hypermutagenic ΔampC Escherichia coli conferring resistance to cefotaxime and β-lactamase inhibitors and increased susceptibility to cefaclor. Antimicrob. Agents Chemother. 47:2551-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glässer, K. 1910. Die fibrinöse Serosen—und Gelenkentzündung der Ferkel, p. 122-125. In C. E. Hill and D. S. Metcalf (ed.), Die Krankheiten des Schweines. M. & H. Schaper, Hannover, Germany.
- 10.Juteau, J. M., M. Sirois, A. A. Medeiros, and R. C. Levesque. 1991. Molecular distribution of ROB-1 beta-lactamase in Actinobacillus pleuropneumoniae. Antimicrob. Agents Chemother. 35:1397-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karlowsky, J. A., G. Verma, G. G. Zhanel, and D. J. Hoban. 2000. Presence of ROB-1 beta-lactamase correlates with cefaclor resistance among recent isolates of Haemophilus influenzae. J. Antimicrob. Chemother. 45:871-875. [DOI] [PubMed] [Google Scholar]
- 12.Kofer, J., F. Hinterdorfer, and M. Awad-Masalmeh. 1992. Occurrence and drug resistance of bacteria pathogenic to the lungs from autopsy material of swine. Tierarztl. Prax. 20:600-604. [PubMed] [Google Scholar]
- 13.Lancashire, J. F., T. D. Terry, P. J. Blackall, and M. P. Jennings. 2005. Plasmid-encoded tet(B) tetracycline resistance in Haemophilus parasuis. Antimicrob. Agents Chemother. 49:1927-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Little, T. W. A. 1970. Haemophilus parasuis infection in pigs. Vet. Rec. 87:399-402. [DOI] [PubMed] [Google Scholar]
- 15.Livermore, D. M., R. Canton, M. Gniadkowski, P. Nordmann, G. M. Rossolini, G. Arlet, J. Ayala, T. M. Coque, I. Kern-Zdanowicz, F. Luzzaro, L. Poirel, and N. Woodford. 2007. CTX-M: changing the face of ESBLs in Europe. J. Antimicrob. Chemother. 59:165-174. [DOI] [PubMed] [Google Scholar]
- 16.NCCLS. 2002. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. Approved standards, 2nd ed. Document M31-A2. NCCLS, Wayne, Pa.
- 17.Oliveira, S., L. Galina, and C. Pijoan. 2001. Development of a PCR test to diagnose Haemophilus parasuis infections. J. Vet. Diagn. Investig. 13:495-501. [DOI] [PubMed] [Google Scholar]
- 18.Oliveira, S., and C. Pijoan. 2004. Haemophilus parasuis: new trends on diagnosis, epidemiology and control. Vet. Microbiol. 99:1-12. [DOI] [PubMed] [Google Scholar]
- 19.San Millan, A., et al. 2006. R1940 beta-lactam resistance in Haemophilus parasuis. Clin. Microbiol. Infect. 12(Suppl. 4):1. [Google Scholar]
- 20.Wissing, A., J. Nicolet, and P. Boerlin. 2001. The current antimicrobial resistance situation in Swiss veterinary medicine. Schweiz. Arch. Tierheilkd. 143:503-510. [PubMed] [Google Scholar]
- 21.Wood, A. R., F. A. Lainson, F. Wright, G. D. Baird, and W. Donachie. 1995. A native plasmid of Pasteurella haemolytica serotype A1: DNA sequence analysis and investigation of its potential as a vector. Res. Vet. Sci. 58:163-168. [DOI] [PubMed] [Google Scholar]