Abstract
Of the 181 unduplicated Escherichia coli strains isolated in nine different hospitals in three Portuguese regions, 119 were extended-spectrum β-lactamase (ESBL)-CTX-M producers and were selected for phenotype and genotype characterization. CTX-M producer strains were prevalent among community-acquired infections (56%), urinary tract infections (76%), and patients ≥60 years old (76%). In MIC tests, all strains were resistant to cefotaxime, 92% were resistant to ceftazidime, 93% were resistant to quinolones, 89% were resistant to aminoglycoside, and 26% were resistant to trimethoprim-sulfamethoxazole; all strains were sensitive to carbapenems, and 92% of the strains had a multidrug resistance phenotype. Molecular methods identified 109 isolates harboring a blaCTX-M-15 gene, 1 harboring the blaCTX-M-32 gene (first identification in the country), and 9 harboring the blaCTX-M-14 gene. All isolates presented the ISEcp1 element upstream from the blaCTX-M genes; one presented the IS903 element (downstream of blaCTX-M-14 gene), and none had the IS26 element; 85% carried blaTEM-1B, and 84% also carried a blaOXA-30. Genetic relatedness analysis based on pulsed-field gel electrophoresis defined five clusters and indicated that 76% of all isolates (from cluster IV) corresponded to a single epidemic strain. Of the 47 strains from one hospital, 41 belonged to cluster IV and were disseminated in three main wards. CTX-M-producing E. coli strains are currently a problem in Portugal, with CTX-M-15 particularly common. This study suggests that the horizontal transfer of blaCTX-M genes, mediated by plasmids and/or mobile elements, contributes to the dissemination of CTX-M enzymes to community and hospital environments. The use of extended-spectrum cephalosporins, quinolones, and aminoglycosides is compromised, leaving carbapenems as the therapeutic option for severe infections caused by ESBL producers.
Production of extended-spectrum β-lactamases (ESBLs) is a major mechanism of resistance to β-lactam antibiotics. First detected in 1986 (27), CTX-M (cefotaximases) is an ESBL family including more than 60 different enzymes in five phylogenetic groups (http://www.lahey.org/studies/webt.htm). CTX-M β-lactamases of group 2be (5), unlike other ESBL, are more active against cefotaxime than against ceftazidime (3). The rapid emergence and worldwide spread of this plasmid-mediated family is associated with mobile elements and particularly insertion sequences (1, 4, 13, 21, 31, 40, 49). This high mobility associated with inefficient antibiotic policies has led to community-acquired and nosocomial infections (24, 51).
Many strains that express CTX-M β-lactamases are multidrug resistant (16, 21, 23, 51). Genes conferring resistance to aminoglycosides and tetracycline and other bla genes have been found on the same plasmids as the blaCTX-M genes (4). Genes conferring plasmid-mediated quinolone resistance have also been associated with blaCTX-M genes (20).
CTX-M-15 appears to have the best dissemination capacity of all of the CTX-M family, probably due to successful genetic rearrangements (48). The corresponding gene is normally associated with an upstream ISEcp1 element. The gene has been detected in Europe (1, 11, 14, 24), Africa (30, 33, 43, 47), North and South America (4, 38), and Asia (21, 22) and has been associated with many community and hospital outbreaks. To our knowledge, only dispersed cases have been reported in Portugal (11, 12, 25, 26, 28).
We investigate the dissemination of CTX-M enzymes among clinical isolates of Escherichia coli recovered in various Portuguese hospitals from cases of community-acquired and nosocomial infections. We describe the phenotypes and genetic characteristics of these isolates.
MATERIALS AND METHODS
Bacterial strains.
A total of 181 unduplicated E. coli strains isolated from patients were collected in nine hospitals in three different regions of Portugal between March 2004 and March 2006 and sent to the Antibiotic Resistance Unit at the National Institute of Health (NIH) Dr. Ricardo Jorge in Lisbon. The bacteriology laboratories of these hospitals collaborate with the NIH as contributors to the Antibiotic Resistance Surveillance Program in Portugal (ARSIP); they sent all E. coli strains identified as ESBL producers by different systems (ATB G-5, VITEK 1, VITEK 2, and Phoenix). Of the 181 strains received, 119 were detected as ESBL-(CTX-M) producers by phenotypic and genotypic characterization at NIH, as stated below. The NIH conducted the main study using 119 of 126 CTX-M producer strains because only the first bacterial isolate from the three patients with multiple isolates was considered (among a total of 181 ESBL producers) (Table 1). The other ESBL producers were not investigated in the present study. Of the 119 strains, 47 were nosocomial and 66 were community acquired, according to Centers for Disease Control and Prevention criteria (18), and 6 were of unknown origin (Table 1). Strains were isolated from urine (n = 90), wounds (n = 13), blood (n = 4), ascitic fluid (n = 3), sputum (n = 3), bronchoalveolar lavage (n = 2) and secretions (n = 3), and gastric fluid (n = 1). E. coli INSRA99 (IRT-2, pI 5.2), E. coli RP4 (TEM-2, pI 5.6), and E. coli SolRl 90 (AmpC, pI 9.2) were used only as control strains for isoelectric focusing (IEF). E. coli R111 (TEM-1, pI 5.4; blaTEM-1 plus ampC), Salmonella enterica serovar Typhimurium (OXA-1, pI 7.4; blaOXA-1), E. coli C600 (SHV-1, pI 7.6; blaSHV-1), and E. coli UA1526 (CTX-M-15, pI 8.9; blaCTX-M-15) were used as control strains for both PCR and IEF. The control strains used for PCR with insertion sequence primers were INSRA5753 (ISEcp1), INSRA5776 (IS903), and Kp125 (IS26).
TABLE 1.
General characterization of the 119 CTX-M-producing E. coli strains studied (one per patient), characterized by hospitals, regions, and origin of isolates
| Hospitala code | Region | Origin and no. of isolates
|
Total no. of isolates | |||||
|---|---|---|---|---|---|---|---|---|
| Community acquired (n = 66)
|
Nosocomial (n = 47)
|
Unknown (n = 6)
|
||||||
| PFGE profile (no. of strains) | Total | PFGE profile (no. of strains) | Total (ward) | PFGE profile (no. of strains) | Total | |||
| A | North | 0009 (1) | 8 | 0009 (1) | 1 (internal medicine) | 9 | ||
| 0010 (7) | ||||||||
| B | South | 0009 (1) | 8 | 0010 (2) | 2 (emergency, internal medicine) | 10 | ||
| 0010 (5) | ||||||||
| 0012 (1) | ||||||||
| 0013 (1) | ||||||||
| C | North | 0010 (27) | 31b | 0010 (14) | 16b (cardiology, surgery, internal medicine, nephrology,c pneumology,c ICU, urology) | 47 | ||
| 0003 (1)c | 0018 (1)c | |||||||
| 0005 (1)c | ND (1)c | |||||||
| 0011 (1) | ||||||||
| ND (1)c | ||||||||
| D | Lisbon and Tagus Valley | 0002 (1)c | 8d | 0007 (1) | 9d (cardiology, surgery, internal medicine, orthopedics, pneumology, gastroenterology, observation) | 0014 (1)c | 1 | 18 |
| 0010 (7) | 0010 (7) | |||||||
| 0016 (1) | ||||||||
| E | Lisbon and Tagus Valley | 0001 (1)c | 8 | 0004 (1) | 9e (emergency, internal medicine, surgery, nephrology, ICU, pediatric, neurology) | 0006 (2) | 4 | 21 |
| 0006 (1) | 0006 (1) | 0010 (1) | ||||||
| 0010 (6) | 0008 (1) | 0019 (1)f | ||||||
| 0010 (4) | ||||||||
| 0017 (1) | ||||||||
| 0019 (1) | ||||||||
| F | Lisbon and Tagus Valley | 0010 (1) | 1 (infectious diseases) | 1 | ||||
| G | Lisbon and Tagus Valley | 0010 (1) | 1 (internal medicine) | 1 | ||||
| H | Lisbon and Tagus Valley | 0010 (5) | 6 (surgery, internal medicine) | 0010 (1) | 1 | 7 | ||
| 0015 (1) | ||||||||
| I | North | 0003 (1)c | 3 | 0010 (2) | 2 (internal medicine) | 5 | ||
| 0010 (1) | ||||||||
| ND (1) | ||||||||
All hospitals were general hospitals.
Two more strains were collected from the same patient: both were noted as community acquired, whereas the first was nosocomial (from the urology ward).
CTX-M-14 producers.
Three more strains were collected from the same patient (two from internal medicine and one from the community).
Two more strains were collected from the same patient, all noted as nosocomial (from the pediatric ward).
CTX-M-32 producers. All other isolates were CTX-M-15 producers excepting those from CTX-M-14 producers.
Susceptibility testing and ESBL confirmation.
The MICs of 23 antibiotics were determined by a broth microdilution method (MicroScan Panel Sólo 1S; Dade Behring, West Sacramento, CA) for all strains. The MICs of antibiotics were determined by the agar dilution method for all transformants obtained as previously (29). The results were interpreted by using CLSI (formerly National Committee for Clinical Laboratory Standards) criteria (10). Isolates were considered multidrug resistant if they had reduced susceptibility to three or more structurally unrelated antibiotics. ESBL production was confirmed by a broth microdilution method: strains with synergy between ceftazidime and ceftazidime-clavulanic acid were suspected of producing ESBL. To confirm ESBL producers, 52 of 119 strains were randomly chosen for further testing by the Etest ESBL method using strips with cefotaxime and ceftazidime both alone and associated with clavulanate (AB Biodisk, Solna, Sweden); findings were interpreted according to the manufacturer's instructions.
Transfer of resistance.
We tested whether the ESBL phenotypes of strains producing CTX-M-14, CTX-M-15, and CTX-M-32 enzymes were transferable. Plasmid DNA was extracted from six producer strains, representative of the different CTX-M-type enzymes detected, and used to transform E. coli DH5α by electroporation. Transformants were selected on Luria broth medium containing 1 μg of cefotaxime/ml. PCR was used to test for the bla gene in transformants as described below for clinical isolates.
IEF.
Cell extracts from all isolates were obtained by ultrasonic treatment, and IEF was used to characterize the pIs of ESBLs as previously described (6). The pIs of each β-lactamase were compared to those produced by control strains.
PCR amplification and gene sequencing.
PCR amplification was used to test for the blaCTX-M gene and ISEcp1, IS26 and IS903 elements, and multiplex PCR was used to test for the blaTEM, blaOXA, blaSHV, and ampC genes in all 119 ESBL producer strains as described previously (26, 29, 42). Specific primers were used for PCR and sequencing (Table 2). A particular specific primer (CTX15i) was used to determine the nucleotides of codon 288, which distinguish blaCTX-M-15 from blaCTX-M-28. PCR products were purified and further sequenced as previously described (29).
TABLE 2.
Primers used for PCR amplification and sequencing
| Gene | Primera | Primer sequence (5′→3′) | PCR product size (bp) | Source or reference |
|---|---|---|---|---|
| blaTEM | P1 | TACGATACGGGAGGGCTTAC | 716 | 2 |
| P2 | TTCCTGTTTTTGCTCACCCA | |||
| FIN | ATTCTTGAAGACGAAAGGGC | 1,091 | 7 | |
| DEB | ATGAGTAAACTTGGTCTGAC | |||
| P3b | TGGGTGAGCAAAAACAGGAA | 7 | ||
| CLBb | AATGAAGCCATACCAAACGA | 2 | ||
| blaSHV | SHVf1 | TCAGCGAAAAACACCTTG | 471 | 32 |
| SHVr2 | TCCCGCAGATAAATCACCA | |||
| blaOXA | oxa1f | TATCTACAGCAGCGCCAGTG | 199 | 17 |
| oxa1r | CGCATCAAATGCCATAAGTG | |||
| oxa1sf | ATGAAAAACACAATACATATC | 816 | 9 | |
| oxa1sr | AATTTAGTGTGTTTAGAATGG | |||
| ampC | ampCf | CCCCGCTTATAGAGCAACAA | 634 | 17 |
| ampCr | TCAATGGTCGACTTCACACC | |||
| blaCTX-M | CTXfc | TTTGCGATGTGCAGTACCAGTAA | 543 | 15 |
| CTXrd | CGATATCGTTGGTGGTGCCATA | |||
| blaCTX-M-15 | CTX15f | AGAATAAGGAATCCCATGGTT | 875 | This study |
| CTX15r | ACCGTCGGTGACGATTTTAG | |||
| CTX15ib | GGAATCTGACGCTGGGTAAA | This study | ||
| blaCTX-M-14 | CTXg3f | CTGATGTAACACGGATTGACC | 871 | This study |
| CTX14r | CGATTTATTCAACAAAACCAG | |||
| ISEcp1 | ISEcp1 | AAAAATGATTGAAAGGTGGT | Variable | 14 |
| IS26 | IS26 | AGCGGTAAATCGTGGAGTGA | Variable | 14 |
| IS903 | IS903 | CGGTTGTAATCTGTTGTCCA | Variable | 14 |
For PCR amplification and sequencing (except those in used only for internal sequencing).
Used only for internal sequencing.
When used with primers ISEcp1, IS26, IS903, this also amplifies the downstream region of the blaCTX-M gene. Various-sized PCR products are expected.
When used with primer ISEcp1, IS26, or IS903, this also amplifies the upstream region of the blaCTX-M gene. Various-sized PCR products are expected.
PFGE.
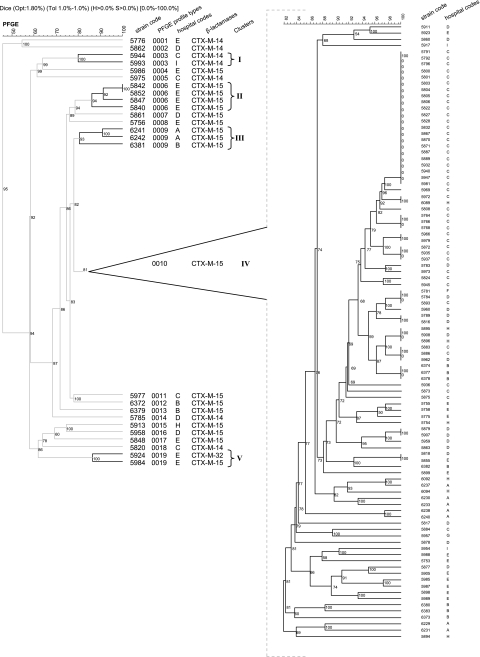
Of the 119 isolates, 116 were analyzed by pulsed-field gel electrophoresis (PFGE). Seven other strains from three patients with multiple isolates in different hospitals were also subjected to PFGE analysis (Table 1). Plugs were prepared from bacterial suspensions (300 μl) with an optical density at 650 nm of 1.2 to 1.4 in phosphate buffer (50 mM). SeaPlaque GTG (Cambrex Bio Science Rockland, Rockland, ME) agarose was added (1.5% in 300 μl of TF buffer); the mixture was immediately taken up in a 1-ml syringe and placed for exactly 4 min at −20°C. The syringe was then incubated at room temperature for 10 min, and its end was cut off. The plugs were sliced into 1-mm-thick sections and incubated in EC buffer (6 mM Tris, 1 M NaCl, 0.1 M EDTA, 0.2% deoxycholic acid, 0.5% N-lauroylsarcosine, and 0.5% Brij 58) for 3 h at 37°C. The EC buffer was replaced with ES lysis buffer (0.5 M EDTA and 1% N-lauroylsarcosine) plus proteinase K (1 mg/strain), and the samples were incubated for 16 h at 55°C. The plugs were then washed with distilled water for 5 min and with TE buffer (100 mM Tris and 10 mM EDTA) four times for 30 min each time. The DNA in the plugs was digested with 30 U of XbaI for 16 h at 37°C. PFGE was performed on a CHEF MAPPER PFGE apparatus (Bio-Rad, Hercules, CA) with 1.2% Seakem Gold agarose (Cambrex) in 0.5× Tris-borate-EDTA at 11°C and 6 V/cm. The duration of the run was 24 h, with initial and final switch times of 0.1 and 36 s, respectively. The gels were stained with ethidium bromide and photographed with Gel Doc 2000 (Bio-Rad). Banding patterns were analyzed by using BioNumerics software (version 3.5; Applied Maths, Sint-Martens-Latem, Belgium). The unweighted-pair-group method was used to construct a dendrogram based on PFGE XbaI restriction patterns of the 123 E. coli isolates. The Dice band-based similarity coefficient, with a band position tolerance of 1.0% and an optimization of 1.8%, was used for clustering. A cutoff value of 80% similarity was determined by the cluster cutoff method according to Bionumerics software. Isolates with a Dice band-based similarity coefficient value of >80% were considered to belong to the same cluster.
RESULTS
Clinical ESBL producer strains and antibiotic susceptibility.
We confirmed that the 119 isolates included in the study were ESBL producers. The CTX-M-32 producer strain was from a wound in a 51-year-old man, and 8 of the 9 CTX-M-14 producers were isolated from urine, and 1 was from a wound. Most of CTX-M-15 producers (92%) were from urine from men or women ≥60 years old (data not shown).
A total of 90% of the isolates were resistant to 11 of the 14 β-lactam antibiotics tested: more than 98% CTX-M group 1 enzyme producers were resistant to ceftazidime and cefotaxime, all CTX-M group 9 producers were resistant to cefotaxime, and 11% resistant to ceftazidime (Table 3). Only 26% CTX-M group 1 producers showed nonsusceptibility to amoxicillin-clavulanate and 12% showed nonsusceptibility to piperacillin-tazobactam. Imipenem and meropenem were the only antibiotics effective against all isolates producing either group 1 or group 9 enzymes (Table 3). A total of 92% of the isolates were multidrug resistant: 50% were isolated in the community, and 37% were isolated from hospitalized patients.
TABLE 3.
MIC50, MIC90, range, percentage of resistant and nonsusceptible E. coli strains (n = 119) producing enzymes, from the CTX-M-1 and CTX-M-9 groups, collected in nine Portuguese hospitals
| Antimicrobial agent | MIC (μg/ml) for strains producing:
|
CLSI breakpointsb (μg/ml)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTX-M group 1a (n = 110)
|
CTX-M group 9 (n = 9)
|
|||||||||||
| MIC50c | MIC90d | Range | Susceptibilitye
|
MIC50 | MIC90 | Range | Susceptibility
|
S | R | |||
| R (%) | IR (%) | R (%) | IR (%) | |||||||||
| Ampicillin | >16 | >16 | 4->16 | 110 (100) | 110 (100) | >16 | >16 | 4->16 | 9 (100) | 9 (100) | ≤8 | ≥32 |
| Ampicillin-sulbactam | >16/8 | >16/8 | 8/4->16/8 | 108 (98) | 109 (99) | >16/8 | >16/8 | 8/4->16/8 | 9 (100) | 9 (100) | ≤8/4 | ≥32/16 |
| Amoxicillin-clavulanic acid | 8/4 | 16/8 | 4/2-16/8 | 3 (3) | 28 (26) | ≤4/2 | 8/4 | ≤4/2-16/8 | 0 (0) | 0 (0) | ≤8/4 | ≥32/16 |
| Piperacillin | >64 | >64 | 16->64 | 110 (100) | 110 (100) | >64 | >64 | 16->64 | 9 (100) | 9 (100) | ≤16 | ≥128 |
| Piperacillin-tazobactam | ≤16 | 32 | ≤16-64 | 7 (6) | 13 (12) | ≤16 | ≤16 | ≤16-64 | 0 (0) | 0 (0) | ≤16/4 | ≥128/4 |
| Ticarcillin | >64 | >64 | 8->64 | 110 (100) | 110 (100) | >64 | >64 | 8->64 | 9 (100) | 9 (100) | ≤16 | ≥128 |
| Cephalothin | >16 | >16 | 8->16 | 110 (100) | 110 (100) | >16 | >16 | 8->16 | 9 (100) | 9 (100) | ≤8 | ≥32 |
| Cefazolin | >16 | >16 | 1->16 | 110 (100) | 110 (100) | >16 | >16 | 1->16 | 9 (100) | 9 (100) | ≤8 | ≥32 |
| Cefuroxime | >16 | >16 | 2->16 | 110 (100) | 110 (100) | >16 | >16 | 2->16 | 9 (100) | 9 (100) | ≤8 | ≥32 |
| Cefotaxime | >32 | >32 | 0.5->32 | 109 (99) | 110 (100) | >32 | >32 | 0.5->32 | 9 (100) | 9 (100) | ≤8 | ≥64 |
| Ceftriaxone | >32 | >32 | 2->32 | 110 (100) | 110 (100) | >32 | >32 | 2->32 | 9 (100) | 9 (100) | ≤8 | ≥64 |
| Aztreonam | >16 | >16 | 1->16 | 110 (100) | 110 (100) | 8 | >16 | 1->16 | 1 (11) | 2 (22) | ≤8 | ≥32 |
| Ceftazidime | >16 | >16 | 0.5->16 | 108 (98) | 109 (99) | 2 | >16 | 0.5->16 | 1 (11) | 1 (11) | ≤8 | ≥32 |
| Cefepime | >16 | >16 | 0.5->16 | 109 (99) | 109 (99) | >16 | >16 | 0.5->16 | 7 (78) | 9 (100) | ≤8 | ≥32 |
| Cefoxitin | ≤4 | 8 | ≤4-16 | 1 (1) | 5 (5) | ≤4 | 16 | ≤4-16 | 0 (0) | 1 (11) | ≤8 | ≥32 |
| Imipenem | ≤0.5 | ≤0.5 | ≤0.5-8 | 0 (0) | 0 (0) | ≤0.5 | ≤0.5 | ≤0.5-8 | 0 (0) | 0 (0) | ≤4 | ≥16 |
| Meropenem | ≤1 | ≤1 | ≤1-8 | 0 (0) | 0 (0) | ≤1 | ≤1 | ≤1-8 | 0 (0) | 0 (0) | ≤4 | ≥16 |
| Amikacin | ≤8 | 16 | ≤8-32 | 4 (4) | 8 (7) | ≤8 | 16 | ≤8-32 | 0 (0) | 0 (0) | ≤16 | ≥64 |
| Gentamicin | >8 | >8 | 2->8 | 87 (79) | 87 (79) | ≤2 | >8 | ≤2->8 | 2 (22) | 2 (22) | ≤4 | ≥16 |
| Tobramycin | >8 | >8 | 2->8 | 103 (94) | 104 (95) | ≤2 | >8 | ≤2->8 | 2 (22) | 2 (22) | ≤4 | ≥16 |
| Ciprofloxacin | >2 | >2 | 0.5->2 | 108 (98) | 108 (98) | ≤0.5 | >2 | ≤0.5->2 | 3 (33) | 3 (33) | ≤1 | ≥4 |
| Norfloxacin | >8 | >8 | 2->8 | 108 (98) | 108 (98) | ≤2 | >8 | ≤2->8 | 3 (33) | 3 (33) | ≤4 | ≥16 |
| Ofloxacin | >4 | >4 | 0.5->4 | 108 (98) | 108 (98) | ≤0.5 | >4 | ≤0.5->4 | 3 (33) | 3 (33) | ≤2 | ≥8 |
| Trimethoprim-sulfamethoxazole | ≤1/19 | >2/38 | ≤1/19->2/38 | 28 (26) | 28 (26) | ≤1/19 | >2/38 | ≤1/19->2/38 | 3 (33) | 3 (33) | ≤2/38 | ≥4/76 |
There were a total of 99 CTX-M-15 producers and 1 CTX-M-32 producer, according to the phylogenetic group 1 defined by Bonnet (3).
As scored according to CLSI guidelines: S, sensitive; R, resistant.
MIC50, MIC at which 50% of the isolates are inhibited.
MIC90, MIC at which 90% of the isolates tested are inhibited.
R, number of resistant isolates; IR, number of nonsusceptible isolates.
Transfer of antibiotic resistance.
We tested whether blaCTX-M genes in six selected isolates were transferable by transformation of E. coli strain DH5α. Only three transformants were obtained: two representative of CTX-M group 1 isolates (one carrying the blaCTX-M-15 plus blaTEM-1B genes and other carrying the blaCTX-M-32 gene) and one representative of CTX-M group 9 isolates (carrying the blaCTX-M-14 gene). Generally, the transformants had antibiotic resistance profiles similar to those of their parental clinical isolates (Table 4). All clinical strains were resistant to ciprofloxacin, but none of the transformants maintained that condition. Transformants of E. coli strains harboring CTX-M-32 or CTX-M-14 enzymes, whose clinical strains were resistant to gentamicin and trimethoprim-sulfamethoxazole, demonstrated susceptibility to those antibiotics.
TABLE 4.
MICs of antibiotics for clinical isolates and E. coli transformants and recipientsa
| Antimicrobial agentb | MIC (μg/ml) of antibiotic for E. coli strain:
|
||||||
|---|---|---|---|---|---|---|---|
| DH5α | INSRA5776 (CTX-M-14 + TEM-1B) | DH5α-URA5776 (CTX-M-14) | INSRA5905 (CTX-M-15 + TEM-1B) | DH5α-URA5905 (CTX-M-15 + TEM-1B) | INSRA5924 (CTX-M-32 + TEM-1B) | DH5α-URA5924 (CTX-M-32) | |
| Amoxicillin | 8 | >4,096 | >4,096 | >4,096 | >4,096 | >4,096 | >4,096 |
| Amoxicillin-clavulanic acid* | 8 | 32 | 32 | 64 | 32 | 128 | 8 |
| Ticarcillin | 4 | >4,096 | >4,096 | >4,096 | >4,096 | >4,096 | >4,096 |
| Piperacillin | 2 | 256 | 256 | >512 | 512 | >512 | 512 |
| Piperacillin-tazobactam† | 1 | 8 | 2 | 2 | 2 | 4 | 2 |
| Mecillinam | 0.125 | 1 | 0.5 | 2 | 2 | 2 | 0.5 |
| Cephalothin | 8 | >1,024 | 1,024 | >1,024 | >1,024 | >1,024 | >1,024 |
| Cefuroxime | 4 | >256 | >256 | >256 | >256 | >256 | >256 |
| Cefoperazone | ≤0.25 | 256 | 16 | >512 | 256 | 256 | 64 |
| Ceftriaxone | 0.03 | 32 | 32 | >512 | 256 | 256 | 64 |
| Ceftriaxone-clavulanic acid* | 0.06 | 0.25 | 0.06 | ≤0.016 | ≤0.016 | 0.125 | 0.06 |
| Cefotaxime | 0.06 | 32 | 32 | 512 | 512 | 256 | 128 |
| Cefotaxime-clavulanic acid* | 0.06 | 0.5 | 0.06 | 0.25 | 0.03 | 0.125 | 0.06 |
| Ceftazidime | 0.25 | 2 | 1 | 128 | 64 | 16 | 16 |
| Ceftazidime-clavulanic acid* | 0.125 | 0.5 | 0.25 | 0.25 | 0.25 | 0.5 | 0.25 |
| Aztreonam | 0.06 | 8 | 8 | 128 | 64 | 64 | 64 |
| Aztreonam-clavulanic acid* | 0.06 | 0.5 | 0.125 | 0.06 | 0.06 | 0.25 | 0.125 |
| Cefepime | 0.03 | 8 | 2 | 32 | 8 | 16 | 4 |
| Cefoxitin | 4 | 32 | 4 | 16 | 4 | 16 | 8 |
| Imipenem | 0.25 | ≤0.06 | 0.5 | 0.25 | 0.25 | 0.125 | 0.125 |
| Ciprofloxacin | ≤0.125 | 256 | ≤0.125 | 1,024 | ≤0.125 | 8 | ≤0.125 |
| Gentamicin | ≤0.125 | 32 | 0.5 | 1 | 0.25 | 16 | ≤0.125 |
| Trimethoprim | ≤0.125 | >64 | ≤0.125 | 0.25 | ≤0.125 | 64 | ≤0.125 |
E. coli DH5α-URA5776 (harboring CTX-M-14 enzymes), E. coli DH5α-URA5905 (harboring TEM-1B and CTX-M-15 enzymes), and E. coli DH5α-URA5924 (harboring CTX-M-32 enzymes) were transformants of E. coli INSRA5776 (harboring TEM-1B and CTX-M-14 enzymes), E. coli INSRA5905 (harboring TEM-1B and CTX-M-15 enzymes), and E. coli INSRA5924 (harboring TEM-1B and CTX-M-32 enzymes), respectively; E. coli DH5α was the recipient.
*, Clavulanic acid, 2 μg/ml; †, tazobactam, 4 μg/ml.
IEF.
As determined by IEF, 82% of the CTX-M group 1 strains (n = 110) contained enzymes with pIs 5.4, 7.4, and 8.9 (Table 5); 9% contained enzymes with pIs of 5.4 and 8.9; and 9% contained enzymes with pIs of 7.4 and 8.9. All CTX-M group 9 strains (n = 9) contained an enzyme of pI 8.1, and 44% contained an enzyme with a pI of 5.4.
TABLE 5.
Phenotypic and genotypic characteristics of 119 E. coli CTX-M producer strains
| Antimicrobial resistance patterna | pI(s)b | Genotype by PCR (gene type by sequencing)c:
|
PFGE profiled (no. of strains) | Hospital code | Total no. of strains | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| blaTEM | blaCTX-M | blaOXA | ISEcp1 | IS26 | IS903 | |||||
| AM A/S AT CF CT FX TX TZ XM KZ PM PI P/T TI CI NX OF AK GM TO | 5.4; 7.4; 8.9 | + (1b) | + (15) | + (30) | + | − | − | 0010 | A | 1 |
| AM A/S A/C AT CF CT TX TZ XM KZ PM PI P/T TI CI NX OF GM TO | 7.4; 8.9 | − | + (15) | + (30) | + | − | − | 0010 | D | 2 |
| AM A/S A/C AT CF CT TX TZ XM KZ PM PI P/T TI CI NX OF AK TO | 5.4; 7.4; 8.9 | + (1b) | + (15) | + (30) | + | − | − | 0006 | E | 1 |
| AM A/S AT CF CT TX TZ XM KZ PM PI P/T TI CI NX OF AK GM TO | 5.4; 7.4; 8.9 | + (1b) | + (15) | + (30) | + | − | − | 0010 | C | 1 |
| AM A/S AT CF CT TX TZ XM KZ PM PI P/T TI CI NX OF GM TO T/S | 5.4; 7.4; 8.9 | + (1b) | + (15) | + (30) | + | − | − | 0010 | C | 1 |
| AM A/S AT CF CT TX TZ XM KZ PM PI TI CI NX OF AK GM TO T/S | 5.4; 7.4; 8.9 | + (1b) | + (15) | + (30) | + | − | − | 0010 | B | 1 |
| AM A/S AT CF CT TX TZ XM KZ PM PI P/T TI CI NX OF GM TO | 5.4; 7.4; 8.9 | + (1b) | + (15) | + (30) | + | − | − | 0010 | C | 1 |
| AM A/S AT CF CT TX TZ XM KZ PM PI TI CI NX OF GM TO T/S | 5.4; 7.4; 8.9 | + (1b) | + (15) | + (30) | + | − | − | 0009 (1), | A, B, C, D, E, F, H | 19 |
| 0010 (13), 0012 (1), | ||||||||||
| 0013 (1), 0015 (1), 0017 (1), ND (1) | ||||||||||
| 5.4; 7.4; 8.9 | + (1)e | + (15) | + (30) | + | − | − | 0010 | C | 1 | |
| 5.4; 8.9 | + (1b) | + (32) | − | + | − | − | 0019 | E | 1 | |
| 5.4; 8.9 | + (1b) | + (15) | − | + | − | − | 0019 | E | 1 | |
| AM A/S AT CF CT TX TZ XM KZ PM PI TI CI NX OF GM TO | 8.1 | − | + (14) | − | + | − | − | 0005 | C | 1 |
| 7.4; 8.9 | − | + (15) | + (30) | + | − | − | 0007 (1), 0010 (3) | B, D, H | 4 | |
| 5.4; 7.4; 8.9 | + (1b) | + (15) | + (30) | + | − | − | 0016 (1), 0009 (2), | A, B, C, D, E, G, H, I | 52 | |
| 0010 (48), 0011 (1) | ||||||||||
| 5.4; 7.4; 8.9 | + (1a) | + (15) | + (30) | + | − | − | 0010 | H | 1 | |
| AM A/S AT CF CT TX TZ XM KZ PM PI TI CI NX OF TO T/S | 5.4; 7.4; 8.9 | + (1b) | + (15) | + (30) | + | − | − | 0010 | I | 1 |
| AM A/S AT CF CT TX TZ XM KZ PM PI TI CI NX OF GM | 5.4; 8.9f | + (1b) | + (15) | + (30) | + | − | − | 0010 | A | 1 |
| AM A/S AT CF CT TX TZ XM KZ PM PI TI CI NX OF TO | 7.4; 8.9 | − | + (15) | + (30) | + | − | − | 0010 | D, E | 3 |
| 5.4; 8.9 | + (1b) | + (15) | − | + | − | − | 0010 | D | 1 | |
| 5.4; 7.4; 8.9 | + (1b) | + (15) | + (30) | + | − | − | 0006 (2), 0008 (1), 0010 (7) | C, D, E, I | 10 | |
| AM A/S AT CF CT TX TZ XM KZ PM PI TI CI NX OF T/S | 5.4; 8.9 | + (1b) | + (15) | − | + | − | − | 0010 | A | 2 |
| AM AT CF CT TX TZ XM KZ PM PI TI CI NX OF TO | 7.4; 8.9 | − | + (15) | + (30) | + | − | − | 0010 | E | 1 |
| AM A/S AT CF CT TX TZ XM KZ PM PI TI CI NX OFg | 5.4; 8.9 | + (1b) | + (15) | − | + | − | − | 0006(1), 0010 (1) | E | 2 |
| AM A/S CF CT TX XM KZ PI TI CI NX OF GM TO T/S | 5.4; 8.1 | + (1b) | + (14) | − | + | − | + | 0001 | E | 1 |
| AM A/S CF CT TX XM KZ PI TI CI NX OF T/S | 5.4; 8.1 | + (1b) | + (14) | − | + | − | − | ND | C | 1 |
| AM A/S AT CF CT TX XM KZ PM PI TI T/Sg | 5.4; 8.9 | + (1b) | + (15) | − | + | − | − | 0004 | E | 1 |
| AM A/S CF CT TX XM KZ PM PI TI T/Sg | 5.4; 8.1 | + (1b) | + (14) | − | + | − | − | 0018 | C | 1 |
| AM A/S CF CT TX XM KZ PM PI TIg | 8.1 | − | + (14) | − | + | − | − | 0002 (1), 0003 (2), ND (1) | C, D, I | 4 |
| 5.4; 8.1 | + (1c) | + (14) | − | + | − | − | 0014 | D | 1 | |
| AM AT CF TX XM KZ PI TIg | 5.4; 8.9 | + (1b) | + (15) | − | + | − | − | 0010 | E | 1 |
AM, ampicillin; A/S, ampicillin-sulbactam; A/C, amoxicillin-clavulanic acid; AT, aztreonam; CF, cephalothin; FX, cefoxitin; CT, cefotaxime; TX, ceftriaxone; TZ, ceftazidime; XM, cefuroxime; KZ, cefazolin; PM, cefepime; PI, piperacillin; P/T, piperacillin-tazobactam; TI, ticarcillin; CI, ciprofloxacin; NX, norfloxacin; OF, ofloxacin; AK, amikacin; GM, gentamicin; TO, tobramycin; T/S, trimethoprim-sulfamethoxazole.
pIs were determined by IEF: 5.4 corresponds to TEM-1A, TEM-1B, or TEM-1C enzyme production; 7.4 corresponds to OXA-30 production; 8.1 corresponds to CTX-M-14; and 8.9 corresponds to CTX-M-15 or CTX-M-32 enzyme production.
blaTEM genes amplified by PCR were identified by sequencing as blaTEM-1A, blaTEM-1B, or blaTEM-1C; blaCTX-M genes were identified by sequencing as blaCTX-M-14, blaCTX-M-15, or blaCTX-M-32; and blaOXA genes were identified by sequencing as blaOXA-30. +, Amplification by PCR; −, no amplification by PCR.
PFGE profile and number of strains with each profile, deduced from the dendrogram presented on Fig. 1. ND, not determined.
More than one blaTEM-1 was present.
β-Lactamase with pI 7.4, corresponding to OXA-30 enzyme production, was not detected by IEF.
Non-multidrug-resistant pattern.
Identification of bla genes and IS elements.
The PCR tests used detected a blaCTX-M gene and the ampC gene in all isolates. A total of 91 of the 119 strains (76%) also contained blaTEM and blaOXA genes, 13 (11%) contained blaCTX-M plus blaTEM but not the blaOXA gene, and 10 (8%) had blaCTX-M and blaOXA genes (Table 5). Sequencing identified the blaCTX-M-15, blaCTX-M-14, and blaCTX-M-32 genes in 109 (92%), 9 (8%), and 1 (1%) strains, respectively; the blaTEM-1B gene was identified in 101 (85%) strains, blaTEM-1C was identified in 1 strain, and blaTEM-1A was identified in 1 strain (note that all of these blaTEM genes had a P3 promoter region). One strain had more than one blaTEM gene sequence. A total of 101 (85%) strains harbored a blaOXA-30 gene. No strain carried the blaSHV gene. In all strains, the ISEcp1 element was found upstream from the blaCTX-M genes. One strain also carried an IS903 element downstream from its blaCTX-M-14 gene (Table 5).
Clonality of CTX-M producer strains.
PFGE analysis was used to establish the genetic relatedness of the 116 CTX-M producer strains. We identified 19 PFGE profile types: 14 included a single clone genetically unrelated to other isolates in the study (types 0001, 0002, 0004, 0005, 0007, 0008, and 0011 to 0018). The five other types, defined as clusters I to V, included numerous related (>80% similarity) or indistinguishable (100% homology) isolates (Fig. 1). Of the 102 (88%) strains in these five clusters, 91 (89%) had profile type 0010 (cluster IV). These 91 isolates included clones from all hospitals (isolated from 53 outpatients, 36 inpatients, and 2 with no information) (Tables 1 and 5 and Fig. 1).
FIG. 1.
Genetic relatedness among 123 E. coli strains by PFGE. PFGE profile types and clusters are shown from left to right. A total of 116 strains were the first isolate from each patient; seven additional isolates were collected from three of the patients. Strains INSRA5966, INSRA5979, INSRA5876, INSRA5879, INSRA5907, INSRA5863, INSRA5758, and INSRA5775 are multiple isolates from three patients. Strains with PFGE profile types 0003, 0006, 0009, 0010, and 0019 were defined as forming clusters I to V, respectively (indicated by vertical bands on the right).
Hospital C provided more isolates than any other hospital, including four single-isolate profile types (0005, 0011, and 0018) and two profile types including numerous isolates (0003 and 0010) (Fig. 1). All isolates from the internal medicine, attending, intensive care unit (ICU), and urology services of hospital C were profile type 0010. Only the nephrology, pediatric, and emergency services of hospital C presented clones belonging to other different profile types (Table 6).
TABLE 6.
Distribution of 47 strains from hospital C with corresponding PFGE profile types, by service, between May 2004 and May 2005
| Service | Profile type (no. of strains)a
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2004
|
2005
|
||||||||||||
| May | June | July | Aug | Sep | Oct | Nov | Dec | Jan | Feb | Mar | Apr | May | |
| Cardiology | 0010 (1) | ||||||||||||
| Attending | 0010 (1) | 0010 (2) | 0010 (3) | 0010 (1) | 0010 (1) | ||||||||
| Emergency room | 0010 (1) | 0010 (1) | 0010 (2) | 0011 (1) | 0010 (1) | ||||||||
| Gastrology | 0010 (1) | ||||||||||||
| ICU | 0010 (1) | ||||||||||||
| Internal medicine | 0010 (1) | 0010 (2) | 0010 (1) | 0010 (3) | 0010 (1) | 0010 (4) | 0010 (2) | 0010 (1) | 0010 (1) | ||||
| Nephrology | 0018 (1) | ND | |||||||||||
| Pediatric | 0003 (1) | 0005 (1) | |||||||||||
| Pneumology | 0010 (1) | 0010 (ND) | |||||||||||
| Surgery | 0010 (1) | ||||||||||||
| Unknown | 0010 (1) | ||||||||||||
| Urology | 0010 (2) | 0010 (3) | |||||||||||
ND, not determined.
Isolates of profile types 0006 to 0013, 0015 to 0017, and 0019 expressed a CTX-M enzyme of group 1 and were multidrug resistant. Isolates of profile types 0001 and 0005 were also multidrug resistant and expressed CTX-M enzyme of group 9. Isolates with profile types 0002, 0003, 0014, and 0018 also produced the CTX-M group 9 enzyme but were not multidrug resistant (Table 5).
Three patients (from three different hospitals, C, D and E) gave multiple isolates; all provided isolates with PFGE profile type 0010, although the patient from hospital D also gave one isolate of PFGE profile type 0007 (Table 1).
DISCUSSION
The prevalence of CTX-M-type β-lactamases has increased substantially since 1992 (3): in Portugal, they have been found in isolates from animals, healthy humans, and patients (12, 26). Here, we investigated the dissemination of these enzymes among clinical strains responsible for community and hospital-acquired infections. In a survey conduced in 1999, no ESBL enzymes of the CTX-M family were detected in clinical E. coli isolates in Portugal (M. Caniça, unpublished data).
We included only strains resistant to extended-spectrum cephalosporins and monobactam-producing CTX-M β-lactamases collected in three different regions of Portugal between March 2004 and March 2006: 119 (66%) CTX-M producer strains out of 181 ESBL producers. The majority of the strains had CTX-M enzymes of group 1, CTX-M-15 (n = 109) and CTX-M-32 (n = 1), and 9 strains had CTX-M-14, an enzyme of group 9. Most of CTX-M-15 producers were isolated in urine from men or women ≥60 years old as in Spain (35).
Strains producing CTX-M of group 1 were more resistant to ceftazidime and aztreonam (100%) than those producing CTX-M of group 9 (11%). This is consistent with previous reports (3). CTX-M-14 differs from its parental enzyme CTX-M-9 by a single substitution (Ala231→Val), which does not confer an extension of resistance (37, 45). CTX-M-32 differs from its parental enzyme CTX-M-1 by a single substitution (Asp240→Gly), which confers high-level resistance to ceftazidime (8). The same substitution is found in CTX-M-15 relative to its parental enzyme CTX-M-3, similarly conferring high-level resistance to ceftazidime (41).
A total of 98% of strains producing CTX-M of group 1 were resistant to quinolones and 95% were resistant to aminoglycosides; only 33 and 22% of isolates producing CTX-M of group 9 were resistant, respectively. Resistance to quinolones (93%) was considerably more prevalent among our isolates than the 21% reported by Edelstein (15); 89% of the isolates in both studies were resistant to aminoglycosides. Resistance to trimethoprim-sulfamethoxazole was more prevalent in isolates carrying CTX-M of group 9 (33%) than CTX-M of group 1 (26%).
Combined production of CTX-M and OXA enzymes by E. coli improved resistance to β-lactamase inhibitors. Less than 24% of the isolates were nonsusceptible to clavulanate and tazobactam, and more than 98% were resistant to sulbactam (Table 3). The usefulness of β-lactams plus a β-lactamase inhibitor is uncertain for patients with treatment failure (24). Ninety-two percent of our strains harboring the blaCTX-M-15 gene (Table 5) also possessed a blaOXA-30 gene, presumably explaining the high percentage of nonsusceptibility to ampicillin-sulbactam. Strains carrying the blaCTX-M of group I enzymes were the only strains not susceptible to amoxicillin-clavulanate and to piperacillin-tazobactam, which may imply a low expression of β-lactamase OXA-30 or even another mechanism of resistance (34).
The genes blaCTX-M-15 plus blaOXA-1 (blaOXA-30) have been found to be associated in the same strain in India (21), the United Kingdom (24), and Canada (4). Note that blaOXA-1 and blaOXA-30 have the same sequence and thus code for the same enzyme, called OXA-1 or OXA-30 (36, 46). The combination of blaCTX-M-15 plus blaOXA-30 (or blaOXA-1) plus blaTEM-1 has been reported in seven strains from Korea (20), in two strains from Senegal (50), and also in Spain (35). An association between blaCTX-M-14 and blaTEM-1B has been described in Korea (22). However, our detection of blaCTX-M-32 in the same strain as blaTEM-1B is, to our knowledge, a first.
There was an ISEcp1 element upstream from all blaCTX-M genes detected. Thus, we add the blaCTX-M-32 gene to the list of blaCTX-M genes associated with the ISEcp1 element (1, 4, 13, 21, 31, 40, 49), which contributes to their mobilization (40). The IS903 element was detected downstream from only the blaCTX-M-14 gene, a finding in agreement with other studies (13, 26). The IS26 element, characteristic of the epidemic strain A from the United Kingdom, was not detected among our strains but was recently detected in clinical isolates from Spain that were collected in the same period as the strains of our study (35, 51).
We tested for gene transfer from six isolates and genes were successfully transferred by electroporation to another E. coli strain from only three. This may suggest a major horizontal transfer by mobile elements. We obtained transformants containing CTX-14 alone, CTX-M-32 alone, and CTX-M-15 plus TEM-1B. The clinical strain containing CTX-M-14 enzyme was the only one not susceptible to cefoxitin; however, the transformant was susceptible, indicating another mechanism of resistance. None of the transformants presented the same resistance to quinolone, aminoglycosides, and trimethoprim as the clinical strains. Nevertheless, plasmid-determined resistance to quinolones and aminoglycosides, recently described in Portugal (25), involving a variant of aminoglycoside acetyltransferase AAC(6′)-Ib (44), was not possible to identify in the three studied strains.
Resistance to quinolones (93%), aminoglycosides (89%), and trimethoprim-sulfamethoxazole (26%) explains why most CTX-M producers were multidrug resistant. Another study reported lower prevalences of nonsusceptibility to quinolones (55%) and aminoglycosides (37%) but more frequent resistance to trimethoprim-sulfamethoxazole (34%) (16), and yet another describes very different percentages of resistance for trimethoprim-sulfamethoxazole (78% resistance) and aminoglycosides (43%) (30). Eckert et al. (14) reported that 58% of isolates were resistant to trimethoprim-sulfamethoxazole and 74% were resistant to aminoglycosides and that 100% of CTX-M enzymes producers were multidrug resistant. In our study, the value was 92% for multidrug resistance, with a higher percentage of isolates from the community (50%) than from hospital environments (37%), which was consistent with the predominance of E. coli expression of the CTX-M enzyme in the community described by Pitout et al. (39). Indeed, the high-level consumption of antibiotics in outpatients in Portugal may be responsible for this resistance (19). A wide diversity of resistance genes of numerous families coding for various antibiotic resistance mechanisms have now been described in CTX-M-15 producer strains.
We used PFGE to classify the strains. Three quarters of the strains clustered together (>80% similarity) in cluster IV, indicating countrywide dissemination of this multidrug-resistant clone. Various factors may have contributed to this dissemination: the small size of the country, the proximity of population areas, and/or an inadequate antibiotic use. Since all strains were susceptible to carbapenems, the use of these or other appropriate drugs could help reduce the prevalence of strains from cluster IV. Note that similar values for susceptibility to carbapenems were reported by others (23, 47, 50). The spread of a single clone in hospital C suggested nosocomial dissemination, especially in the internal medicine service. Nevertheless, the genetic similarity of clones in community services, in particular the attending and emergency rooms, also suggest dissemination within the community. The hospital-community and community-hospital dissemination of the blaCTX-M-15 enzyme, mainly in hospital C (Tables 1 and 6), suggest the presence of an epidemic strain. The analysis of the three patients with multiple isolates shows that nosocomial infections due to E. coli CTX-M-15 producer strains are persistent (hospitals D and E), that nosocomial infection is easily transferred among services (hospital D), and that nosocomial infection can become a community infection (hospitals C and D) (data not shown).
Our study suggests that E. coli CTX-M producers are widespread in at least three regions of Portugal, possibly a consequence of the dissemination of major clones between hospitals and community and between regions and of the horizontal transfer of plasmids or mobile elements. Our findings also illustrate the potential dangers that the misuse of antibiotic can cause and the importance of measures to control infection. Nevertheless, rational use of other drugs may improve the situation. As previously noted (24), the use of extended-spectrum cephalosporins, quinolones, and aminoglycosides could be replaced with the use of carbapenems for treating infections in which ESBL-producing strains are likely to emerge.
Acknowledgments
This study was supported financially by the POCTI/2001/ESP/43037 grant from the Fundação para a Ciência e a Tecnologia. N.M. received a grant from NIH Dr. Ricardo Jorge, Lisbon, Portugal.
We thank D. Louro for excellent technical support. We also thank the laboratories participating in the Antibiotic Resistance Surveillance Program in Portugal (ARSIP) for sending E. coli isolates and laboratory records to the Antibiotic Resistance reference laboratory at the NIH: Centro Hospitalar Vila Nova de Gaia, Vila Nova de Gaia (I. Calheiros); Centro Hospitalar do Barlavento Algarvio, Portimão (T. Vaz); Centro Hospitalar do Alto Minho, Viana do Castelo (A. Santos); Hospital Garcia de Orta, Almada (J. Diogo); Hospital Militar de Belém, Lisboa (J. Lago); Hospital São Pedro, Vila Real (A. P. Castro); Hospital Fernando da Fonseca, Amadora (L. Sancho); Hospital São Francisco Xavier, Lisboa (F. Martins); and Hospital Reynaldo dos Santos, Vila Franca de Xira (M. Vasconcelos).
Footnotes
Published ahead of print on 19 March 2007.
REFERENCES
- 1.Baraniak, A., J. Fiett, W. Hryniewicz, P. Nordmann, and M. Gniadkowski. 2002. Ceftazidime-hydrolyzing CTX-M-15 extended-spectrum β-lactamase (ESBL) in Poland. J. Antimicrob. Chemother. 50:393-396. [DOI] [PubMed] [Google Scholar]
- 2.Belaaouaj, A., C. Lapoumeroulie, M. M. Caniça, G. Vedel, P. Névot, R. Krishnamoorthy, and G. Paul. 1994. Nucleotide sequences of the genes coding for the TEM-like β-lactamases IRT-1 and IRT-2 (formerly called TRI-1 and TRI-2). FEMS Microbiol. Lett. 120:75-80. [DOI] [PubMed] [Google Scholar]
- 3.Bonnet, R. 2004. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd, D. A., S. Tyler, S. Christianson, A. McGeer, M. P. Muller, B. M. Willey, E. Bryce, M. Gardam, P. Nordmann, M. R. Mulvey, et al. 2004. Complete nucleotide sequence of a 92-kilobase plasmid harboring the CTX-M-15 extended-spectrum beta-lactamase involved in an outbreak in long-term-care facilities in Toronto, Canada. Antimicrob. Agents Chemother. 48:3758-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caniça, M. M., M. Barthelemy, L. Gilly, R. Labia, R. Krishnamoorthy, and G. Paul. 1997. Properties of IRT-14 (TEM-45), a newly characterized mutant of TEM-type β-lactamases. Antimicrob. Agents Chemother. 41:374-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caniça, M. M., C. Y. Lu, R. Krishnamoorthy, and G. Paul. 1997. Molecular diversity and evolution of blaTEM genes encoding β-lactamases resistant to clavulanic acid in clinical Escherichia coli. J. Mol. Evol. 44:57-65. [DOI] [PubMed] [Google Scholar]
- 8.Cartelle, M., M. M. Tomas, F. Molina, R. Moure, R. Villanueva, and G. Bou. 2004. High-Level resistance to ceftazidime conferred by a novel enzyme, CTX-M-32, derived from CTX-M-1 through a single Asp240-Gly substitution. Antimicrob. Agents Chemother. 48:2308-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casin, I., J. Breuil, A. Brisabois, F. Moury, F. Grimont, and E. Collatz. 1999. Multidrug-resistant human and animal Salmonella typhimurium isolates in France belong predominantly to a DT104 clone with the chromosome- and integron-encoded β-lactamases PSE-1. J. Infect. Dis. 179:1173-1182. [DOI] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. 2007. Performance standards for antimicrobial susceptibility testing; 17th informational supplement. M100-S17. CLSI, Wayne, PA.
- 11.Conceição, T., A. Brízio, A. Duarte, L. M. Lito, J. M. Cristino, and M. J. Salgado. 2005. First description of CTX-M-15-producing Klebsiella pneumoniae in Portugal. Antimicrob. Agents Chemother. 49:477-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costa, D., P. Poeta, L. Brinas, Y. Saenz, J. Rodrigues, and C. Torres. 2004. Detection of CTX-M-1 and TEM-52 beta-lactamases in Escherichia coli strains from healthy pets in Portugal. J. Antimicrob. Chemother. 54:960-961. [DOI] [PubMed] [Google Scholar]
- 13.Eckert, C., V. Gautier, and G. Arlet. 2006. DNA sequence analysis of the genetic environment of various blaCTX-M genes. J. Antimicrob. Chemother. 57:14-23. [DOI] [PubMed] [Google Scholar]
- 14.Eckert, C., V. Gautier, M. Saladin-Allard, H. Hidri, C. Verdet, Z. Ould-Hocine, G. Barnaud, F. Delisle, A. Rossier, T. Lambert, A. Philippon, and G. Arlet. 2004. Dissemination of CTX-M-type β-lactamases among clinical isolates of Enterobacteriaceae in Paris, France. Antimicrob. Agents Chemother. 48:1249-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edelstein, M., M. Pimkin, I. Edelstein, and L. Stratchounski. 2003. Prevalence and molecular epidemiology of CTX-M extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Russian hospitals. Antimicrob. Agents Chemother. 47:3724-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisner, A., E. J. Fagan, G. Feierl, H. H. Kessler, E. Marth, D. M. Livermore, and N. Woodford. 2006. Emergence of Enterobacteriaceae Isolates producing CTX-M extended-spectrum β-lactamases in Austria. Antimicrob. Agents Chemother. 50:785-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Féria, C., E. Ferreira, J. D. Correia, J. Gonçalves, and M. Caniça. 2002. Patterns and mechanisms of resistance to β-lactams and β-lactamase inhibitors in uropathogenic Escherichia coli isolated from dogs in Portugal. J. Antimicrob. Chemother. 49:77-85. [DOI] [PubMed] [Google Scholar]
- 18.Garner J. S., W. R. Jarvis, T. G. Emori, T. C. Horan, and J. M. Hughes. CDC definitions for nosocomial infections, p. A1-A20. In R. N. Olmsted (ed.), APIC infection control and applied epidemiology: principles and practice. Mosby, St. Louis, MO.
- 19.Goossens, H., M. Ferech, R. V. Stichele, M. Elseviers, et al. 2005. Outpatient antibiotic use in Europe and association with resistance: a cross-national database study. Lancet 365:579-587. [DOI] [PubMed] [Google Scholar]
- 20.Jacoby, G. A., K. E. Walsh, D. M. Mills, V. J. Walker, H. Oh, A. Robicsek, and D. C. Hooper. 2006. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob. Agents Chemother. 50:1178-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karim, A., L. Poirel, S. Nagarajan, and P. Nordmann. 2001. Plasmid-mediated extended-spectrum β-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS Microbiol. Lett. 201:237-241. [DOI] [PubMed] [Google Scholar]
- 22.Kim, J., Y.-M. Lim, Y.-S. Jeong, and S.-Y. Seol. 2005. Occurrence of CTX-M-3, CTX-M-15, CTX-M-14, and CTX-M-9 extended-spectrum β-lactamases in Enterobacteriaceae clinical isolates in Korea. Antimicrob. Agents Chemother. 49:1572-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livermore, D. M. 2005. Minimising antibiotic resistance. Lancet Infect. Dis. 5:450-459. [DOI] [PubMed] [Google Scholar]
- 24.Livermore, D. M., and P. M. Hawkey. 2005. CTX-M: changing the face of ESBLs in the UK. J. Antimicrob. Chemother. 56:451-454. [DOI] [PubMed] [Google Scholar]
- 25.Machado, E., T. M. Coque, R. Canton, F. Baquero, J. C. Sousa, L. Peixe, et al. 2006. Dissemination in Portugal of CTX-M-15-, OXA-1-, and TEM-1-producing Enterobacteriaceae strains containing the aac(6′)-Ib-cr gene, which encodes an aminoglycoside- and fluoroquinolone-modifying enzyme. Antimicrob. Agents Chemother. 50:3220-3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Machado, E., T. M. Coque, R. Cantón, J. C. Sousa, and L. Peixe. 2004. Emergence of CTX-M β-lactamase-producing Enterobacteriaceae in Portugal: report of an Escherichia coli isolate harbouring blaCTX-M-14. Clin. Microbiol. Infect. 10:755-757. [DOI] [PubMed] [Google Scholar]
- 27.Matsumoto, Y., F. Ikeda, T. Kamimura, Y. Yokota, and Y. Mine. 1988. Novel plasmid-mediated beta-lactamase from Escherichia coli that inactivates oxyimino-cephalosporins. Antimicrob. Agents Chemother. 32:1243-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mendonça, N., D. Louro, A. P. Castro, J. Diogo, and M. Caniça. 2006. CTX-M-15, OXA-30 and TEM-1-producing Escherichia coli in two Portuguese regions. J. Antimicrob. Chemother. 57:1014-1016. [DOI] [PubMed] [Google Scholar]
- 29.Mendonça, N., E. Ferreira, D. Louro, and M. Caniça. 2006. Occurrence of a novel SHV-type enzyme (SHV-55) among isolates of Klebsiella pneumoniae from Portuguese origin in a comparison study for extended-spectrum β-lactamase-producing evaluation. Diagn. Microbiol. Infect. Dis. 56:415-420. [DOI] [PubMed] [Google Scholar]
- 30.Moubareck, C., Z. Daoud, N. I. Hakime, M. Hamze, N. Mangeney, H. Matta, J. E. Mokhbat, R. Rohban, D. K. Sarkis, and F. Doucet-Populaire. 2005. Countrywide spread of community- and hospital acquired extended-spectrum beta-lactamase (CTX-M-15)-producing Enterobacteriaceae in Lebanon. J. Clin. Microbiol. 43:3309-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munday, C. J., J. Xiong, C. Li, D. Shen, and P. M. Hawkey. 2004. Dissemination of CTX-M type β-lactamases in Enterobacteriaceae isolates in the People's Republic of China. Int. J. Antimicrob. Agents 23:175-180. [DOI] [PubMed] [Google Scholar]
- 32.M'Zali, F., D. M. Gascoyne-Binzi, J. Heritage, and P. M. Hawkey. 1996. Detection of mutations conferring extended-spectrum activity on SHV β-lactamases using polymerase chain reaction single strand conformational polymorphism (PCR-SSCP). J. Antimicrob. Chemother. 37:797-802. [DOI] [PubMed] [Google Scholar]
- 33.Ndugulile, F., R. Jureen, S. Harthug, W. Urassa, and N. Langeland. 2005. Extended spectrum beta-lactamases among gram-negative bacteria of nosocomial origin from an intensive care unit of a tertiary health facility in Tanzania. BMC Infect. Dis. 5:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oliver, A., M. Pérez-Vázquez, M. Martínez-Ferrer, F. Baquero, L. Rafael, and R. Cantón. 1999. Ampicillin-sulbactam and amoxicillin-clavulanate susceptibility testing of Escherichia coli isolates with different β-lactam resistance phenotypes. Antimicrob. Agents Chemother. 43:862-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oteo, J., C. Navarro, E. Cercenado, A. Delgado-Iribarren, I. Wilhelmi, B. Orden, C. Garcia, S. Miguelañez, M. Pérez-Vázquez, S. García-Cobos, B. Aracil, V. Bautista, and J. Campos. 2006. Spread of Escherichia coli strains with high-level cefotaxime and ceftazidime resistance between the community, long-term care facilities, and hospital institutions. J. Clin. Microbiol. 44:2359-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ouellette, M., L. Bissonnette, and P. Roy. 1987. Precise insertion of antibiotic resistance determinants into Tn21-like transposons: nucleotide sequence of the OXA-1 β-lactamase gene. Proc. Natl. Acad. Sci. USA 84:7378-7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pai, H., E.-H. Choi, H.-J. Lee, J. Y. Hong, and G. A. Jacoby. Identification of CTX-M-14 extended-spectrum β-lactamase in clinical isolates of Shigella sonnei, Escherichia coli, and Klebsiella pneumoniae in Korea. J. Clin. Microbiol. 39:3747-3749. [DOI] [PMC free article] [PubMed]
- 38.Pallecchi, L., M. Malossi, A. Mantella, E. Gotuzzo, C. Trigoso, A. Bartoloni, F. Paradisi, G. Kronvall, and G. M. Rossolini. 2004. Detection of CTX-M-type β-lactamase genes in fecal Escherichia coli from healthy children in Bolivia and Peru. Antimicrob. Agents Chemother. 48:4556-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pitout, J. D. D., P. Nordmann, K. B. Laupland, and L. Poirel. 2005. Emergence of Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs) in the community. J. Antimicrob. Chemother. 56:52-59. [DOI] [PubMed] [Google Scholar]
- 40.Poirel, L., M.-F. Lartigue, J.-W. Decousser, and P. Nordmann. 2005. ISEcp1B-mediated transposition of blaCTX-M in Escherichia coli. Antimicrob. Agents Chemother. 49:447-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poirel, L., M. Gniadkowski, and P. Nordmann. 2002. Biochemical analysis of the ceftazidima-hydrolyzing extended-spectrum β-lactamase CTX-M-15 and of its structurally related β-lactamase CTX-M-3. J. Antimicrob. Chemother. 50:1031-1034. [DOI] [PubMed] [Google Scholar]
- 42.Pomba, C., N. Mendonça, M. Costa, D. Louro, B. Baptista, M. Ferreira, J. D. Correia, and M. Caniça. 2005. Evaluation of multiplex PCR for the detection of beta-lactamase Escherichia coli producer strains isolated from animals. Diagn. Microbiol. Infect. Dis. 56:103-106. [DOI] [PubMed] [Google Scholar]
- 43.Ramdani-Bouguessa, N., N. Mendonça, J. Leitão, E. Ferreira, M. Tazir, and M. Caniça. 2006. The spread of CTX-M β-lactamases among Escherichia coli isolates in Mustapha Pacha hospital, Algiers. J. Clin. Microbiol. 44:4584-4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robicsek, A., J. Strahilevitz, G. A. Jacoby, M. Macielag, D. Abbanat, C. H. Park, K. Bush, and D. C. Hooper. 2006. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 2:83-88. [DOI] [PubMed] [Google Scholar]
- 45.Sabaté, M., R. Tarragó, F. Navarro, E. Miro, C. Vergés, J. Barbe, and G. Prats. 2000. Cloning and sequence of the gene encoding a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-9) from Escherichia coli in Spain. Antimicrob. Agents Chemother. 44:1970-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siu, L. K., J. Y. Lo, K. Y. Yuen, P. Y. Chau, M. H. Nq, and P. L. Ho. 2000. β-lactamases in Shigella flexneri isolates from Hong Kong and Shanghai and a novel OXA-1-like β-lactamase, OXA-30. Antimicrob. Agents Chemother. 44:2034-2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soge, O. O., A. M. Queenan, K. K. Ojo, B. A. Adeniyi, and M. C. Roberts. 2005. CTX-M-15 extended-spectrum (beta)-lactamase from Nigerian Klebsiella pneumoniae. J. Antimicrob. Chemother. 57:24-30. [DOI] [PubMed] [Google Scholar]
- 48.Walsh, T. R. 2006. Combinatorial genetic evolution of multiresistance. Curr. Opin. Microbiol. 9:476-482. [DOI] [PubMed] [Google Scholar]
- 49.Walther-Rasmussen, J., and N. Høiby. 2004. Cefotaximases (CTX-M-ases), an expanding family of extended-spectrum β-lactamases. Can. J. Microbiol. 50:137-165. [DOI] [PubMed] [Google Scholar]
- 50.Weill, F.-X., J.-D. Perrier-Gros-Claude, M. Demartin, S. Coignard, and P. A. D. Grimont. 2004. Characterization of extended-spectrum-β-lactamase (CTX-M-15)-producing strains of Salmonella enterica isolated in France and Senegal. FEMS Microbiol. Lett. 238:353-358. [DOI] [PubMed] [Google Scholar]
- 51.Woodford, N., M. E. Ward, M. E. Kaufmann, J. Turton, E. J. Fagan, D. James, A. P. Johnson, R. Pike, M. Warner, T. Cheasty, A. Pearson, S. Harry, J. B. Leach, A. Loughrey, J. A. Lowes, R. E. Warren, and D. M. Livermore. 2004. Community and hospital spread of Escherichia coli producing CTX-M extended-spectrum β-lactamases in the UK. J. Antimicrob. Chemother. 54:735-743. [DOI] [PubMed] [Google Scholar]