Abstract
Fourteen Aspergillus fumigatus clinical isolates that exhibited a pattern of reduced susceptibility to triazole drugs were analyzed. The sequences of the cyp51A gene from all isolates showed the presence of a point mutation at t364a, which led to the substitution of leucine 98 for histidine (L98H), together with the presence of two copies of a 34-bp sequence in tandem in the promoter of the cyp51A gene. Quantitative expression analysis (real-time PCR) showed up to an eightfold increase in the level of expression of the cyp51A gene compared to that by the susceptible strain. Three PCR fragments of one azole-resistant strain (strain CM2627) that included the promoter with the tandem repeat and part of cyp51A with the t364a mutation or PCR fragments with only one of the modifications were used to replace the cyp51A gene of an azole drug-susceptible A. fumigatus wild-type strain (strain CM237). Only transformants which had incorporated the tandem repeat in the promoter of the cyp51A gene and the L98H amino acid substitution exhibited similarly reduced patterns of susceptibility to all triazole agents and similarly increased levels of cyp51A expression, confirming that the combination of both alterations was responsible for the azole-resistant phenotype.
Infections with Aspergillus fumigatus, one of the most prevalent airborne fungal pathogens causing invasive aspergillosis worldwide, result in high rates of mortality and morbidity in immunocompromised hosts (20, 21). A. fumigatus is intrinsically resistant to fluconazole, but other triazole drugs like itraconazole and voriconazole are active both in vitro and in vivo against this species. Also, the new triazoles, such as posaconazole and ravuconazole, have been shown to have good in vitro activities against this species (5, 7, 17, 37, 45). In many centers voriconazole has become a common choice for the primary therapy of invasive aspergillosis, as the drug has been shown to have superior efficacy compared with those of other licensed antifungal therapies, most notably, amphotericin B (19). Nevertheless, more than half of patients still fail to respond to antifungal therapy. Although this is mainly due to factors related to the host, the resistance of Aspergillus to triazole drugs might play a role (22, 47). Indeed, a number of A. fumigatus isolates with in vitro itraconazole resistance have been described (3, 8, 13, 15, 24, 31), and strains that show high MICs against the new triazole agents are being reported (29, 31, 37). We recently observed primary and breakthrough cases of invasive aspergillosis due to A. fumigatus isolates with a phenotype characterized by high MICs of itraconazole, voriconazole, and ravuconazole and elevated MICs of posaconazole (51). There was no apparent epidemiological link between the cases.
In filamentous fungi, azole drugs inhibit ergosterol biosynthesis by targeting the enzyme 14-α-sterol demethylase (Cyp51). So far, the prevalence of mechanisms for azole drug resistance in A. fumigatus seems to be very different from that in Candida spp. In A. fumigatus, there are two different but related Cyp51 proteins, and these are encoded by cyp51A and cyp51B (28). Targeted disruption of the cyp51A gene in different itraconazole-resistant A. fumigatus strains resulted in strains with similar patterns of decreased susceptibility to azole drugs, confirming that Cyp51A is the target of these antifungal agents (30). In itraconazole-resistant A. fumigatus strains, two molecular mechanisms of resistance to azole drugs have been described: first, azole drug resistance in A. fumigatus seems to be mostly related to point mutations in Cyp51A (2, 15, 25, 29, 32); and second, reduced intracellular accumulation, due to either increased expression of efflux pumps (13, 46) or reduced penetration of the drug (24), have also been proposed.
Regarding the modification of A. fumigatus Cyp51, specific mutations in cyp51A have been associated with two different susceptibility profiles: (i) cross-resistance to itraconazole and posaconazole has been associated with amino acid substitutions at glycine 54 (G54) (15, 25, 32); and (ii) a pattern of itraconazole resistance, characterized by different patterns of elevated MICs for azole drugs, has been linked to different amino acid substitutions at methionine 220 (M220) (2, 29).
Here we describe the analysis of a new molecular mechanism responsible for a phenotype of A. fumigatus cross-resistance to azole drugs. We have determined that a base change causing an amino acid substitution in Cyp51A (L98H) in combination with the duplication in tandem of a 34-bp sequence in the cyp51A promoter, which is responsible for the increased level of cyp51A gene expression, accounted for the resistant phenotype.
(Part of this study was presented at the 45th Interscience Conference on Antimicrobial Agents and Chemotherapy, Washington, DC, December 2005.)
MATERIALS AND METHODS
Fungal strains and media.
The fungal strains used in the study included (i) A. fumigatus strain CM237 (CM refers to the fungal collection of the Mycology Reference Laboratory at the Centro Nacional de Microbiologia, ISCIII) was used to describe the cyp51A and cyp51B gene sequences and was also the control strain (28); (ii) A. fumigatus strain akuBKU80 (10) was used as the recipient strain in some experiments of fungal transformation; and (iii) 14 clinical A. fumigatus strains were isolated from 10 patients (1 from Spain and 13 from the collection of the Department of Medical Microbiology, Radboud University Nijmegen Medical Center, Nijmegen, The Netherlands) (Table 1). The fungi were grown at 37°C in potato dextrose agar (Oxoid, Madrid, Spain), GYEP broth (2% glucose, 0.3% yeast extract, 1% peptone), and minimal medium (38). Conidial stocks were preserved in sterile distilled water (15).
TABLE 1.
Clinical data for patients infected or colonized with azole-resistant A. fumigatus, genotypes, and ranges of MICs of different antifungals for the isolatesa
| Sex (age) | Underlying disease | Azole exposure prior to aspergillosis diagnosis (time) | A. fumigatus isolate | Date of isolation (day-mo-yr) | Genotype | MIC (μg/ml)
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| AMB | ITC | VRC | RVC | PSC | ||||||
| F | Unknown | Unknown | M03/669 - CM2627 | 21-10-2003 | A | 0.25 | >8 | 4-8 | 4-8 | 0.5 |
| M (15) | X-linked CGD | ITC (6 yr) | V13/02 - CM3271 | 04-04-2002 | B | 0.25 | >8 | 2 | >8 | 1 |
| V13/03 - CM3272 | 05-04-2002 | B | 0.25 | >8 | 2 | >8 | 1 | |||
| M (73) | No | No | V27/28 - CM3273 | 03-12-2003 | C | 0.25 | >8 | 4 | 4-8 | 1 |
| F (77) | AML | VRC (77 days) | V28/78 - CM3275 | 02-03-2004 | D | 0.25 | >8 | 4 | 8 | 0.5-1 |
| M (16) | Hyper-IgE syndrome | VRC (2 yr) | V34/75 - CM3276 | 19-11-2004 | E | 0.25 | >8 | 4 | 4 | 0.5 |
| V34/76 - CM3277 | E | 0.25 | >8 | 2-4 | 4 | 0.5-1 | ||||
| V34/77 - CM3278 | E | 0.25 | >8 | 2 | 4 | 0.5 | ||||
| V34/78 - CM3279 | E | 0.25 | >8 | 2 | 4 | 0.5 | ||||
| F (76) | Pulmonary fibrosis | No | V41/26 - CM3820 | 26-06-2005 | F | 0.25 | >8 | 4-8 | 4-8 | 0.5-1 |
| M (31) | CGD | ITC (>10 yr) | V45/07 - CM3819 | 01-11-2005 | G | 0.25 | >8 | >8 | 8 | 1 |
| F (68) | AML | No | V48/27 - CM3936 | 14-02-2006 | H | 0.25 | >8 | 4 | 8 | 4 |
| F (62) | COPD | No | V49/09 - CM4023 | 05-04-2006 | I | 0.25 | >8 | 4 | 8 | 4 |
| M (45) | AML | ITC (>2 yr) | V49/77 - CM4050 | 11-05-2006 | J | 0.25 | >8 | 4 | 4-8 | 1 |
IgE, immunoglobulin E; CGD, chronic granulomatous disease; AML, acute myeloid leukemia; COPD, chronic obstructive pulmonary disease; AMB, amphotericin B; ITC, itraconazole; VRC, voriconazole; RVC, ravuconazole; PSC, posaconazole; F, female; M, male.
Antifungal susceptibility testing: MICs.
Broth microdilution susceptibility testing was performed as described in CLSI (formerly NCCLS) document M38-A (33), with modifications (6, 35, 40). Itraconazole (Janssen Pharmaceutical S.A., Madrid, Spain), voriconazole (Pfizer, S.A., Madrid, Spain), ravuconazole (Bristol-Myers Squibb, Madrid, Spain), posaconazole (Schering Plough, Madrid, Spain), and amphotericin B (Sigma, Madrid, Spain) were tested. Susceptibility tests were performed at least three times with each strain on different days. Aspergillus flavus ATCC 204304 and A. fumigatus ATCC 204305 were used as quality control strains for susceptibility testing.
PCR amplification and sequence analysis of cyp51A and cyp51B genes.
Conidia from each strain were inoculated in 3 ml GYEP broth and grown overnight at 37°C. Mycelial mats were recovered and subjected to a DNA extraction protocol (28). The full coding sequences of cyp51A and cyp51B were amplified by PCR and both strands were sequenced, as described previously (15). To rule out the possibility that any sequence changes identified were due to PCR-induced errors, each isolate was independently fully analyzed twice. The nucleotide sequences of the cyp51A and cyp51B genes in strains CM237 and akuBKU80 are identical.
Aspergillus fumigatus transformations.
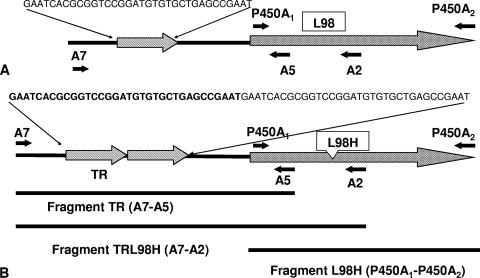
Three linear DNA fragments were used throughout this work for azole-susceptible A. fumigatus (CM237 and akuBKU80) strain transformations: (i) a fragment containing the coding sequence of the cyp51A gene with a single nucleotide change (t364a) that substitutes leucine for histidine (named the L98H fragment), (ii) a fragment that includes the cyp51A promoter sequence of the gene with the 34-bp tandem repeat (named the TR fragment), and (iii) a fragment containing the coding sequence of cyp51A gene with a t364a change that results in the L98H substitution in combination with the 34-bp TR in the cyp51A promoter (named the TRL98H fragment) (Fig. 1). Linear fragments were obtained by PCR amplification of cyp51A by using genomic DNAs from A. fumigatus strain CM2627 and the following primer sets: (i) primer set P450A1-P450A2, (ii) primer set A7-A5, and (iii) primer set A7-A2 (28). First, the conidia of azole-susceptible wild-type strain CM237 were used as the recipients for transformation by electroporation by separately using each of the PCR-amplified linear fragments of cyp51A.
FIG. 1.
Diagram of the A. fumigatus cyp51A gene and promoter showing the locations of the gene modifications and the primers used for PCR amplification of the different cyp51A gene fragments: (A) Wild-type strain CM237; (B) azole-resistant strain CM2627. The primer sets were as follows: (i) primer set A7-A2, which contains the TR and the t364a base change; (ii) primer set A7-A2, which contains only the 34-bp TR; and (iii) primer set P450A1-P450A2, which amplifies the full cyp51A gene, including only the t364a base change (L98H).
Electroporation was then carried out by a protocol previously described for Aspergillus nidulans (41) and subsequently adapted for A. fumigatus (52). Transformants were selected on medium containing itraconazole (8 μg/ml or 1 μg/ml), as described previously (15). Itraconazole-resistant transformants appeared after 2 to 7 days of incubation.
Using these conditions, we were unable to recover any transformant carrying the L98H mutation at cyp51A. A different azole-susceptible A. fumigatus strain (akuBKU80), which has an increased frequency of homologous recombination (10), was used as the recipient strain for electroporation by using the PCR-amplified linear fragments of cyp51A that included the TR or the L98H fragment independently. Transformants were selected on medium containing itraconazole (0.5 to 1.0 μg/ml), as described above. The transformants were labeled with “TR,” “L98H,” or “TRL98H,” followed by a number, depending on the DNA fragment used for the electroporation, and preceded by the identification number of the recipient strain (CM237 or akuBKU80).
RNA isolation and reverse transcription-PCR.
Total RNA was extracted from the A. fumigatus azole-susceptible strains (strains CM237 and akuBKU80), two of the clinical isolates (isolates CM2627 and CM3275), the transformants obtained from strain CM237 (transformants CM237-TRL98H1 and CM237-TR7), and the transformants obtained from strain akuBKU80 (transformants akuBKU80-TR2, akuBKU80-TR3, akuBKU80-L98H1, and akuBKU80-L98H3). The reverse transcription reactions were performed as described before (1). First, we evaluated the cyp51A transcription level differences between azole-susceptible A. fumigatus CM237 and the akuBKU80 strains used for the transformation experiments. Then, we evaluated the cyp51A transcription level differences between the transformants and their parental strains. For transcription level determination, a real-time PCR assay was performed as follows: the levels of expression of the target gene (cyp51A) were quantified for each strain by comparison with their respective control or parental strain and by use of the A. fumigatus β-tubulin housekeeping gene (tub1) as a reference for gene expression. The primer set A6-CypA2 (28) was used to amplify the cDNA from the cyp51A gene. The primer set Tub5 (5′-TGACCCAGCAGATGTT-3′) and Tub6 (5′-GTTGTTGGGAATCCACTC-3′) was used for the amplification of a fragment of the A. fumigatus β-tubulin gene (GenBank accession number AY048754). Real-time PCRs were set up with the FastStart DNA Master SYBR green (Roche Diagnostic). Each assay was repeated in duplicate and with RNA from at least two different RNA extractions. Each experiment included standard curves for the target gene (cyp51A) and for the reference gene. The PCR efficiencies of β-tubulin and cyp51A cDNA amplification were calculated from the slopes of the curves given by LightCycler software (Roche Diagnostic), and the efficiency values were used to validate each experiment and also for relative quantification by the method of Pfaffl (36).
Genotyping of Aspergillus isolates.
The A. fumigatus isolates were genotyped by using a panel of short tandem repeats (STRs). Two sets of three STRs (M3 and M4) consisting of tri- and tetranucleotide motifs were used in separate multicolor multiplex PCRs to analyze both A. fumigatus azole-resistant and azole-susceptible (control) isolates. For each of the multiplex PCRs with STRs M3 and M4, the forward primers were labeled with carboxyfluorescein, hexachlorofluorescein, or tetrachlorofluorescein at the 5′ end. The primer sequences were used exactly as described previously (14). Each PCR mixture contained 1 μM all amplification primers, 3.0 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, 1 U of FastStart Taq DNA polymerase, and 1 ng of genomic DNA in 1× reaction buffer. The thermoprofile was 10 min of denaturation at 95°C, followed by 30 cycles of 30 s of denaturation at 95°C, 30 s of annealing at 60°C, and 1 min of extension at 72°C. Before the reaction mixtures were cooled to room temperature, an additional incubation for 10 min at 72°C was performed. The fragments obtained were combined with the ET400-R size standard (GE Healthcare, Roosendaal, The Netherlands) and analyzed on a MegaBACE 500 automated DNA platform (GE Healthcare).
Data analysis.
The significance of the differences in MICs was determined by Student's t test after logarithmic conversion of the values (unpaired, unequal variance). For statistical evaluation of the crossing point and relative expression variations, the data were analyzed by analysis of variance for significant differences. Statistical analysis was done with the SPSS package (version 14.0; SPSS S.L., Madrid, Spain). A P value of <0.01 was considered significant.
RESULTS
Antifungal susceptibility testing: MICs.
All 14 strains were obtained from patients under different clinical conditions, and only half of the patients had a history of exposure to azole drugs (Table 1). To investigate whether these mutations originated from a clonal Aspergillus strain, the different isolates were genotyped by using a panel of microsatellites or STRs. STR analyses yield highly reproducible, exact typing results. Each multiplex reaction amplified three trinucleotide repeats. For each different STR, between 6 and 85 alleles were found. The isolates from all patients showed unique genetic profiles, indicating no genetic relatedness between the different A. fumigatus isolates (Table 1). The 14 strains exhibited a common pattern of reduced susceptibility to all four triazoles tested with various MICs, depending on the antifungal drugs, with posaconazole being the most active compound in vitro. The MICs for amphotericin B showed no variations between any strains (Table 1).
PCR amplification and sequence analysis of cyp51A and cyp51B genes.
Sequence analysis of cyp51A showed the same t364a point mutation in all strains, resulting in an amino acid substitution with histidine (L98H) at the leucine 98 codon. Two isolates (both from the same patient) had two extra base changes: these were t960a and t1554a, which were responsible for amino acid changes of serine for threonine at codon 297 (S297T) and phenylalanine for isoleucine at codon 495 (F495I), respectively (Table 2). All strains also contained three different point mutations in the cyp51B gene that were not conserved across the 14 isolates, although all of them were silent (Table 2). The sequence alignments of the region encompassing L98 revealed that this amino acid residue is not totally conserved in all yeasts and molds. However, the fact that all azole-resistant strains carried the same mutation strongly suggested that the substitution was associated with azole drug resistance. Moreover, all strains had a duplication in tandem of a 34-bp sequence in the cyp51A promoter (5′-GAATCACGCGGTCCGGATGTGTGCTGAGCCGAAT3-′) located at positions −288 and −322 from the cyp51A start codon. A BLAST database search based on the repeated sequence unit identified no striking similarities other than similarity to the cyp51A promoter region of A. fumigatus deposited in the GenBank database (accession numbers AF338659 and AF222068).
TABLE 2.
Nucleotide and amino acid substitutions in cyp51A and cyp51B genes from A. fumigatus clinical isolates
| Isolate | TR promoter | Nucleotide substitutiona
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
|
cyp51A
|
cyp51B
|
||||||||
| Codon 54 | Codon 98 | Codon 220 | Codon 297 | Codon 495 | Codon 35 | Codon 394 | Codon 464 | ||
| CM237 (wild type) | (−) | GGG | CTC | ATG | TCG | TTT | TCT | CCT | ATT |
| akuBKU80b | (−) | GGG | CTC | ATG | TCG | TTT | TCT | CCT | ATT |
| M03/669 - CM2627 | (+) | GGG | 364CAC | ATG | TCG | TTT | TCT | 1285CCG | ATT |
| V13/02 - CM3271 | (+) | GGG | 364CAC | ATG | 960ACG | 1554ATT | TCT | 1285CCG | 1495ATA |
| V13/03 - CM3272 | (+) | GGG | 364CAC | ATG | 960ACG | 1554ATT | TCT | 1285CCG | 1495ATA |
| V27/28 - CM3273 | (+) | GGG | 364CAC | ATG | TCG | TTT | TCT | 1285CCG | ATT |
| V28/78 - CM3275 | (+) | GGG | 364CAC | ATG | TCG | TTT | 105TCC | 1285CCG | ATT |
| V34/75 - CM3276 | (+) | GGG | 364CAC | ATG | TCG | TTT | TCT | 1285CCG | ATT |
| V34/76 - CM3277 | (+) | GGG | 364CAC | ATG | TCG | TTT | TCT | 1285CCG | ATT |
| V34/77 - CM3278 | (+) | GGG | 364CAC | ATG | TCG | TTT | TCT | 1285CCG | ATT |
| V34/78 - CM3279 | (+) | GGG | 364CAC | ATG | TCG | TTT | TCT | 1285CCG | ATT |
| V41/26 - CM3820 | (+) | GGG | 364CAC | ATG | TCG | TTT | 105TCC | 1285CCG | ATT |
| V45/07 - CM3819 | (+) | GGG | 364CAC | ATG | TCG | TTT | TCT | 1285CCG | ATT |
| V48/27 - CM3936 | (+) | GGG | 364CAC | ATG | TCG | TTT | 105TCC | 1285CCG | ATT |
| V49/09 - CM4023 | (+) | GGG | 364CAC | ATG | TCG | TTT | TCT | 1285CCG | ATT |
| V49/77 - CM4050 | (+) | GGG | 364CAC | ATG | TCG | TTT | TCT | 1285CCG | ATT |
| Amino acids | (G54) | (L98H) | (M220) | (S297T) | (F495I) | (S35S) | (P394P) | (I464I) | |
Nucleotides are numbered from the translation start codon ATG of cyp51A and cyp51B. The numbers indicate the position at which a base change occurs (in boldface).
The akuBKU80 mutant strain (10) is a genetically modified A. fumigatus strain with a high frequency of homologous recombination.
The region encompassing the mutation and the repeat in the promoter were sequenced in 25 A. fumigatus azole-susceptible clinical strains, as well as in several A. fumigatus azole-resistant strains with different known resistance mechanisms. None of the strains had either the variation at codon L98 or the 34-bp tandem duplication.
Levels of Cyp51A expression by A. fumigatus azole-resistant strains.
The levels of accumulation of cyp51A mRNA in two azole-resistant clinical strains (strains CM2627 and CM3275) in comparison with those in the A. fumigatus azole-susceptible CM237 control strain were analyzed. The values presented in Table 3 are the differences in the levels of transcription of cyp51A between the azole-resistant strains and azole-susceptible strain CM237 normalized to the levels of transcription of the reference β-tubulin gene for each strain included in the assay.
TABLE 3.
A. fumigatus cyp51A mRNA transcription levels of different clinical and mutant strains with respect to that for wild-type strain CM237
| Strain |
cyp51A mRNA transcription levela
|
VC (%)b | |
|---|---|---|---|
| Geometric mean | SD | ||
| CM2627 | 7.70 | 1.45 | 18.8 |
| CM3275 | 8.05 | 1.25 | 15.5 |
| CM237-L98HTR1 | 8.10 | 1.36 | 16.8 |
| CM237-TR7 | 4.23 | 1.02 | 24.0 |
| akuBKU80 recipient | 1.48 | 0.10 | 6.9 |
| akuBKU80-TR2 | 4.74 | 0.91 | 19.2 |
| akuBKU80-TR3 | 8.23 | 1.96 | 23.8 |
| akuBKU80-L98H1 | 0.65 | 0.15 | 22.9 |
| akuBKU80-L98H3 | 1.32 | 0.14 | 17.8 |
Geometric means for at least two different RNAs and six cDNAs. SD, standard deviation.
VC, coefficient of variation
Replacement of wild-type cyp51A gene with different gene modifications.
Therefore, the cyp51A gene fragments which had been amplified from strain CM2627 by PCR and which carried the different cyp51A modifications were electroporated into wild-type (azole-susceptible) A. fumigatus strain CM237. The different PCR fragments (L98H, TR, and TRL98H) were individually electroporated into wild-type A. fumigatus strain CM237 (Fig. 1). When itraconazole (8 μg/ml) was used for selection, mutants (n = 15) were obtained only when TRL98H PCR fragment was used for transformation (strains CM237-TRL98H1 to CM237-TRL98H15). However, when 1 μg/ml of itraconazole was used as the selective concentration, only one mutant (strain CM237-TR7) arose when the TR fragment obtained by PCR was used for transformation. On the other hand, after several experiments with the L98H fragment and itraconazole at concentrations of 1 μg/ml and 8 μg/ml, we were unable to obtain any mutant showing only the t364a base change. Therefore, a different azole-susceptible A. fumigatus strain (strain akuBKU80) was used as the recipient for electroporation with the linear fragments obtained by PCR which included only one of the cyp51A modifications (the TR fragment or the L98H fragment). Transformants were selected on medium containing itraconazole (0.5 to 1.0 μg/ml). By using this strain, two mutants (strains akuBKU80-TR2 and akuBKU80-TR3) were obtained when the PCR-generated TR fragment was used for transformation and five mutants (strains akuBKU80-L98H1 to akuBKU80-L98H5) were obtained when the PCR-generated L98H fragment was used for transformation.
Mutant analysis.
Several different aspects of the A. fumigatus transformants were analyzed: (i) antifungal drug susceptibility; (ii) the DNA sequence, to verify the incorporation of sequence changes; and (iii) cyp51A mRNA transcription levels.
In order to rule out the possibility of the integration of extra copies of cyp51A in the transformation procedure, DNAs from every transformant were digested with a restriction enzyme, blotted, and hybridized (29) with a labeled fragment of cyp51A (data not shown).
The cyp51A gene including the promoter area and the cyp51B genes from all transformants were sequenced. All mutants analyzed (nine) had incorporated the mutated cyp51A allele corresponding to the fragment that was introduced (Table 4). With the exception of the L98H mutation and/or duplication of the repeat in tandem, none of the transformants had any other mutation in either cyp51A or cyp51B compared to the sequence of wild-type strain CM237. The A. fumigatus transformants' susceptibilities to triazole drugs were determined as described above. All the transformants with both cyp51A alterations (TR and L98H) exhibited susceptibility profiles similar to those of the clinical isolates (Table 4). However, transformant CM237-TR7 had a slight but not significant increase in azole drug MICs (P > 0.01) that did not correspond to an azole-resistant phenotype. Similarly, transformants obtained with the akuBKU80 strain (containing either the TR or the L98H modification independently) had a slight but not significant increase in azole drug MICs (P > 0.01) that did not correspond to an azole-resistant phenotype (Table 4).
TABLE 4.
Nucleotide and amino acids substitutions in A. fumigatus cyp51A and cyp51B genes and ranges of MICs of different antifungal drugs for A. fumigatus mutant strains with the TR present in the cyp51A promoter and/or the Cyp51A amino acid substitution at L98H
| Isolate or straina |
cyp51A gene and promoterb
|
cyp51B geneb
|
MIC range (μg/ml)c
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Codon 54 | Codon 98 | Codon 220 | TR | Codon 35 | Codon 394 | AMB | ITC | VRC | RVC | PSC | |
| CM237 (wild type) | GGG | CTC | ATG | (−) | TCT | CCT | 0.25 | 0.25 | 0.5 | 0.5 | 0.12 |
| M03/669 - CM2627 | GGG | 364CAC | ATG | (+) | 105TCT | 1285CCG | 0.25 | 8.0 | 4.0-8.0 | 4.0-8.0 | 0.5 |
| VC28-78 - CM3275 | GGG | 364CAC | ATG | (+) | 105TCC | 1285CCG | 0.50 | 8.0 | 4.0 | 4.0 | 0.5 |
| CM237-TRL98H1 | GGG | 364CAC | ATG | (+) | TCT | CCT | 0.5 | 8.0 | 4.0 | 4.0-8.0 | 1.0 |
| CM237-TRL98H3 | GGG | 364CAC | ATG | (+) | TCT | CCT | 0.25 | 8.0 | 4.0 | 8.0 | 0.5-1.0 |
| CM237-TRL98H4 | GGG | 364CAC | ATG | (+) | TCT | CCT | 0.25 | 8.0 | 4.0 | 4.0 | 0.5-1.0 |
| CM237-TRL98H5 | GGG | 364CAC | ATG | (+) | TCT | CCT | 0.25 | 8.0 | 4.0 | 4.0-8.0 | 0.5-1.0 |
| CM237-TR7 | GGG | CTC | ATG | (+) | TCT | CCT | 0.50 | 0.5 | 1.0 | 1.0 | 0.25 |
| akuBKU80 (recipient) | GGG | CTC | ATG | (−) | TCT | CCT | 0.25 | 0.12-0.25 | 0.25-0.5 | 0.5 | 0.03-0.06 |
| akuBKU80-TR2 | GGG | CTC | ATG | (+) | TCT | CCT | 0.50 | 0.5 | 1.0 | 2.0 | 0.12 |
| akuBKU80-TR3 | GGG | CTC | ATG | (+) | TCT | CCT | 0.25-0.50 | 0.5 | 1.0 | 1.0-2.0 | 0.12 |
| akuBKU80-L88H1 | GGG | 364CAC | ATG | (−) | TCT | CCT | 0.25-0.50 | 0.5 | 1.0-2.0 | 1.0-2.0 | 0.12 |
| akuBKU80-L88H3 | GGG | 364CAC | ATG | (−) | TCT | CCT | 0.25-0.50 | 0.5 | 1.0-2.0 | 1.0-2.0 | 0.12 |
| Amino acids | (G54) | (L98H) | (M220) | (S35S) | (P394P) | ||||||
Mutant nomenclature: strains CM237-TRL98H1 to CM237-TRL98H5, gene replacement of the TRL98H fragment from strain CM2627 into azole-susceptible strain CM237; strain CM237-TR7, gene replacement of the TR fragment from strain CM2627 into strain CM237; strains akuBKU80-L98H1 and akuBKU80-L98H3, gene replacement of the L98H fragment from strain CM2627 into the akuBKU80 azole-susceptible strain; strains akuBKU80-TR2 and akuBKU80-TR3, gene replacement of the TR fragment from strain CM2627 into strain akuBKU80.
Nucleotides are numbered from the translation ATG start codon of cyp51A and cyp51B. The numbers indicate the position at which a base change occurs (in boldface). TR (+), presence of the 34-bp fragment duplication in tandem in the cyp51A promoter; TR (−), absence of the 34-bp fragment duplication in tandem in the cyp51A promoter.
AMB, amphotericin B; ITC, itraconazole; VRC, voriconazole; RVC, ravuconazole; PSC, posaconazole.
The levels of cyp51A mRNA expression in two transformants (strains CM237-TRL98H1 and CM237-TR7) in comparison with those in A. fumigatus azole-susceptible recipient strain CM237 and two azole-resistant clinical strains (strains CM2627 and CM3275) were analyzed. The relative increases in the levels of cyp51A expression obtained in CM237-TRL98H1 were eightfold compared to the level of expression in the wild-type strain, whereas the levels of cyp51A transcription in CM237-TR7 were increased only fourfold compared to the level of transcription in the wild type (Table 3). Also, the levels of cyp51A mRNA expression in four mutants (strains akuBKU80-TR2, akuBKU80-TR3, akuBKU80-L98H1, and akuBKU80-L98H3) in comparison with that in their A. fumigatus recipient akuBKU80 strain were analyzed. The relative increases in the levels of cyp51A expression obtained in strains akuBKU80-TR2 and akuBKU80-TR3 were between five- and eightfold those in the akuBKU80 parental strain, whereas the levels of cyp51A transcription in strains akuBKU80-L98H1 and akuBKU80-L98H3 were similar to that in the parental akuBKU80 strain (Table 3).
DISCUSSION
In Candida spp., the majority of reports on azole drug resistance are related to the increased efflux of azole drugs due to the overexpression of efflux pumps of different types (4, 39, 42). In addition, a number of reports have identified polymorphisms in the erg11 gene (which is homologous to cyp51A) from clinical Candida albicans isolates that are responsible for and/or that are associated with fluconazole resistance (16, 26, 43). On the other hand, the resistance of filamentous fungi to different demethylase inhibitors (DMIs) used for agricultural purposes has mainly been correlated with one amino acid change (Y136F) in the azole target, Cyp51 (11, 12, 53), or with its overexpression (18, 23, 44, 46). In A. fumigatus, azole drug resistance has been described for both laboratory mutants and clinical strains and has mainly been attributed to alterations in the target enzyme (Cyp51A) (2, 15, 25, 28, 32). Even though analysis of the A. fumigatus genome has shown the existence of more than 40 ATP binding cassette transporter (ABC) homologs and more than 100 major facilitator transporter genes (five times more than the number in yeast) (49), isolates that overexpress efflux pumps as an azole resistance mechanism have rarely been described. Nevertheless, there are a number of reports of efflux pump overexpression related to azole resistance in A. fumigatus, although these have been A. fumigatus mutant strains generated in the laboratory and not clinical isolates (9, 32). Only one clinical strain (strain AF72) has been found to overexpress (by five times) the atrF gene (which encodes an ABC transporter efflux pump), but only when it is grown in the presence of itraconazole (46).
Up to now, decreases in the susceptibility of A. fumigatus to itraconazole and posaconazole only (15, 25, 32) or to all azole drugs (2, 29) have exclusively been linked to specific mutations in cyp51A. The 14 A. fumigatus clinical strains described in this study present a similar triazole cross-resistance phenotype but have none of the known Cyp51A amino acid substitutions. A new resistance mechanism that consisted of the presence of a duplication of a TR in the cyp51A promoter and a L98H substitution in the cyp51A-coding sequence was found in all isolates. This fact strongly suggests that both alterations were related to the azole-resistant phenotype. Nevertheless, we conducted experiments to analyze each of the cyp51A modifications individually.
The results obtained with the mutants (strains CM237-TR7, akuBKU80-TR2, and akuBKU80-TR3) suggest that the duplication of the 34-bp TR by itself seems to be insufficient for reproduction of the resistant phenotype, even though there was a four- to eightfold increase in the level of cyp51A expression which corresponded to a slight increase in azole MICs. Previous reports have related azole resistance to the high level of cyp51/erg11 expression in both yeast and filamentous fungi. The mechanisms that have been associated with increased expression include gene or chromosome duplication and promoter modification. The first mechanism, known in mammals as gene amplification, was described in Candida glabrata (27) and in an engineered laboratory strain of Aspergillus niger (50). The latter mechanism has been seen in an A. nidulans triazole-resistant strain obtained by amplification or overexpression of the A. nidulans cyp51 gene expressed in a high-copy-number plasmid (34). An increased level of cyp51 expression has been suggested as a possible mechanism for sterol DMI resistance in the plant pathogen Mycosphaerella graminicola (48). Similarly, a DMI resistance mechanism has been described for another plant pathogenic mold, Venturia inaequalis. The high level of cyp51 expression was correlated with the presence of 553-bp insertion located in the gene promoter (44). Also, just recently, the analysis of 59 DMI-resistant isolates of Bumeriella jaapii has shown 5- to 12-fold increased levels of cyp51 expression related to various forms of a truncated non-long terminal direct repeat, long interspersed nuclear retrotransposon element (23). In Penicillium digitatum a mechanism similar to the one that we describe has been reported for strains that have a 126-bp sequence tandemly repeated in the cyp51 promoter and that is directly related to a pattern of resistance to different DMIs. Nevertheless, the number of tandem repeats proved to be essential for the production of a high level of DMI resistance (18). Therefore, the results obtained in this study suggest that the 34-bp sequence in the promoter region of cyp51A might be a transcriptional enhancer and that its duplication is responsible for the increased level of expression of cyp51A and the higher A. fumigatus MICs for azole drugs. However, these results also suggest that the TR duplication by itself is not enough to confer the azole cross-resistant phenotype shown by these strains. On the other hand, the introduction of the L98H amino acid substitution at Cyp51A alone seems to be responsible for only a slight increase in the A. fumigatus MICs for azole drugs without influencing the cyp51A expression levels. In the A. fumigatus Cyp51A protein, the leucine at position 98 is close to the first substrate recognition site (SRS1). This residue is conserved throughout yeast and filamentous fungi but not in plant or human Cyp51 proteins. An homology model based on the X-ray crystal structure of Cyp51 from Mycobacterium tuberculosis was used to predict the specific Cyp51A residues within A. fumigatus Cyp51A in combination with azole drugs (54). However, the specific residue L98 has not been implicated in any azole drug interactions with the protein (54). The search for Cyp51A mutations at L98 in A. fumigatus strains with intermediate MICs of azole drugs will help to clarify whether this amino acid substitution is present in strains isolated in the clinical setting and/or in environmental isolates (these experiments are already under way in our laboratory).
Finally, the results obtained with the mutants incorporating both cyp51A alterations (TR and L98H) confirmed that the combination of both modifications is needed to reproduce the azole-resistant phenotype shown by the clinical strains.
It is quite remarkable that since itraconazole-resistant A. fumigatus strains were first described, A. fumigatus strains with phenotypes of resistance to single or multiple azoles have emerged in subsequent years. The recent isolation (2002 to 2006) of a number of A. fumigatus strains, all unrelated, with a novel mechanism of azole resistance suggests that these molds might be exposed to the selective pressure of azole compounds either in the patient or in the environment. This aspect requires further studies, especially in areas where 14-α-demethylase inhibitors are extensively used in agriculture.
Acknowledgments
This work was funded in part by grants from the European Commission STREP Project (LSHM-CT-2005-518199) and SAF2005-06541 from the Ministerio de Educacion y Ciencia (Spain). L. Alcazar-Fuoli has a postdoctoral fellowship from project LSHM-CT-2005-518199.
We thank J. P. Latge (Institute Pasteur) for providing the A. fumigatus akuBKU80 strain and for helpful suggestions. We are grateful to G. del Rio for technical support. We thank C. H. Klaassen for help with the genotypic analysis of the isolates and Fiona Westbury for the English revision of the manuscript.
Footnotes
Published ahead of print on 19 March 2007.
REFERENCES
- 1.Alcazar-Fuoli, L., E. Mellado, G. Garcia-Effron, M. J. Buitrago, J. F. Lopez, J. O. Grimalt, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2006. Aspergillus fumigatus C-5 sterol desaturases Erg3A and Erg3B: role in sterol biosynthesis and antifungal drug susceptibility. Antimicrob. Agents Chemother. 50:453-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, J., H. Li, R. Li, D. Bu, and Z. Wan. 2005. Mutations in the cyp51A gene and susceptibility to itraconazole in Aspergillus fumigatus serially isolated from a patient with lung aspergilloma. J. Antimicrob. Chemother. 55:31-37. [DOI] [PubMed] [Google Scholar]
- 3.Chryssanthou, E. 1997. In vitro susceptibility of respiratory isolates of Aspergillus species to itraconazole and amphotericin B acquired resistance to itraconazole. Scand. J. Infect. Dis. 29:509-512. [DOI] [PubMed] [Google Scholar]
- 4.Cowen, L. E., A. Nantel, M. S. Whiteway, D. Y. Thomas, DC Tessier, L. M. Kohn, and J. B. Anderson. 2002. Population genomics of drug resistance in Candida albicans. Proc. Natl. Acad. Sci. USA 99:9284-9289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuenca-Estrella, M., A. Gomez-Lopez, E. Mellado, M. J. Buitrago, A. Monzon, and J. L. Rodriguez-Tudela. 2006. Head-to-head comparison of the activities of currently available antifungal agents against 3,378 Spanish clinical isolates of yeasts and filamentous fungi. Antimicrob. Agents Chemother. 50:917-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuenca-Estrella, M., C. B. Moore, F. Barchiesi, J. Bille, E. Chryssanthou, D. W. Denning, J. P. Donnelly, F. Dromer, B. Dupont, J. H. Rex, M. D. Richardson, B. Sancak, P. E. Verweij, J. L. Rodriguez-Tudela, et al. 2003. Multicenter evaluation of the reproducibility of the proposed antifungal susceptibility testing method for fermentative yeasts of the Antifungal Susceptibility Testing Subcommittee of the European Committee on Antimicrobial Susceptibility Testing (AFST-EUCAST). Clin. Microbiol. Infect. 9:467-474. [DOI] [PubMed] [Google Scholar]
- 7.Cuenca-Estrella, M., J. L. Rodriguez-Tudela, E. Mellado, J. V. Martinez-Suarez, and A. Monzon. 1998. Comparison of the in vitro activity of voriconazole (UK-109,496), itraconazole and amphotericin B against clinical isolates of Aspergillus fumigatus. J. Antimicrob. Chemother. 42:531-533. [DOI] [PubMed] [Google Scholar]
- 8.Dannaoui, E., F. Persat, M. F. Monier, E. Borel, M. A. Piens, and S. Picot. 1999. In-vitro susceptibilities of Aspergillus spp. isolates to amphotericin B and itraconazole. J. Antimicrob. Chemother. 44:553-555. [DOI] [PubMed] [Google Scholar]
- 9.da Silva Ferreira, M. E., J. L. Capellaro, E. dos Reis Marques, I. Malavazi, D. Perlin, S. Park, J. B. Anderson, A. L. Colombo, B. A. Arthington-Skaggs, M. H. Goldman, and G. H. Goldman. 2004. In vitro evolution of itraconazole resistance in Aspergillus fumigatus involves multiple mechanisms of resistance. Antimicrob. Agents Chemother. 48:4405-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.da Silva Ferreira, M. E., M. R. Crees, M. Savoldi, M. H. Goldman, A. Hartl, T. Heinekamp, A. A. Brakhage, and G. H. Goldman. 2006. The akuBKU80 mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot. Cell 5:207-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Délye, C., L. Bousset, and M. F. Corio-Costet. 1998. PCR cloning and detection of point mutations in the eburicol 14α-demethylase (CYP51) gene from Erysiphe graminis f. sp. hordei, a “recalcitrant” fungus. Curr. Genet. 34:399-403. [DOI] [PubMed] [Google Scholar]
- 12.Délye, C., F. Laigret, and M. F. Corio-Costet. 1997. A mutation in the 14α-demethylase gene of Uncinula necator that correlates with resistance to a sterol biosynthesis inhibitor. Appl. Environ. Microbiol. 63:2966-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denning, D. W., K. Venkateswarlu, K. L. Oakley, M. J. Anderson, N. J. Manning, D. A. Stevens, D. W. Warnock, and S. L. Kelly. 1997. Itraconazole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 41:1364-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Valk, H. A., J. F. Meis, I. M. Curfs, K. Muehlethaler, J. W. Mouton, and C. H. Klaassen. 2005. Use of a novel panel of nine short tandem repeats for exact and high-resolution fingerprinting of Aspergillus fumigatus isolates. J. Clin. Microbiol. 43:4112-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diaz-Guerra, T. M., E. Mellado, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2003. A point mutation in the 14-alpha sterol demethylase gene cyp51A contributes to itraconazole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 47:1120-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Favre, B., M. Didmon, and N. S. Ryder. 1999. Multiple amino acid substitutions in lanosterol 14α-demethylase contribute to azole resistance in Candida albicans. Microbiology 145:2715-2725. [DOI] [PubMed] [Google Scholar]
- 17.Fortun, J., P. Martin-Davila, M. A. Sanchez, V. Pintado, M. E. Alvarez, A. Sanchez-Sousa, and S. Moreno. 2003. Voriconazole in the treatment of invasive mold infections in transplant recipients. Eur. J. Clin. Microbiol. Infect. Dis. 22:408-413. [DOI] [PubMed] [Google Scholar]
- 18.Hamamoto, H., K. Hasegawa, R. Nakaune, Y. J. Lee, Y. Makizumi, K. Akutsu, and T. Hibi. 2000. Tandem repeat of a transcriptional enhancer upstream of the sterol 14alpha-demethylase gene (CYP51) in Penicillium digitatum. Appl. Environ. Microbiol. 66:3421-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herbrecht, R. 2004. Voriconazole: therapeutic review of a new azole antifungal. Expert Rev. Anti.-Infect. Ther. 2:485-497. [DOI] [PubMed] [Google Scholar]
- 20.Kontoyiannis, D. P., and G. P. Bodey. 2002. Invasive aspergillosis in 2002: an update. Eur. J. Clin. Microbiol. Infect. Dis. 21:161-172. [DOI] [PubMed] [Google Scholar]
- 21.Latgé J.-P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin, S. J., J. Schranz, and S. M. Teutsch. 2001. Aspergillosis case-fatality rate: systematic review of the literature. Clin. Infect. Dis. 32:358-366. [DOI] [PubMed] [Google Scholar]
- 23.Ma, Z., T. J. Proffer, J. L. Jacobs, and G. W. Sundin. 2006. Overexpression of the 14 alpha-demethylase target gene (CYP51) mediates fungicide resistance in Blumeriella jaapii. Appl. Environ. Microbiol. 72:2581-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manavathu, E. K., J. A. Vazquez, and P. H. Chandrasekar. 1999. Reduced susceptibility in laboratory-selected mutants of Aspergillus fumigatus to itraconazole due to decreased intracellular accumulation of the antifungal agent. Int. J. Antimicrob. Agents 12:213-219. [DOI] [PubMed] [Google Scholar]
- 25.Mann, P. A., R. M. Parmegiani, S. Q. Wei, C. A. Mendrick, X. Li, D. Loebenberg, B. DiDomenico, R. S. Hare, S. S. Walker, and P. M. McNicholas. 2003. Mutations in Aspergillus fumigatus resulting in reduced susceptibility to posaconazole appear to be restricted to a single amino acid in the cytochrome P450 14-alpha demethylase Antimicrob. Agents Chemother. 47:577-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marichal, P., L. Koymas, S. Willemsens, D. Bellens, P. Verhasselt, W. Luyten, M. Borgers, F. C. S. Ramaekers, F. C. Odds, and H. Vanden Bossche. 1999. Contribution of mutations in the cytochrome P450 14 α-demethylase (Erg11p, Cyp51p) to azole resistance in Candida albicans. Microbiology 145:2701-2713. [DOI] [PubMed] [Google Scholar]
- 27.Marichal, P., H. V. Bossche, F. C. Odds, G. Nobels, D. W. Warnock, V. Timmerman, C. van Broeckhoven, S. Fay, and P. Mose-Larsen. 1997. Molecular biological characterization of an azole-resistant Candida glabrata isolate. Antimicrob. Agents Chemother. 41:2229-2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mellado, E., T. M. Diaz-Guerra, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2001. Identification of two different 14-α sterol demethylase-related genes (cyp51A and cyp51B) in Aspergillus fumigatus and other Aspergillus species. J. Clin. Microbiol. 39:2431-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mellado, E., G. Garcia-Effron, L. Alcazar-Fuoli, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2004. Substitutions at methionine 220 in the 14-alpha sterol demethylase (Cyp51A) of Aspergillus fumigatus are responsible for resistance in vitro to azole antifungal drugs. Antimicrob. Agents Chemother. 48:2747-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mellado, E., G. Garcia-Effron, M. J. Buitrago, L. Alcazar-Fuoli, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2005. Targeted gene disruption of the 14-alpha sterol demethylase (cyp51A) in Aspergillus fumigatus and its role in azole drug susceptibility. Antimicrob. Agents Chemother. 49:2536-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mosquera, J., and D. W. Denning. 2002. Azole cross-resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 46:556-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nascimento, A. M., G. H. Goldman, S. Park, S. A. Marras, G. Delmas, U. Oza, K. Lolans, M. N. Dudley, P. A. Mann, and D. S. Perlin. 2003. Multiple resistance mechanisms among Aspergillus fumigatus mutants with high-level resistance to itraconazole. Antimicrob. Agents Chemother. 47:1719-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.National Committee for Clinical Laboratory Standards. 2002. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi: approved standard M38-A. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 34.Osherov, N., D. P. Kontoyannis, A. Romans, and G. S. May. 2001. Resistant to itraconazole in Aspergillus nidulans and Aspergillus fumigatus is conferred by extra copies of the A. nidulans P-450 14-alpha-demethylase gene, pdmA. J. Antimicrob. Chemother. 48:75-81. [DOI] [PubMed] [Google Scholar]
- 35.Petrikkou, E., J. L. Rodriguez-Tudela, M. Cuenca-Estrella, A. Gomez, A. Molleja, and E. Mellado. 2001. Inoculum standardization for antifungal susceptibility testing of filamentous fungi pathogenic for humans. J. Clin. Microbiol. 39:1345-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfaffl, M. W. 2001. A new mathematical model for relative quantification on real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfaller, M. A., S. A. Messer, R. J. Hollis, and R. N. Jones. 2002. Antifungal activities of posaconazole, ravuconazole, and voriconazole compared to those of itraconazole and amphotericin B against 239 clinical isolates of Aspergillus spp. and other filamentous fungi: report from SENTRY Antimicrobial Surveillance Program, 2000. Antimicrob. Agents Chemother. 46:1032-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pontecorvo, G., J. A. Roper, L. M. Hemmons, K. D. MacDonald, and A. W. Bufton. 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5:141-238. [DOI] [PubMed] [Google Scholar]
- 39.Prasad, R., and K. Kapoor. 2005. Multidrug resistance in yeast Candida. Int. Rev. Cytol. 242:215-248. [DOI] [PubMed] [Google Scholar]
- 40.Rodriguez-Tudela, J. L., E. Chryssanthou, E. Petrikkou, J. Mosquera, D. W. Denning, and M. Cuenca-Estrella. 2003. Interlaboratory evaluation of hematocytometer method of inoculum preparation for testing antifungal susceptibilities of filamentous fungi. J. Clin. Microbiol. 41:5236-5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanchez, O., and J. Aguirre. 1996. Efficient transformation of Aspergillus nidulans by electroporation of germinated conidia. Fungal Genet. Newsl. 43:48-51. [Google Scholar]
- 42.Sanglard, D., and F. C. Odds. 2002. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect. Dis. 2:73-85. [DOI] [PubMed] [Google Scholar]
- 43.Sanglard, D., F. Ischer, K. Koymans, and J. Bille. 1998. Amino acid substitutions in the cytochrome P450 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob. Agents Chemother. 42:241-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schnabel, G., and A. L. Jones. 2001. The 14 α-demethylase (CYP51A1) gene is overexpressed in Venturia inaequalis strains resistant to myclobutanil. Phytopathology 91:102-110. [DOI] [PubMed] [Google Scholar]
- 45.Segal, B. H., and T. J. Walsh. 2005. Current approaches to diagnosis and treatment of invasive aspergillosis. Am. J. Respir. Crit. Care Med. 173:707-717. [DOI] [PubMed] [Google Scholar]
- 46.Slaven, J. W., M. J. Anderson, D. Sanglard, G. K. Dixon, J. Bille, I. S. Roberts, and D. W. Denning. 2002. Increased expression of a novel Aspergillus fumigatus ABC transporter gene, atrF, in the presence of itraconazole in an itraconazole resistant clinical isolate. Fungal Genet. Biol. 36:199-206. [DOI] [PubMed] [Google Scholar]
- 47.Steinbach, W. J., and D. A. Stevens. 2003. Review of newer antifungal and immunomodulatory strategies for invasive aspergillosis. Clin. Infect. Dis. 37(Suppl. 3):S157-S187. [DOI] [PubMed] [Google Scholar]
- 48.Stergiopoulos, I., J. G. van Nistelrooy, G. H. Kema, and M. A. De Waard. 2003. Multiple mechanisms account for variation in base-line sensitivity to azole fungicides in field isolates of Mycosphaerella graminicola. Pest. Manag. Sci. 59:1333-1343. [DOI] [PubMed] [Google Scholar]
- 49.Tekaia, F., and J. P. Latge. 2005. Aspergillus fumigatus: saprophyte or pathogen? Curr. Opin. Microbiol. 8:385-392. [DOI] [PubMed] [Google Scholar]
- 50.van den Brink, H. J., H. J. van Nistelrooy, M. A. de Waard, C. A. van den Hondel, and R. F. van Gorcom. 1996. Increased resistance to 14 alpha-demethylase inhibitors (DMIs) in Aspergillus niger by coexpression of the Penicillium italicum eburicol 14 alpha-demethylase (cyp51) and the A. niger cytochrome P450 reductase (cprA). J. Biotechnol. 49:13-18. [DOI] [PubMed] [Google Scholar]
- 51.Verweij, P. E., E. Mellado, and W. J. G. Melchers. 2007. Multiple-triazole-resistant aspergillosis. N. Engl. J. Med. 356:1481-1483. [DOI] [PubMed] [Google Scholar]
- 52.Weidner, G., C. d'Enfert, A. Koch, P. C. Mol, and A. A. Brakhage. 1998. Development of a homologous transformation system for the human pathogenic fungus Aspergillus fumigatus based on the pyrG gene encoding orotidine 5′-monophosphate decarboxylase. Curr. Genet. 33:378-385. [DOI] [PubMed] [Google Scholar]
- 53.Wyand, R. A., and J. K. Brown. 2005. Sequence variation in the CYP51 gene of Blumeria graminis associated with resistance to sterol demethylase inhibiting fungicides. Fungal Genet. Biol. 42:726-735. [DOI] [PubMed] [Google Scholar]
- 54.Xiao, L., V. Madison, A. S. Chau, D. Loebenberg, R. E. Palermo, and P. M. McNicholas. 2004. Three-dimensional models of wild-type and mutated forms of cytochrome P450 14alpha-sterol demethylases from Aspergillus fumigatus and Candida albicans provide insights into posaconazole binding. Antimicrob. Agents Chemother. 48:568-574. [DOI] [PMC free article] [PubMed] [Google Scholar]