Abstract
Structures of yeast Mediator complex, of a related complex from mouse cells and of thyroid hormone receptor-associated protein complex from human cells have been determined by three-dimensional reconstruction from electron micrographs of single particles. All three complexes show a division in two parts, a “head” domain and a combined “middle-tail” domain. The head domains of the three complexes appear most similar and interact most closely with RNA polymerase II. The middle-tail domains show the greatest structural divergence and, in the case of the tail domain, may not interact with polymerase at all. Consistent with this structural divergence, analysis of a yeast Mediator mutant localizes subunits that are not conserved between yeast and mammalian cells to the tail domain. Biochemically defined Rgr1 and Srb4 modules of yeast Mediator are then assigned to the middle and head domains.
Keywords: RNA polymerase II holoenzyme, transcriptional regulation, TRAP, electron microscopy
A Mediator complex with ≈20 polypeptide components is involved in regulation of RNA polymerase II transcription in the yeast Saccharomyces cerevisiae (1, 2). A number of complexes containing subunits homologous to those of the yeast Mediator have been identified in higher organisms. These related complexes include a murine Mediator (3), human thyroid hormone receptor-associated protein (TRAP) complex, a coactivator associated with the thyroid hormone nuclear receptor, and other human complexes (4–9). Mediator and related complexes interact with RNA polymerase II to form holoenzymes and confer on the polymerase both enhanced activity in basal transcription and responsiveness to transcriptional activators. Evidence of direct Mediator–activator interaction has come from the physical isolation of TRAP as a complex with thyroid hormone receptor from hormone-induced but not uninduced cells. Other human Mediators have been isolated as complexes with activators or isolated by activator-affinity chromatography.
Initial structural analysis of yeast, murine Mediators, and holoenzymes was performed by averaging a small number (<100) of electron-microscope images of the complexes viewed in a single orientation in projection. Despite rather limited sequence homology, the yeast and murine complexes appeared remarkably alike in size, shape, and conformational changes associated with Mediator–polymerase interaction (10). Here we present three-dimensional (3-D) structures of yeast and murine Mediators and of human TRAP complex, obtained by averaging hundreds of electron-microscope images from views in random orientations. Beyond confirming the overall similarity of yeast and murine Mediators and extending this result to TRAP, the structures reveal details of surface and internal organization, disclosing further similarities and also notable differences.
Materials and Methods
Sample Preparation and Data Collection.
Yeast Mediator purified from commercial yeast as described (11), mouse Mediator purified as described (3), and human TRAP complex immunopurified from HeLa cells as described (12) were diluted to a concentration of 15–25 μg/ml and applied to specimen grids covered with a thin amorphous carbon film. Because of their relatively low abundance, the amounts of murine Mediator and of the human TRAP complex available for our structural studies were not sufficient for the preparation of unstained specimens; thus, individual particles were imaged in negative stain. The carbon film substrate was glow discharged before preparation of the samples to facilitate adhesion of the molecules and aid staining by making the film surface more hydrophilic. The particles were negatively stained by using a 1% solution of uranyl acetate. After staining, a second, prestained carbon layer was placed over the specimen to ensure complete coverage of the particles by the stain. Samples were examined by using a Philips CM120 electron microscope, equipped with a LaB6 filament. Fields of particles were imaged at 0°, 45°, and 55° degrees to the incident electron beam, at a nominal magnification of 45,000. Samples of a Δsin4 mutant yeast holoenzyme were prepared in the same manner, by using mutant holoenzyme purified as described (13), but the particles were only imaged at 0°. All micrographs were digitized by using a Zeiss SCAI scanner, and transferred to a Silicon Graphics workstation for analysis of the images.
Data Analysis.
All data analysis was carried out by using the spider suite of programs (14). The occurrence of a preferred orientation for both yeast and mouse Mediator and for human TRAP complex particles, made possible the application of the random conical tilt reconstruction method implemented in spider. ≈2,500 particles were included in each data set, and after careful classification and alignment, ≈700 yeast Mediator particles, ≈500 mouse Mediator particles, and ≈300 TRAP particles were used to calculate the reported structures. The initial volume calculated by the back-projection method (15, 16) was refined by centering the images of tilted particles used to calculate the initial volume against corresponding projections of the 3-D structure. Subsequently, the 3-D structure was refined further by cross-correlating each individual particle in the input data set against a set of reference projections calculated from the 3-D structure. This refinement allowed for optimization of the shift and rotation parameters assigned to each individual particle and increased significantly the final resolution of the structure. The final resolution was calculated by dividing each data set in half, calculating two independent reconstructions, and then applying the Fourier ring cross-correlation test (17, 18). For the calculation of the projection map of the Δsin4 mutant holoenzyme, images of individual particles were aligned by using a reference-free algorithm, to ensure that the final map was not biased because of the selection of a particular reference image.
Results
3-D Structures of Mediator Complexes.
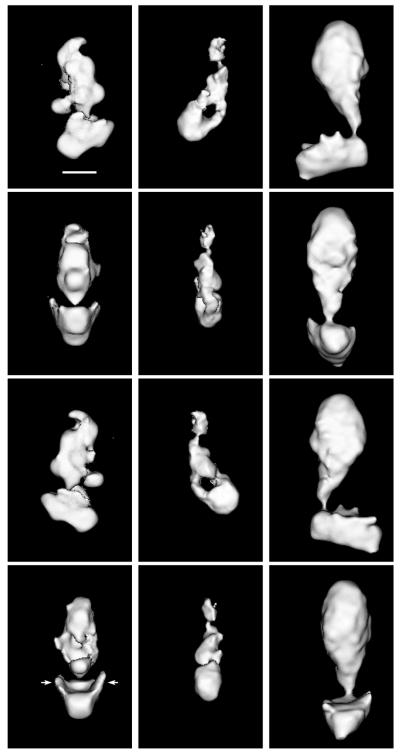
The 3-D structures of yeast and murine Mediators and of TRAP complex are shown in Fig. 1. All three complexes are formed by two separate domains, with the bottom portion of the structures joined at approximately right angles to a larger domain on top. The bottom parts of the structures correspond to the head domain previously identified in yeast Mediator (10). Of particular interest is the presence in the head domain of the yeast Mediator complex of extended, thin, side flaps (arrows in Fig. 1), which are not evident in images of individual particles or in projection maps of yeast Mediator, appear in lower-resolution reconstructions as short protrusions, much like the ones observed along the sides of the head domain in the murine Mediator and TRAP structures. A close interaction between the Mediator head domain and RNA polymerase II has been noted in the structure of the Mediator/RNA polymerase II yeast holoenzyme complex (10). The concave shape of the internal face of the head domains and the presence of side flaps seem tailored to maximize and enhance contact at the Mediator–polymerase interface.
Figure 1.
Shown are 3-D structures of yeast Mediator (Left), murine Mediator (Center), and of the human TRAP complex (Right). The orientation of each complex in consecutive rows differs by 90°. (Top) Complexes in an orientation corresponding to the preferred orientation of the respective particles on the grid. (Bar = 100 Å.) The arrows near the head domain of the yeast Mediator indicate the “flaps” that must wrap around RNA polymerase II when the holoenzyme complex is formed. Cut-off levels for displaying the different maps were chosen to emphasize their overall domain organization. The resolution of the maps is 30–35 Å, as determined by the Fourier-ring correlation method (17), although a loss of resolution in the direction perpendicular to the plane of Top results from incomplete sampling of the structures in that direction.
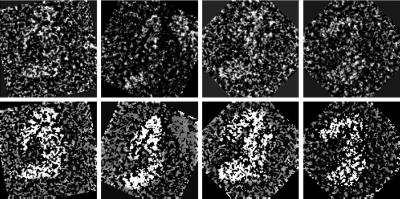
Previous analyses of yeast and murine Mediators also revealed a transition from a compact conformation like that shown in Fig. 1 to an extended conformation in the holoenzyme complex (10), in which the top portion of the Mediator structure separates into clearly distinguishable middle and tail domains. Close examination of single TRAP particles (Fig. 2) reveals a similar characteristic: the top portion of a small percentage (<5%) of TRAP molecules (that were not selected for inclusion in the data set used to calculate the TRAP structure shown in Fig. 1) appears partially extended, indicating that TRAP also undergoes a conformational transition (in the case of yeast Mediator, the fully extended conformation is also observed only in a small percentage of molecules in the absence of RNA polymerase). Although we have not studied the interaction of TRAP with human polymerase, the presence of these partially extended TRAP particles indicates that the top portion of the TRAP structure shown in Fig. 1 is formed by a combination of two (middle- and tail-like) domains.
Figure 2.
Original (Upper), and contrast-enhanced (Lower) images of individual TRAP particles that appear partially extended, revealing middle- and tail-like domains that form the top portion of the TRAP structure shown in Fig. 1. The contrast-enhanced images were produced by applying a threshold to the corresponding original image and then partially obscuring the background around the particle.
Morphological differences among the three Mediator structures were greatest in the middle-tail domains. The order of sizes of these regions corresponded with that of the total masses of the three complexes—greatest for TRAP and smallest for murine Mediator. Although the length of the middle-tail region was similar for the three complexes, the detailed organization of this region appeared completely different.
Structure of a Yeast Mediator Mutant.
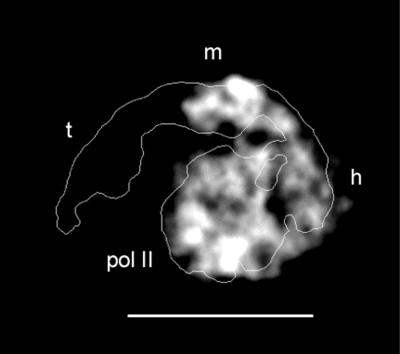
To relate structural features of Mediator complexes to conserved and variable polypeptides, we have undertaken the structural analysis of yeast Mediator-deletion mutants. We began with a sin4 deletion strain, because Mediator and holoenzyme isolated from this strain lack not only Sin4 but also Gal11, Med2, and Pgd1 (“Sin4 module”; refs. 13 and 19). An average (projection map) calculated from ≈50 images of a single view of the Δsin4 holoenzyme is shown in Fig. 3, superimposed on an outline of the projection map of the corresponding wild-type complex. In the extended conformation of Mediator seen in the wild-type holoenzyme, the head, middle, and tail domains of Mediator are all clearly distinguishable; they surround a central, globular, RNA polymerase II. In the Δsin4 holoenzyme, all features are present except for the tail domain. In all likelihood, the components of the Sin4 module—Sin4, Gal11, Med2, and Pgd1—constitute the tail domain.
Figure 3.
Gray-scale projection map of a ΔSin4 mutant yeast holoenzyme (lacking subunits Sin4, Gal11, Med2, and Pgd1), calculated after alignment and averaging of ≈50 molecules preserved in uranyl acetate. (Bar = 200 Å.) An outline of the wild-type holoenzyme complex is shown for comparison (10). Mediator in the mutant holoenzyme appears wrapped around polymerase (pol II) in a extended conformation similar to that observed for the wild-type complex, but comparison of the structures reveals that the “tail” domain (labeled t in the wild-type holoenzyme outline) is missing, whereas the head (h) and middle (m) domains are present and interact with RNA polymerase as they do in the wild-type holoenzyme.
Discussion
Our success in computing 3-D structures of Mediator complexes from images of randomly oriented, individual particles attests to the discrete nature and homogeneity of the complexes. The similarities between the yeast and mammalian complexes in both domain organization and details of head-domain morphology strengthen the proposition that the mammalian complexes are true counterparts of yeast Mediator (10). Morphological differences are concentrated in the region where the nonconserved Sin4 module is located in the yeast complex.
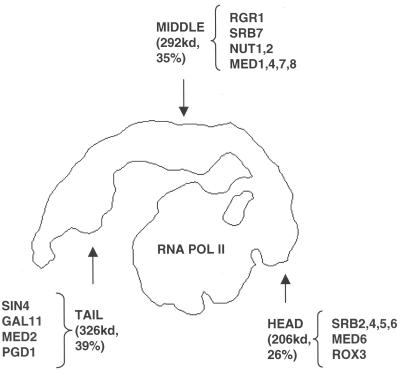
From the location of the Sin4 module, we may extrapolate to the arrangement of other Mediator components in the RNA polymerase II holoenzyme structure. The Sin4 and Rgr1 subunits are physically associated in vivo (20), and the Sin4 module is known to interact with the rest of the Mediator complex through the carboxyl-terminal domain (CTD) of the Rgr1 subunit, because deletion of this domain causes the loss of the entire module (13). These observations, together with the absence of physical interaction between head and tail domains in the extended conformation of Mediator (and in the compact conformation as well, based on inspection of the yeast Mediator structure in Fig. 1), indicate that Rgr1 is part of the middle domain.
Biochemical studies have shown a subdivision of yeast Mediator into an Rgr1 module (with an associated Sin4 module) and an Srb4 module (21). With the Sin4 module now relegated to the tail and Rgr1 relegated to the middle domain, the Srb4 module (21, 22) would plausibly correspond to the head domain (Fig. 4). This assignment of the Srb4 module is consistent with the genetic characterization of Srb proteins and the biochemistry of Mediator–RNA polymerase II interaction. Genes for the Srb2, Srb4, Srb5, and Srb6 components of the Srb4 module were identified as dominant, gain-of-function suppressors of Rpb1 CTD truncation phenotypes (23, 24). This evidence of Mediator–CTD interaction in vivo is supported by a requirement for the CTD for Mediator–polymerase interaction in vitro (25). Srb proteins in the head domain may, therefore, make a major contribution to CTD binding, but they are not solely responsible for polymerase interaction. Previous structural studies revealed species specificity of holoenzyme formation (10), despite the evolutionary conservation of the CTD. This finding and the appearance of additional contacts between the middle domain and the polymerase suggest multiple points of Mediator–polymerase interaction.
Figure 4.
Outline of the wild-type yeast holoenzyme and proposed location of Mediator subunit modules identified by biochemical studies. The combined mass of the subunits assigned to each Mediator domain (shown in parenthesis along with the respective percentage of the total mass of the complex) appears consistent with the apparent relative size of the different domains.
What correlation between the biochemical and structural characteristics of Mediator can be made in light of the proposed subunit mapping? All components of the Sin4 module, which form the Mediator tail domain, are dispensable for cell viability (25). Deletion of the Sin4 module results in a holoenzyme that is competent in basal transcription but unresponsive to activation by Gal4-VP16 or Gcn4 (19, 26), a limitation that can be bypassed by artificial recruitment of the mutant holoenzyme to a promoter (26). The components of the Srb4 module seem to play a more general role. For example, although it can bind activators as well as the wild-type enzyme, an Srb5 deletion mutant holoenzyme is incompetent in both basal and activated transcription (24), and this condition cannot be alleviated by artificial recruitment of the mutant holoenzyme to a promoter (26). Involvement of the Srb4 module in control of basal-transcription activity is in keeping with its proposed assignment to the Mediator head domain, which interacts directly with RNA polymerase II (10).
Yeast, murine, and human Mediator complexes are similar in overall organization despite their evolutionary divergence (homologous subunits are only marginally related and account for at most 10–15% of the total mass). We suggest that, although only distantly related biochemically, different Mediator-like complexes share a basic mechanism for transcription regulation that imposes a set of common structural requirements. Our observations indicate significant structural similarities in domains directly involved in interactions with the well conserved RNA polymerase, and structural differences in domains that might be involved in interactions with species-specific activator and repressor proteins (e.g., the Sin4 module that forms the tail domain in yeast Mediator). Our findings are consistent with previous proposals (19, 26) that suggest an activation mechanism involving more than simple recruitment of the transcription machinery to a promoter (27). The structure and organization of Mediator complexes seem to enable the transmission of regulatory information from modules that interact directly with activators and repressors to modules that interact with RNA polymerase and affect its function. The extensive conformational transition that Mediator must undergo to interact with polymerase in the holoenzyme could also play a role in regulation, and studying the effect of different activators and repressors on the conformations of Mediator and holoenzyme might reveal its significance.
Acknowledgments
This work was supported by National Institutes of Health Grant GM-60607 (to F.J.A.).
Abbreviations
- 3-D
three-dimensional
- CTD
carboxyl-terminal domain
- TRAP
thyroid hormone receptor-associated protein
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.260489497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.260489497
References
- 1.Flanagan P M, Kelleher R J, Sayre M H, III, Tschochner H, Kornberg R D. Nature (London) 1991;350:436–438. doi: 10.1038/350436a0. [DOI] [PubMed] [Google Scholar]
- 2.Kim Y J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 3.Jiang Y W, Veschambre P, Erdjument-Bromage H, Tempst P, Conaway J W, Conaway R C, Kornberg R D. Proc Natl Acad Sci USA. 1998;95:8538–8543. doi: 10.1073/pnas.95.15.8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito M, Yuan C X, Malik S, Gu W, Fondell J D, Yamamura S, Fu Z Y, Zhang X, Qin J, Roeder R G. Mol Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- 5.Naar A M, Beaurang P A, Zhou S, Abraham S, Solomon W, Tjian R. Nature (London) 1999;398:828–832. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- 6.Rachez C, Lemon B D, Suldan Z, Bromleigh V, Gamble M, Naar A M, Erdjument-Bromage H, Tempst P, Freedman L P. Nature (London) 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 7.Sun X, Zhang Y, Cho H, Rickert P, Lees E, Lane W, Reinberg D. Mol Cell. 1998;2:213–222. doi: 10.1016/s1097-2765(00)80131-8. [DOI] [PubMed] [Google Scholar]
- 8.Ryu S, Zhou S, Ladurner A G, Tjian R. Nature (London) 1999;397:446–450. doi: 10.1038/17141. [DOI] [PubMed] [Google Scholar]
- 9.Boyer T G, Martin M E, Lees E, Ricciardi R P, Berk A J. Nature (London) 1999;399:276–279. doi: 10.1038/20466. [DOI] [PubMed] [Google Scholar]
- 10.Asturias F J, Jiang Y W, Myers L C, Gustafsson C M, Kornberg R D. Science. 1999;283:985–987. doi: 10.1126/science.283.5404.985. [DOI] [PubMed] [Google Scholar]
- 11.Myers L C, Leuther K, Bushnell D A, Gustafsson C M, Kornberg R D. Methods. 1997;12:212–216. doi: 10.1006/meth.1997.0473. [DOI] [PubMed] [Google Scholar]
- 12.Fondell J D, Ge H, Roeder R G. Proc Natl Acad Sci USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Bjorklund S, Jiang Y W, Kim Y-J, Lane W S, Stillman D J, Kornberg R D. Proc Natl Acad Sci USA. 1995;92:10864–10868. doi: 10.1073/pnas.92.24.10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 15.Harauz G, van Heel M. Optik. 1986;73:146–156. [Google Scholar]
- 16.Radermacher M. J Electron Microsc Technol. 1988;9:359–394. doi: 10.1002/jemt.1060090405. [DOI] [PubMed] [Google Scholar]
- 17.Saxton W O, Baumeister W. J Microsc (Oxford) 1982;127:127–138. doi: 10.1111/j.1365-2818.1982.tb00405.x. [DOI] [PubMed] [Google Scholar]
- 18.van Heel M. Ultramicroscopy. 1987;21:95–100. doi: 10.1016/0304-3991(87)90078-7. [DOI] [PubMed] [Google Scholar]
- 19.Myers L C, Gustafsson C M, Hayashibara K C, Brown P O, Kornberg R D. Proc Natl Acad Sci USA. 1999;96:67–72. doi: 10.1073/pnas.96.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang Y W, Dohrmann P R, Stillman D J. Genetics. 1995;140:47–54. doi: 10.1093/genetics/140.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee Y C, Kim Y J. Mol Cell Biol. 1998;18:5364–5370. doi: 10.1128/mcb.18.9.5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koh S S, Ansari A Z, Ptashne M, Young R A. Mol Cell. 1998;1:895–904. doi: 10.1016/s1097-2765(00)80088-x. [DOI] [PubMed] [Google Scholar]
- 23.Nonet M L, Young R A. Genetics. 1989;123:715–724. doi: 10.1093/genetics/123.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson C M, Koleske A J, Chao D M, Young R A. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 25.Myers L C, Gustafsson C M, Bushnell D A, Lui M, Erdjument-Gromage H, Tempst P, Kornberg R D. Genes Dev. 1998;12:45–54. doi: 10.1101/gad.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee Y C, Park J M, Min S, Han S J, Kim Y. Mol Cell Biol. 1999;19:2967–2976. doi: 10.1128/mcb.19.4.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ptashne M, Gann A. Nature (London) 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]