Abstract
Exposure-response analyses were performed for the microbiological and clinical efficacy of tigecycline in the treatment of complicated skin and skin-structure infections, where Staphylococcus aureus and streptococci are the predominant pathogens. A prospective method was developed to create homogeneous patient populations for PK-PD analyses. Evaluable patients from three clinical trials were pooled for analysis. Patients received a tigecycline 100-mg loading dose/50 mg every 12 h or a 50-mg loading dose/25 mg every 12 h. At the test-of-cure visit, microbiologic and clinical responses were evaluated. Patients were prospectively evaluated and classified into cohorts based on baseline pathogens: S. aureus only (cohort 1), monomicrobial S. aureus or streptococci (cohort 2), two gram-positive pathogens (cohort 3), polymicrobial (cohort 4), or other monomicrobial infections (cohort 5). A prospective procedure for combining cohorts was used to increase the sample size. Logistic regression evaluated steady-state 24-h area under the concentration-time curve (AUC24)/MIC ratio as a predictor of response, and classification and regression tree (CART) analyses were utilized to determine AUC/MIC breakpoints. Analysis began with pooled cohorts 2 and 3, the focus of these analyses, and included 35 patients with 40 S. aureus and/or streptococcal pathogens. CART analyses identified a significant AUC/MIC breakpoint of 17.9 (P = 0.0001 for microbiological response and P = 0.0376 for clinical response). The continuous AUC/MIC ratio was predictive of microbiological response based on sample size (P = 0.0563). Analysis of all pathogens combined decreased the ability to detect exposure-response relationships. The prospective approach of creating homogeneous populations based on S. aureus and streptococci pathogens was critical for identifying exposure-response relationships.
The global increase in bacterial resistance to existing agents has created a critical priority for antimicrobial discovery. Tigecycline, a new glycylcycline antimicrobial agent with an expanded broad spectrum of activity against both sensitive and multiple-drug resistant aerobic and anaerobic gram-positive and -negative microorganisms, was recently approved by the U.S. Food and Drug Administration (FDA) for the treatment of complicated skin and skin-structure infections (cSSSI) and complicated intra-abdominal infections (cIAI) (Tygacil product insert; Wyeth Pharmaceuticals, Inc., Philadelphia, PA). Phase 3 clinical trials for the treatment of both community-acquired and hospital-acquired pneumonia are still ongoing.
Bacteria that carry any of the classical tetracycline resistance genes, conferring either ribosomal protection or a tetracycline efflux pump, remain susceptible to tigecycline (17). Tigecycline has demonstrated in vitro activity against important resistant organisms, including methicillin-resistant Staphylococcus aureus, penicillin-resistant Streptococcus pneumoniae, and vancomycin-resistant enterococcal species, as well as extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Pseudomonas aeruginosa and Proteus spp. are notable exceptions to the broad spectrum of activity of tigecycline. As a result of the activity against emerging multidrug-resistant pathogens, tigecycline has the potential to be an important addition to the therapeutic options available for complicated infectious diseases, including cSSSI and cIAI (2, 6, 15, 23).
Comprehensive exposure-response analyses for the clinical and microbiological efficacy of tigecycline in the treatment of patients with cSSSI, where Staphylococcus aureus and streptococci are the predominant pathogens, have been performed. Such analyses have become fundamental in defining drug efficacy (7). Identification of relationships between pharmacokinetic (PK) parameters and pharmacodynamic (PD) response in patient populations is critical in dose justification and contributes to the establishment of in vitro MIC susceptibility breakpoints by regulatory and clinical agencies (i.e., the Clinical and Laboratory Standards Institute and the European Committee on Antimicrobial Susceptibility Testing) (5). Two critical steps in exposure-response analyses include the development of a population PK model (22) to characterize the kinetic profile of tigecycline in the target patient population and the identification of the PK-PD target and the magnitude of that target associated with optimal clinical and microbiological outcomes.
In vivo PK-PD infection models have long been used to study the activity of antimicrobial agents and to identify PK-PD targets and thresholds (3, 11). Furthermore, results from these models have been shown to correlate with clinical findings (1, 8). The data from in vitro studies have shown that tigecycline exhibits a time-dependent pattern of bactericidal activity against several gram-positive and -negative organisms, including S. pneumoniae, Haemophilus influenzae, and Neisseria gonorrhoeae (17). The PK-PD index often associated with drugs that display a time-dependent pattern of killing is the time above the MIC of the organism. However, for antimicrobial agents with moderate to prolonged postantibiotic effects (PAE), the time of exposure is less important and the area under the concentration-time curve (AUC)/MIC ratio (AUC/MIC) is the important determinant of efficacy (3). As a result of substantial PAEs associated with the tetracycline antibiotics, for example, the AUC/MIC ratio is the PK-PD index characteristically associated with these agents (2a, 3).
In a neutropenic mouse-thigh infection model conducted by van Ogtrop et al., the PK-PD index required for optimal activity of tigecycline against selected gram-negative and gram-positive organisms (Streptococcus pneumoniae, Staphylococcus aureus, Escherichia coli, and Klebsiella pneumoniae) was the time above a certain factor times the MIC for five of six organisms tested (21). The AUC, however, was also reasonably predictive, with only slightly lower r2 values. The results from the present study also revealed a prolonged in vivo PAE with tigecycline against S. pneumoniae and E. coli (8.9 and 4.9 h, respectively). Therefore, the prolonged persistent effect, in combination with the relatively long half-life of tigecycline in humans (13), would again suggest that the AUC/MIC ratio is likely to be predictive of therapeutic efficacy.
PK-PD analyses of tigecycline, using the AUC/MIC ratio as the optimal index, may allow for a better understanding of different responses among subgroups of patients in clinical trials. Variability in drug exposure and/or patient covariates may potentially explain such observed differences. The objectives of our analyses were to assess the relationships between tigecycline drug exposure and microbiological and clinical responses and to determine patient demographic characteristics, drug exposure measurements, and other covariates predictive of clinical and microbiological outcome in the treatment of patients with cSSSI.
MATERIALS AND METHODS
Clinical and PK data acquisition.
A single phase 2 randomized, open-label, dose comparison study and two phase 3 randomized, double-blind comparison trials of the safety and efficacy of intravenous (i.v.) tigecycline in the treatment of hospitalized patients with cSSSI were conducted. Prior to analyses, these studies were assessed for the appropriateness of pooling data. Protocols were reviewed for trial design and inclusion and exclusion criteria to determine whether the patients included in these trials were homogeneous in nature. Clinical and microbiological endpoints and study designs were deemed to be comparable, with the notable exception of two different dosing regimens in the phase 2 study. Baseline demographic data were also comparable between phases.
Patients with clinical signs and symptoms of cSSSI, defined as infections involving deeper soft tissue, requiring surgical intervention, or occurring in patients with significant underlying diseases, were enrolled. This included clinical entities such as infected ulcers, major abscesses, and deep or extensive cellulitis. Patients with osteomyelitis, necrotizing fasciitis, gas gangrene, infected diabetic foot ulcers, or chronic decubitus ulcers were excluded. In addition, patients with neutropenia or hepatic disease and those with a calculated creatinine clearance <30 ml/min were not enrolled. In the phase 3 program, patients with known or suspected Pseudomonas aeruginosa infections were also excluded. Concomitant antibiotic therapy was not allowed per protocol; however, select topical antiseptics could be administered. Operative procedures and daily debridements required per standard of care were permissible.
In the phase 2 study, patients were randomly assigned (in a 1:1 ratio) to receive one of two dosage regimens of i.v. tigecycline for a period of 7 to 14 days: a 100-mg loading dose, followed by 50 mg every 12 h (100/50 mg), or a 50-mg loading dose, followed by 25 mg every 12 h (50/25 mg). The dosing regimen in both phase 3 trials was tigecycline 100/50 mg plus placebo versus i.v. vancomycin (1 g) plus i.v. aztreonam (2 g) both given every 12 h and administered for up to 14 days. For all patients, PK samples were collected predose and at 1 h (end of infusion), 3 h, and 6 h after the start of infusion.
Patient- and disease-related descriptors were recorded during the screening visit. It was assumed the values of demographic characteristics recorded at baseline remained constant for the duration of the trial. Baseline microorganisms were collected and sent to a central laboratory (Covance Laboratories) for identification and susceptibility testing. Clinical and Laboratory Standards Institute guidelines were followed, using broth dilution in fresh media.
Efficacy was assessed using both clinical and microbiological criteria at the test-of-cure (TOC) visit, which was at least 2 weeks after the last dose of study medication. Clinically evaluable patients received at least 5 days of tigecycline, unless the patient was declared a failure after at least four doses. Clinical efficacy was graded by the principal investigator as the global response to therapy as follows: 1, clinical cure, i.e., the improvement or resolution of all signs and symptoms of the infection noted at enrollment; 2, clinical failure, i.e., the persistence of presenting signs and symptoms requiring additional or alternate antibacterial treatment, initial recovery from the infection followed by deterioration, death due to infection, or new unfavorable findings related to efficacy measures subsequent to study entry; or 3, indeterminate, i.e., extenuating circumstances that precluded classification as either cure or failure. Only patients with a clinical response of cure or failure were included in the clinically evaluable patient population. Per protocol, patients with baseline P. aeruginosa infections (phase 3 only) were excluded from the clinically evaluable population.
Microbiological efficacy was evaluated at both the patient and pathogen levels. At the patient level, one of the following microbiological responses was assigned: eradication (documented or presumed), persistence (documented or presumed), superinfection, or indeterminate (death, lost to follow-up, or no baseline pathogen). A response of eradication, persistence, or indeterminate was also assigned for each isolated baseline pathogen. Patients who were clinically evaluable and had a baseline culture from the infected site with at least one identified causative pathogen susceptible to study drug were classified as microbiologically evaluable. Patients with an indeterminate response were excluded from the microbiologically evaluable population.
Population PK model.
PK samples were collected, and the tigecycline dose, date, and time of each infusion were recorded. Serum was collected and frozen at −70°C until analyzed for tigecycline concentrations by using a validated liquid chromatography-tandem mass spectrometry assay with a lower limit of quantitation of 10 ng/ml (Wyeth Research, data on file).
Individual tigecycline exposure measures for the patients in the cSSSI trials were generated by using a previously developed population PK model (22). This model was developed with data from 174 subjects and 195 patients (a total of 3,056 serum concentrations) in all three phases of development. The data was best fit by a two-compartment model with zero-order input and first-order elimination. Tigecycline clearance was related to body weight, creatinine clearance as calculated by using the method proposed by Jelliffe (10), and gender. Using Bayesian estimation, the 24-h steady-state AUC (AUC24), the critical exposure measure of interest, was predicted for each patient in this analysis.
Exposure-response analysis of efficacy.
Patients with available tigecycline exposure measurements and those classified as both clinically and microbiologically evaluable at the TOC visit were included in this analysis. For the microbiological analysis, patients found to have a superinfection (clinical infection due to a new pathogen not present at baseline) were reviewed. If all baseline pathogens were eradicated, the patient was categorized as a patient-level microbiological success. Conversely, if at least one of the baseline pathogens persisted at the TOC visit, the patient was categorized as a patient-level microbiological failure. For patients with more than one baseline pathogen, the organism with the highest MIC was used in the analysis of microbiological and clinical efficacy. Bayesian parameter estimates from the final tigecycline PK model were used to generate individual AUC24/MIC ratios (22).
In order to create a homogeneous population, each patient was clinically reviewed and classified into one of five predefined cohorts based on identified baseline organisms (Table 1) . Cohorts were established prior to conducting the statistical analyses and focused on gram-positive organisms as the primary causes of cSSSI. For the purposes of this analysis, coagulase-negative staphylococci, viridans streptococci, Candida spp., and Corynebacterium spp. were not considered pathogenic microorganisms. Cohort 1 included patients with monomicrobial infections due to S. aureus, a significant pathogen in cSSSI. Cohort 2 included patients in cohort 1 plus patients with monomicrobial infections due to a pathogenic streptococcal species. Patients with two gram-positive pathogens isolated at baseline (S. aureus plus a pathogenic streptococcal species or two streptococcal species) were grouped into cohort 3. Patient classification included the following additional cohorts: patients with a mixed gram-positive and gram-negative polymicrobial infection (cohort 4) and patients with a monomicrobial gram-negative or anaerobic infection (cohort 5). The sample size and the distribution of outcomes within each cohort was prospectively evaluated to determine whether exploratory or statistical analyses could be performed or if cohorts needed to be combined to create a more robust patient population. Within these cohorts, the presence of a baseline anaerobe or P. aeruginosa was analyzed as a covariate. The following patient- and disease-related descriptors were also evaluated as potential predictors of microbiological and clinical efficacy: age, weight, country or region of treatment, monomicrobial or polymicrobial infection status, and preexisting diabetes or peripheral vascular disease (PVD).
TABLE 1.
Baseline pathogen classification system
| Cohort | Baseline pathogen status | MIC range (μg/ml) | No. of patients |
|---|---|---|---|
| 1 | Patients with monomicrobial S. aureus infections | 0.12-0.5 | 19 |
| 2 | Patients with monomicrobial S. aureus or Streptococcus spp. infections (includes cohort 1) | 0.06-0.5 | 28 |
| 3 | Patients with polymicrobial infections with S. aureus plus Streptococcus spp. or patients with two Streptococcus spp. | 0.06-0.5 | 7 |
| 4 | Patients with polymicrobial gram-negative infections with or without gram-positive infections | 0.06-16 | 14 |
| 5 | Patients with monomicrobial gram-negative or anaerobic infections | 0.25-1 | 8 |
Statistical analysis.
All data processing, data clean-up, database creation, and statistical analyses were performed by using SAS software, version 8.2 (18). Classification and regression tree (CART) analysis was performed by using S-Plus, version 6.2 (12). Exploratory analyses of microbiological and clinical response were conducted to identify relationships between outcome, exposure measurements, patient demographic characteristics, and comorbidities. CART analyses were performed to determine breakpoints in exposure measures stratified by response for each cohort. The number of cures and failures within each CART-identified category were computed. Categories with less than five cures or failures were combined with adjacent categories to allow the groups to be tested as a predictor of the probability of response by using logistic regression. Categories that could not be combined were evaluated by using the χ2 test.
Logistic regression analyses were used to determine whether exposure measures and patient covariates were statistically significant predictors of clinical and microbiological response. In the case of multiple observations per patient, generalized estimating equations were used. Univariate analyses were followed by multivariable modeling, utilizing a backward elimination procedure with a level of significance defined by sample size, to identify predictor variables with a statistically significant influence on outcomes. The level of significance for the analyses of cohorts 2 and 3 was 0.07 due to small sample size; for all other analyses it was 0.05. The goodness-of-fit of the logistic regression model was assessed by using the Hosmer-Lemeshow test. Predictive ability of the model was assessed using the area under the receiver operating characteristic (ROC) curve.
RESULTS
Population pharmacokinetic parameters.
Exposure estimates were generated by using a population PK model for tigecycline, the details of which have been recently reported (22). Differences between the PK parameters for patients enrolled in the phase 2 and phase 3 trials were not observed. Further examination of the patient descriptor variables revealed the ranges of age, weight, and creatinine clearance for the phase 3 patients were within the ranges studied in phase 2. Although the phase 3 patient population was predominantly Caucasian (88%) and the phase 2 population was almost equally split between Caucasians and Hispanics, ethnic origin was not determined to be a significant predictor of clearance in the final PK model.
Individual predicted AUC24 values were calculated for each evaluable cSSSI patient. For all patients in the efficacy analyses who were administered the 50/25-mg dosage regimen (n = 26), the mean (standard deviation [SD]) AUC24 was 2.66 (SD = 0.997) μg·h/ml and ranged from 1.48 to 4.98 μg·h/ml. For patients administered the 100/50-mg regimen (n = 34), the mean AUC24 was 5.39 (SD = 1.59) μg·h/ml and ranged from 2.81 to 9.36 μg·h/ml. Similar ranges of AUC24 values were observed across each cohort.
Exposure-response analysis of efficacy. (i) Data.
The analysis data set included 60 evaluable patients with 90 pathogens: 42 (70%) patients from phase 2 and 18 (30%) patients from the phase 3 trials. One patient, a 50-year-old Caucasian female with a past medical history of hernia repair with insertion of nonabsorbable prolene mesh and recurrent S. aureus abscesses at the surgical site, was considered a medical outlier. Foreign body infections were specifically excluded per protocol. The analyses of cohorts 2 and 3 were performed both with and without (data not shown) the medical outlier.
Patient demographic characteristics are presented in Table 2. The mean age and weight were 46 (SD = 15) years and 81 (SD = 22) kg, respectively, and 27% of the patients were female. Approximately 62% of patients were Caucasian, and 22% were Hispanic. A total of 73% of the patients were enrolled in trials conducted in the United States; the remaining patients were enrolled in European trials. PVD was reported in 10% of the patients, and 27% had a past medical history of diabetes. Monomicrobial baseline infections occurred in 55% of patients, 32% had two baseline pathogens, and 13% had three or more baseline organisms. Only five (8%) patients had an anaerobe at baseline, and four (7%) patients had P. aeruginosa isolated at baseline, of which all were eradicated at the end of treatment. The mean AUC24/MIC ratio was 13.3 (SD = 13.5) and ranged from 0.09 to 54.1 for patients given tigecycline (50/25 mg), and the mean AUC24/MIC ratio was 32.5 (SD = 24.1) and ranged from 0.21 to 102 for patients administered the 100/50-mg tigecycline regimen.
TABLE 2.
Summary statistics of patient demographic characteristics
| Demographic characteristic | Summary statistics (n = 60)a |
|---|---|
| Age (yr) | |
| Mean (SD) | 45.7 (15.4) |
| Median | 46 |
| Range | 21-78 |
| Wt (kg) | |
| Mean (SD) | 81.2 (21.8) |
| Median | 78.8 |
| Range | 54.1-164 |
| Gender (n [%]) | |
| Male | 44 (73) |
| Female | 16 (27) |
| Ethnicity (n [%]) | |
| Caucasian | 37 (61.6) |
| Black | 7 (11.6) |
| Hispanic | 13 (21.7) |
| Other | 3 (5.00) |
| Region of treatment (n [%]) | |
| Europe | 16 (26.7) |
| United States | 44 (73.3) |
n, number of subjects.
(ii) Microbiological response.
Table 3 provides a summary of baseline pathogens included in the analyses. Cohort 1 included a total of 19 patients classified as having S. aureus as a single baseline pathogen. Approximately 53, 42, and 5% of the patients had baseline S. aureus MICs of 0.12, 0.25, and 0.5 μg/ml, respectively. Fifteen (79%) patients in this group had a successful microbiological response. Cohort 2 was comprised of all patients in cohort 1 plus nine additional patients with monomicrobial streptococcal infections. MICs in this cohort ranged from 0.06 to 0.5 μg/ml. Of these 28 patients, eradication of the baseline pathogens was achieved in 24 (86%) patients. Seven patients, each with two baseline gram-positive pathogens, were included in cohort 3. Of the 14 pathogens (2 without reported MICs) represented in this group, 12 (86%) were eradicated. Combining cohorts 2 and 3 created a population of 35 patients with 40 baseline gram-positive pathogens with MICs, of which only 5 (12%) pathogens persisted at the TOC visit. This combined group had an adequate distribution of cures and failures, as well as sufficient sample size for analysis. Since the majority of patients in this group had monomicrobial infections, standard logistic regression analysis (one observation per patient defined by a patient-level response) was used to examine microbiological response. For patients with more than one baseline pathogen, the highest MIC was used in the analysis.
TABLE 3.
Baseline pathogens
| Organism | No. of observations | MIC (μg/ml)a |
|---|---|---|
| Gram positive | ||
| E. faecalis | 5 | 0.12-0.25 |
| E. faecium | 1 | 0.06 |
| S. aureus | 35 | 0.06-0.5 |
| S. agalactiae | 5 | 0.06 |
| S. anginosus | 1 | 0.06 |
| S. constellatus | 2 | 0.12 |
| S. dysgalactiae | 1 | 0.12 |
| S. equisimilis | 1 | 0.06 |
| S. intermedius | 4 | 0.06-0.12 |
| S. pyogenes | 6 | 0.06-0.12 |
| Gram negative | ||
| A. calcoaceticus-A. baumannii | 1 | 0.25 |
| A. haemolyticus | 1 | 0.5 |
| C. koseri | 1 | 0.5 |
| C. freundii | 1 | 0.5 |
| E. cloacae | 1 | 1 |
| E. coli | 8 | 0.25-0.5 |
| K. oxytoca | 2 | 0.5 |
| K. pneumoniae | 2 | 0.5-1 |
| P. mirabilis | 4 | 2-16 |
| P. aeruginosa | 4 | 8-16 |
| S. marcescens | 3 | 1-2 |
Ranges are given where applicable.
Due to the sensitivity of the CART procedure, two terminal breakpoints in the AUC24/MIC for each cohort were identified at 11.4 and 17.9. The breakpoints were then evaluated for the numbers of microbiological outcomes above and below each point. The number of microbiological cures and failures within each category of AUC24/MIC defined by the breakpoints was computed. Since this analysis included a total of ten failures, categories with fewer than five failures in each group could not be collapsed. The breakpoint at 11.4 occurred more frequently and had a more favorable distribution of cures and failures: two (40%) microbiological failures occurred below the CART-identified breakpoint, and three (10%) occurred above the breakpoint. No statistically significant difference in the proportion of failures was detected below the breakpoint at 11.4. Five (50%) microbiological failures occurred below the CART-identified breakpoint of 17.9, and none occurred above this breakpoint. A significantly higher proportion of failures occurred below the breakpoint of 17.9 (P = 0.0001). These breakpoints were subsequently utilized in the logistic regression analyses. However, since there were no patients with microbiological failures above the breakpoint at 17.9, this breakpoint could only be evaluated for clinical response in the logistic regression analyses.
Neither the indicator variables for a baseline anaerobic pathogen or P. aeruginosa nor the indicator variable for monomicrobial infection status could be evaluated in the logistic regression analysis due to the small sample size. Preexisting diabetes and PVD also could not be evaluated as categorical covariates because of the distribution of outcomes. Due to the limited sample size, multivariable logistic regression models were not evaluated.
Univariate logistic regression models assessing the impact of tigecycline exposure and patient covariates on the probability of microbiological response were assessed. For cohorts 2 and 3 combined, the AUC24/MIC ratio as a continuous covariate was found to be predictive of microbiological response based on sample size (P = 0.0563). The CART-identified breakpoint at 11.4 was not a significant predictor of a successful microbiological response (P = 0.1024). However, AUC24 as a continuous variable was predictive of outcome (P = 0.0595).
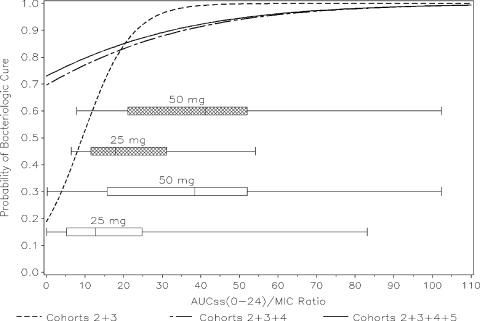
The AUC24/MIC ratio as a continuous covariate was considered the most informative model for these data (Fig. 1). As the AUC24/MIC ratio increased, the model-predicted probability of microbiological success increased. A patient was 17.1% more likely to have a successful microbiological response for each one-unit increase in the AUC24/MIC ratio. The Hosmer-Lemeshow goodness-of-fit statistic was 5.25 with 6 df (P = 0.5121), and the area under the ROC curve was 0.89, indicating a highly predictive model. At the median exposure values of AUC24/MIC ratios of 13.5 and 29 for the 50/25- and 100/50-mg dose groups, the model-predicted probabilities of microbiological success were 0.660 and 0.957, respectively. The final model for cohorts 2 and 3 combined is depicted in Fig. 1 (dashed line).
FIG. 1.
Final logistic regression model of microbiological response versus AUC24/MIC ratio. The boxplots indicate the AUC24/MIC ratio range for the 25- and 50-mg dose groups. The shaded boxes represent the distribution of cohorts 2 and 3, and the open boxes represent the distribution from cohorts 2, 3, 4, and 5. The solid line represents cohorts 2, 3, 4, and 5 combined; the dashed-dotted line represents cohorts 2, 3, and 4 combined; the dashed line represents cohorts 2 and 3 combined.
Additional analyses including all pathogens (cohorts 2, 3, 4, and 5) were performed to justify the use of the cohort classification methodology. The results are summarized in Table 4. To evaluate the impact of adding polymicrobial infections into the analyses, cohort 4 was combined with cohorts 2 and 3 (Fig. 1, dashed-dotted line). This increased the sample size to 49 patients with 79 pathogens. The range of MICs was dramatically increased to include values as high as 16 μg/ml, and the majority of MICs added by this group were at the higher end of the range. Since 58% of the patients in this group had polymicrobial infections, longitudinal logistic regression analyses defined by multiple pathogens using generalized estimating equations were required.
TABLE 4.
Microbiological response rates stratified by combined cohorts
| Cohorts | No. of patients/no. of pathogens | Pathogen-level microbiological response (no. of patients [%])
|
Odds ratio for AUC24/MIC | Area under the ROC curve | |
|---|---|---|---|---|---|
| Eradication | Persistence | ||||
| 2 + 3 | 35/40 | 35 (88) | 5 (12) | 1.171 | 0.89 |
| 2 + 3 + 4 | 49/79 | 66 (84) | 13 (16) | 1.038 | 0.65 |
| 2 + 3 + 4 + 5 | 57/87 | 74 (85) | 13 (15) | 1.036 | 0.64 |
The CART-identified breakpoint at 11.4 was still not a statistically significant predictor with the addition of cohort 4 (P = 0.1245). The AUC24/MIC ratio (as a continuous covariate) was marginally significant (P = 0.0520). The Hosmer-Lemeshow goodness-of-fit statistic was 7.43 with 8 df (P = 0.4907), and the area under the ROC curve was reduced to 0.65, indicating a less adequate model fit with some lack of predictive ability. At the median AUC24/MIC exposure values of 13.5 and 29 for the 50/25- and 100/50-mg dose groups, the model-predicted probabilities of microbiological success were 0.797 and 0.874, respectively. The addition of cohort 4 increased sample size and dramatically increased the range of MICs, with MICs up to 16 μg/ml. Since AUC24 values were similar between cohorts 2, 3, and 4, this resulted in a large proportion of observations with cures at the lower AUC24/MIC ratios.
The addition of cohort 5 in an all-pathogen analysis was the least informative model (Fig. 1, solid line). This added eight patients with monomicrobial infections due to gram-negative or anaerobic pathogens to the analysis, with MICs ranging from 0.25 to 1 μg/ml, and all of the patients experienced a microbiological cure. Neither the CART-identified breakpoint at 11.4 nor the AUC24/MIC ratio as a continuous covariate were significant predictors of microbiological outcome (P = 0.1176 and 0.0662, respectively). Combining all pathogens together, i.e., gram-positive, gram-negative, and anaerobic organisms, obscured the ability to detect a continuous exposure-response relationship.
(iii) Clinical response.
Of the 19 patients in cohort 1, 17 (89%) patients had a successful clinical response. We found that 24 (86%) of the 28 patients in cohort 2 and 6 (86%) of the 7 patients in cohort 3 were clinically cured. Only 71% of the 14 patients with polymicrobial infections in cohort 4 had a successful clinical response, whereas all 8 patients in cohort 5 were clinically cured.
Univariate logistic regression models assessing the impact of tigecycline exposure and patient covariates on the probability of clinical response were evaluated. Of the 35 patients in combined cohorts 2 and 3, 30 (86%) had a successful clinical response. The AUC24/MIC ratio as a continuous covariate was not statistically significant (P = 0.1960), and AUC24 was marginally significant as a predictor of clinical response (P = 0.0708). The CART-identified AUC24/MIC breakpoint at 17.9, however, was a significant predictor of clinical response (P = 0.0376), whereas the breakpoint at 11.4 was not significant (P = 0.1662).
Although the AUC24/MIC ratio as a continuous covariate did not reach statistical significance, it was an informative model for these data. As the AUC24/MIC ratio increased, the model-predicted probability of clinical success increased, and a patient was 3.7% more likely to have a successful clinical response for every one-unit increase in AUC24/MIC ratio value. The Hosmer-Lemeshow goodness-of-fit statistic was 6.40 with 7 df (P = 0.4936), and the area under the ROC curve was 0.68, indicating an adequate model fit with some lack of predictive ability. At the median AUC24/MIC exposure values of 13.5 and 29 for the 50/25- and 100/50-mg dose groups, the model-predicted probabilities of clinical success were 0.66 and 0.96, respectively.
AUC24 as a continuous covariate was also an informative model for these data. The probability of clinical success increased as AUC24 values increased and a patient was twice as likely to achieve a clinical cure. The Hosmer-Lemeshow goodness-of-fit statistic was 10.8 with 7 df (P = 0.1473) and the area under the ROC curve was 0.76, indicating an adequate and predictive model. At the median exposure values for AUC24 of 2.3 and 5.2 μg·h/ml for the 50/25- and 100/50-mg dose groups, the model-predicted probabilities of clinical success were 0.65 and 0.94, respectively. As with the microbiological analyses, none of the evaluated patient characteristics were predictors of clinical success.
DISCUSSION
The detection of an exposure-response relationship in heterogeneous patient and/or pathogen populations has proven to be challenging, especially when small datasets or infectious diseases that are polymicrobial in nature are being analyzed. Previous analyses have been unsuccessful in identifying relationships when all pathogens were considered together. A novel prospective method, with consideration for the predominant skin and skin-structure pathogens, was developed to create more homogeneous patient populations for the microbiological and clinical exposure-response analysis of tigecycline in the treatment of cSSSI.
cSSSI encompass infections that involve deeper soft tissue, require surgical intervention, or occur in patients with significant underlying diseases, including diabetes and PVD. These infections include clinical entities such as infected ulcers, major abscesses, and deep or extensive cellulitis. Although a mixture of gram-positive and -negative aerobic or anaerobic bacteria can cause these infections, gram-positive organisms, specifically Staphylococcus aureus and streptococci, are the predominant pathogens (4, 14). Infections caused by nonfermentative gram-negative bacilli are generally associated with nosocomial infections and are infrequently associated with community-acquired infection (9, 19).
In an effort to create a more homogeneous patient population for the microbiological exposure-response analyses, each patient eligible for analysis was evaluated and patients were grouped into cohorts depending upon baseline pathogen(s), as previously described. Patients grouped into cohorts 1, 2, and 3 were considered to be the most homogeneous since they had monomicrobial S. aureus, monomicrobial streptococcal infections, or two baseline gram-positive pathogens. In order to achieve sufficient sample size, analyses were performed on cohorts 2 (which included cohort 1) and 3 combined. To further validate the cohort approach, cohort 4, the true polymicrobial infections with gram-negative pathogens, and cohort 5 were added for an “all-pathogen” analysis.
When we compared all microbiological response models (Fig. 1), the most discernible exposure-response relationship was detected when we evaluated the combined cohorts 2 and 3. The models evaluating the AUC24/MIC ratio as a continuous covariate provided the most information about the nature of the exposure-response relationship. CART analyses identified two breakpoints in AUC24/MIC ratio distribution.
The prospective approach taken to evaluate this cohort methodology proved to be a useful tool in detecting an exposure-response relationship. When we compared the AUC24/MIC ratio models as continuous covariates across analyses, the addition of cohort 4 with polymicrobial infections with gram-negative pathogens, and thus higher MICs, added variability and noise to the analyses. The model resulted in a much shallower exposure-response curve. The addition of the cohort 5 patients (i.e., the “all-pathogen” analysis) further decreased the ability to detect an exposure-response relationship in these data. Although combining cohorts increased the sample size and potentially the ability to detect relationships, the magnitude of the effect decreased due to increased heterogeneity in the patient population.
When patients with S. aureus and/or streptococcal infections (cohorts 2 and 3 combined) were included in the analysis, the AUC24/MIC ratio as a categorical variable at the identified breakpoint at 17.9 (P = 0.0001) was highly predictive of microbiological success. An exposure-response relationship was noted when the AUC24/MIC ratio was treated as a continuous variable (P = 0.0563). The final logistic regression model had a high predictive ability, as evidenced by an area under the ROC curve value of 0.89.
Patient demographic factors, as well as several patient covariates, were also examined in the microbiological efficacy analyses. Age, gender, and documented evidence of PVD and/or diabetes as comorbidities were not statistically significant predictors of microbiological response. Three other patient covariates were considered in this analysis: the presence of an anaerobe at baseline, the presence of P. aeruginosa at baseline, and the monomicrobial or polymicrobial infection status. Anaerobes can play an important role in skin infections, especially in diabetic foot infections (9) and bite wounds (20). The role of anaerobic bacteria in the pathogenesis of polymicrobial cellulitis in some patient populations, however, has been questioned (16). Since P. aeruginosa is intrinsically resistant to tigecycline, the presence of this organism at baseline was considered to determine whether there was a direct effect on outcome. None of these covariates, however, could be evaluated due to the small sample size or distribution of responses.
Exposure-response analyses for clinical outcome were also performed with results very similar to those of the microbiologic analysis. The CART-identified AUC24/MIC breakpoint of 17.9 was a statistically significant predictor of clinical outcome (P = 0.0376), whereas the breakpoint at 11.4 was not significant (P = 0.1662). It is important to note that the CART-identified AUC24/MIC breakpoints were remarkably consistent across both clinical and microbiological outcomes and across all cohorts. Moreover, the breakpoints identified in these analyses are consistent with those identified in animal infection models (breakpoint range of 15 to 20) (William A. Craig, unpublished data). It should be noted however, that the breakpoints identified in the current analysis reflect those obtained in patients with cSSSI and therefore should not be extrapolated to other disease conditions.
Understanding the relationship between the pharmacokinetics and pharmacodynamics of new antimicrobial agents has become increasingly important for supporting dose selection and in setting susceptibility breakpoints. When all pathogens were analyzed together, the ability to detect exposure-response relationships was decreased. The use of an approach to categorize cSSSI patients treated with tigecycline into cohorts based on the pathogens encountered most often in this infectious disease, S. aureus and/or streptococci, resulted in the identification of an exposure-response relationship for microbiological and clinical outcome. The strongest exposure-response relationship, as measured by the AUC24/MIC ratio, was associated with the most homogeneous patient and pathogen population.
Acknowledgments
This study was supported, in part, by a grant from Wyeth Research.
Footnotes
Published ahead of print on 12 March 2007.
REFERENCES
- 1.Ambrose, P. G., D. M. Grasela, T. H. Grasela, J. Passarell, H. B. Mayer, and P. F. Pierce. 2001. Pharmacodynamics of fluoroquinolones against Streptococcus pneumoniae in patients with community-acquired respiratory tract infections. Antimicrob. Agents Chemother. 45:2793-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babinchak, T., E. Ellis-Grosse, N. Dartois, G. M. Rose, and E. Loh. 2005. The efficacy and safety of tigecycline for the treatment of complicated intra-abdominal infections: analysis of pooled clinical trial data. Clin. Infect. Dis. 41(Suppl.):S354-S367. [DOI] [PubMed] [Google Scholar]
- 2a.Christianson, J., D. Andes, and W. Craig. 2001. Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. A-1103.
- 3.Craig, W. A. 1998. Pharmacokinetic/pharmacodynamic parameters: rationale for antibacterial dosing of mice and men. Clin. Infect. Dis. 26:1-10. [DOI] [PubMed] [Google Scholar]
- 4.Doern, G. V., R. N. Jones, M. A. Pfaller, K. C. Kugler, and M. L. Beach. 1999. Bacterial pathogens isolated from patients with skin and soft tissue infections: frequency of occurrence and antimicrobial susceptibility patterns from the SENTRY Antimicrobial Surveillance Program (United States and Canada, 1997). Diagn. Microbiol. Infect. Dis. 34:65-72. [DOI] [PubMed] [Google Scholar]
- 5.Drusano, G. L., S. L. Preston, C. Hardalo, R. Hare, C. Banfield, D. Andes, O. Vesga, and W. A. Craig. 2001. Use of preclinical data for selection of a phase II/III dose for evernimicin and identification of a preclinical MIC breakpoint. Antimicrob. Agents Chemother. 45:13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellis-Grosse, E. J., T. Babinchak, N. Dartois, G. Rose, and E. Loh. 2005. The efficacy and safety of tigecycline in the treatment of skin and skin-structure infections: results of 2 double-blind phase 3 comparison studies with vancomycin-aztreonam. Clin. Infect. Dis. 41(Suppl. 5):S341-S353. [DOI] [PubMed] [Google Scholar]
- 7.Exposure-Response Working Group. 2003. FDA guidance for industry exposure-response relationships: study design, data analysis, and regulatory applications. [Online.] http://www.fda.gov/cber/gdlns/exposure.htm. U.S. Department of Health and Human Services, Rockville, MD.
- 8.Forrest, A., D. E. Nix, C. H. Ballow, T. F. Goss, M. C. Birmingham, and J. J. Schentag. 1993. Pharmacodynamics of intravenous ciprofloxacin in seriously ill patients. Antimicrob. Agents Chemother. 37:1073-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerding, D. N. 1995. Foot infections in diabetic patients: the role of anaerobes. Clin. Infect. Dis. 20(Suppl. 2):S283-S288. [DOI] [PubMed] [Google Scholar]
- 10.Jelliffe, R. W. 1973. Creatinine clearance: bedside estimate. Ann. Intern. Med. 79:604-605. (Letter.) [DOI] [PubMed] [Google Scholar]
- 11.MacGowan, A., C. Rogers, and K. Bowker. 2001. In vitro models, in vivo models, and pharmacokinetics: what can we learn from in vitro models? Clin. Infect. Dis. 33(Suppl. 3):S214-S220. [DOI] [PubMed] [Google Scholar]
- 12.Mathsoft, Inc. 2000. S-Plus 6.0 for UNIX guide to statistics, vol. 1 and 2. MathSoft, Inc., Seattle, WA.
- 13.Muralidharan, G., M. Micalizzi, J. Speth, D. Raible, and S. Troy. 2005. Pharmacokinetics of tigecycline after single and multiple doses in healthy subjects. Antimicrob. Agents Chemother. 49:220-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nichols, R. L. 1999. Optimal treatment of complicated skin and skin structure infections. J. Antimicrob. Chemother. 44(Suppl. A):19-23. [DOI] [PubMed] [Google Scholar]
- 15.Oliva, M. E., A. Rekha, A. Yellin, J. Pasternak, M. Campos, G. M. Rose, T. Babinchak, E. J. Ellis-Grosse, and E. Loh. 2005. A multicenter trial of the efficacy and safety of tigecycline versus imipenem/cilastatin in patients with complicated intra-abdominal infections. BMC Infect. Dis. 5:88-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orangio, G. R., S. D. Pitlick, P. Della Latta, L. J. Mandel, C. Marino, J. J. Guarneri, J. A. Giron, and I. B. Margolis. 1984. Soft tissue infections in parenteral drug abusers. Ann. Surg. 199:97-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petersen, P. J., N. V. Jacobus, W. J. Weiss, P. E. Sum, and R. T. Testa. 1999. In vitro and in vivo antibacterial activities of a novel glycylcycline, the 9-t-butylglycylamido derivative of minocycline (GAR-936). Antimicrob. Agents Chemother. 43:738-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.SAS Institute, Inc. 1999. SAS/STAT user's guide, version 8.2. SAS Institute, Inc., Cary, NC.
- 19.Summanen, P. H., D. A. Talan, C. Strong, M. McTeague, R. Bennion, J. E. Thompson, Jr., M. L. Vaisanen, G. Moran, M. Winer, and S. M. Finegold. 1995. Bacteriology of skin and soft-tissue infections: comparison of infections in intravenous drug users and individuals with no history of intravenous drug use. Clin. Infect. Dis. 20(Suppl. 2):S279-S282. [DOI] [PubMed] [Google Scholar]
- 20.Talan, D. A., D. M. Citron, F. M. Abrahamian, G. J. Moran, and E. J. Goldstein. 1999. Bacteriologic analysis of infected dog and cat bites. N. Engl. J. Med. 340:85-92. [DOI] [PubMed] [Google Scholar]
- 21.van Ogtrop, M. L., D. Andes, T. J. Stamstad, B. Conklin, W. J. Weiss, W. A. Craig, and O. Vesga. 2000. In vivo pharmacodynamic activities of two glycylcyclines (GAR-936 and WAY 152,288) against various gram-positive and gram-negative bacteria. Antimicrob. Agents Chemother. 44:943-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Wart, S. A., J. S. Owen, E. A. Ludwig, A. K. Meagher, J. M. Korth-Bradley, and B. B. Cirincione. 2006. Population pharmacokinetics of tigecycline in patients with complicated intra-abdominal or skin and skin-structure infections. Antimicrob. Agents Chemother. 50:3701-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilcox, M. H. 2005. Efficacy of tigecycline in complicated skin and skin structure infections and complicated intra-abdominal infections. J. Chemother. 17(Suppl. 1):23-29. [DOI] [PubMed] [Google Scholar]