Abstract
A novel class A β-lactamase (SCO-1) encoded by an 80-kb self-transferable plasmid from Escherichia coli is described. The interaction of SCO-1 with β-lactams was similar to that of the CARB-type enzymes. Also, SCO-1 exhibited a 51% amino acid sequence identity with the RTG subgroup of chromosomal carbenicillinases (RTG-1, CARB-5, and CARB-8).
Production of β-lactamases is the main mechanism of resistance to β-lactam antibiotics, particularly among gram-negative microorganisms (19). Based on their amino acid sequences, β-lactamases have been divided into the molecular classes A, C, and D that include active serine enzymes and the molecular class B enzymes that require zinc ions for activity. Class A comprises numerous clavulanate-inhibited enzymes from a wide range of bacterial species. Despite their structural similarities, class A β-lactamases exhibit an ample diversity of substrate spectra and have been classified into various functional groups (3). The CARB- and PSE-type β-lactamases represent a distinct functional group (2c) of class A penicillinases that preferentially hydrolyze carbenicillin (the carbenicillinases). These β-lactamases are encountered mostly in Pseudomonas aeruginosa (6, 18), as well as in other obligate aerobes such as Acinetobacter baumannii (5, 11), Vibrio cholerae (4, 15, 17), and Alcaligenes xylosoxidans (7). Carbenicillinases also occur in members of the family Enterobacteriaceae at relatively low frequencies (20, 24). In this study, we describe SCO-1, a novel plasmid-mediated class A enzyme with carbenicillinase characteristics, produced by Escherichia coli.
Selection and properties of SCO-1-producing Escherichia coli.
From a PCR-based screening using primers specific for various bla genes, including blaTEM, blaSHV, blaCARB, blaCTX-M, blaCMY, blaACC, and blaOXA of oxyimino-cephalosporin-resistant E. coli isolates recovered from patients in Athens hospitals during 2000 to 2004, an isolate (EC-3521r) positive for blaACC and also blaTEM was identified. E. coli EC-3521r was isolated in 2002 from a urine sample from a patient treated in a general hospital. Partial sequencing of the PCR products confirmed the identity of the bla genes (data not shown). A blaACC-type cephalosporinase gene was detected for the first time in this setting. Thus, E. coli EC-3521r was studied further.
MICs of β-lactam antibiotics were determined by an agar dilution technique. E. coli EC-3521r exhibited a β-lactam resistance phenotype consistent with the production of an ACC-type enzyme (2) (Table 1). Analysis of the β-lactamase content by isoelectric focusing (IEF) of sonicated cell extracts indicated that the isolate produced three main β-lactamase species with pIs of 7.8 (corresponding to ACC-1 [2]), 5.4 (corresponding to TEM-1), and 5.8. The last band, along with TEM-1, was inhibited in situ by clavulanic acid (3 μM).
TABLE 1.
MICs of β-lactam antibiotics for clinical and laboratory E. coli strains carrying SCO-1-encoding plasmids
| β-Lactama | MIC (μg/ml)
|
|||
|---|---|---|---|---|
| E. coli 3521r(pR3521) | E. coli K-12(pR3521) | E. coli DH5α(pSCO-1) | E. coli DH5α | |
| Amoxicillin | ≥256 | ≥256 | ≥256 | 4 |
| Amoxicillin + CL | 128 | ≥256 | 8 | 2 |
| Ticarcillin | ≥256 | ≥256 | ≥256 | 2 |
| Ticarcillin + CL | 128 | 64 | 16 | 2 |
| Carbenicillin | 128 | 64 | 128 | 1 |
| Piperacillin | 128 | 64 | 32 | 2 |
| Piperacillin + TZ | 64 | 32 | 2 | 0.5 |
| Cephalothin | ≥256 | ≥256 | 8 | 2 |
| Cefoxitin | 8 | 8 | 2 | 2 |
| Cefuroxime | 32 | 32 | 1 | 1 |
| Cefotaxime | 8 | 4 | ≤0.12 | ≤0.12 |
| Ceftazidime | 32 | 32 | ≤0.12 | ≤0.12 |
| Aztreonam | 1 | 1 | ≤0.12 | ≤0.12 |
| Cefepime | 0.5 | 0.25 | ≤0.12 | ≤0.12 |
| Imipenem | 0.25 | 0.25 | ≤0.12 | ≤0.12 |
CL, clavulanate at a fixed concentration of 2 μg/ml; TZ, tazobactam at a fixed concentration of 4 μg/ml.
Conjugal transfer of blaSCO-1 and plasmid characterization.
Mating experiments were performed in mixed broth cultures as described previously (10), using a rifampin-resistant E. coli K-12 lac mutant strain (29R793) as the recipient. β-Lactam-resistant transconjugants were selected with Mueller-Hinton agar containing rifampin (150 μg/ml) and ampicillin (40 μg/ml). The β-lactam resistance phenotype of EC-3521r was readily transferred (Table 1) at a frequency of approximately 10−4 per donor cell. Transconjugant clones also acquired resistance to streptomycin and chloramphenicol, as found by a disk diffusion test. Analysis of plasmid DNA content of the donor and the transconjugant clones by an alkaline lysis technique (10) indicated transfer of an 80-kb plasmid designated pR3521. Purified DNA preparations from pR3521 were positive for blaTEM and blaACC by PCR assays. As also found by IEF, pR3521-harboring transconjugants produced the same three main β-lactamases as the donor strain did. These data showed that pR3521 encoded multiple β-lactamases, including an unidentified enzyme that was inhibited by clavulanic acid and that had an apparent pI of 5.8.
Cloning and sequencing of blaSCO-1.
Plasmid pR3521 was partially digested with the endonuclease Sau3A, and the fragments were ligated into the polycloning site of the pBCSK(+) vector (Stratagene, La Jolla, CA). The resulting recombinant plasmids were used to transform E. coli DH5α competent cells by electroporation. Transformants were selected with medium supplemented with chloramphenicol (20 μg/ml) and ampicillin (40 μg/ml). β-Lactam-resistant clones were analyzed by IEF as well as by PCR for blaTEM and blaACC. A clone that produced only the β-lactamase species with a pI of 5.8 and that was negative for blaTEM and blaACC was identified. Determination of MICs of β-lactams showed that this clone was resistant to penicillins but not to cephalosporins. MICs of penicillins were decreased by clavulanic acid and tazobactam (Table 1). The respective recombinant plasmid (pSCO-1) was purified with a Plasmid Midi kit (QIAGEN, Hilden, Germany), and the nucleotide sequence of the bla-carrying insert was determined on both strands by using an ABI 377 sequencer (Applied Biosystems, Foster City, CA). pSCO-1 carried a 3,833-bp Sau3A fragment containing an open reading frame (ORF) of 867 bp (from nucleotide [nt] 1494 to nt 2360; GenBank accession no. EF104648), homologous to the orf1 recently observed in ACC-1-encoding plasmids from Klebsiella pneumoniae and Salmonella enterica serovar Bareilly, isolated in France and The Netherlands, respectively (from nt 2809 to nt 3675; GenBank accession no. AJ870922) (8). This ORF did not exhibit significant homology with any known sequence. However, the deduced polypeptide (288 amino acid [aa] residues) possessed the typical motifs of a class A β-lactamase (14). Therefore, orf1 was identified as a bla gene and designated blaSCO-1.
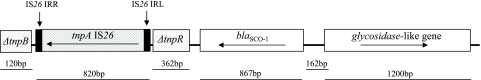
The 867-bp-long blaSCO-1 gene had a G+C content of 51.9%. The codon adaptation index (CAI) (21) was 0.657. Putative −35 (TTTAGA) and −10 (TAAATT) promoter regions, separated by 19 bp, were found in the sequence preceding blaSCO-1. An ORF of 1,200 bp (G+C content, 48.7%; CAI, 0.632) in the opposite orientation was located 162 bp upstream of blaSCO-1. The deduced polypeptide (399 aa) contained motifs typical for the family 5 glycosidases (cellulases) and was 53% similar to a glycosidase from Pseudomonas putida (GenBank accession no. NC002947). Homologies of the remaining right-hand sequence (113 bp) of the Sau3A insert with published sequences were not detected. At a 547-bp distance downstream of blaSCO-1, an intact IS26 element (820 bp), including tnpA and the respective inverted repeats IRL and IRR, was identified. The IS26 sequence was bracketed by sequences of 120 bp (on the left) and 362 bp (on the right), corresponding to internal fragments of a putative transposase (tnpB) and a resolvase (tnpR) gene, respectively. Both genes were described in the strA-strB-containing regions from S. enterica serovar Typhimurium DT193 (GenBank accession no. AY524415) and the Tn5393 transposon from Erwinia amylovora (GenBank accession no. M96392), respectively (Fig. 1).
FIG. 1.
Schematic representation of the 3,833-bp Sau3A insert of the recombinant plasmid pSCO-1 encoding the β-lactamase SCO-1. Arrows indicate the translational orientation of the respective genes. Sizes of the respective nucleotide sequences and intervening regions are indicated below the schematic.
Characteristics of SCO-1.
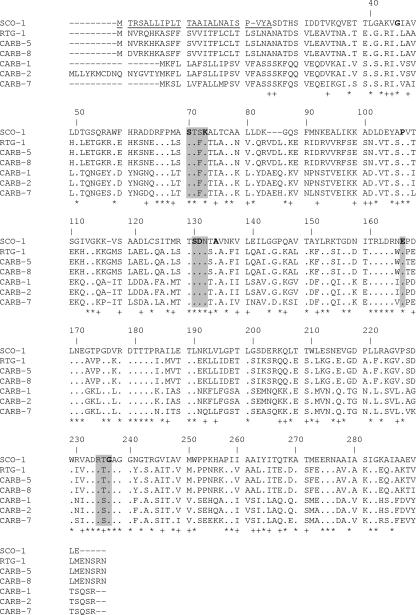
The likely secretory signal sequence of SCO-1 comprised 25 amino acid residues. The mature β-lactamase (263 aa) would have a molecular mass of 28,097 kDa. The calculated pI (5.8) matched the apparent pI of the native form. SCO-1 contained the conserved amino acid residues of class A β-lactamases, including Ser-70, Lys-73, Ser-130, Asn-132, and Glu-166 (Ambler et al.'s numbering scheme [1]) (14). A BLASTP search showed that SCO-1 was distantly related to CARB-type β-lactamases, exhibiting the highest similarity scores with the chromosomal carbenicillinases RTG-1 from Proteus mirabilis GN79 (51% identity) (20), CARB-5 from A. calcoaceticus subsp. anitratus (51% identity) (5), and CARB-8 from Oligella urethralis (51% identity) (13). Identities with other group 2c β-lactamases ranged from 45 to 47%. Position 234 was occupied by an arginine residue, which is characteristic for the CARB-type β-lactamases and is considered important for carbenicillinase activity (12). Additionally, SCO-1 contained Thr-235 as RTG-1, CARB-5 (RTG-2), and CARB-8 (RTG-3) (Fig. 2). The latter carbenicillinases are evolutionarily distinct from the remaining CARB enzymes and constitute the so-called RTG subgroup (5). A potential phylogenetic relationship of SCO-1 with the RTG carbenicillinases was also indicated in a dendrogram constructed by the neighbor-joining method based on a ClustalW multiple alignment (not shown).
FIG. 2.
Amino acid sequence alignment of SCO-1 and six carbenicillinases, including those of the RTG subgroup (RTG-1, CARB-5, and CARB-8). The dashes indicate gaps introduced to optimize alignment. Asterisks show positions occupied by identical amino acid residues. Conservative amino acid substitutions are also indicated (+). Residues that are strictly conserved in class A β-lactamases are shown in boldface type. Shaded segments correspond to the structural elements characteristic of class A β-lactamases. The putative secretory peptide of SCO-1 is underlined. Ambler et al.'s numbering scheme was followed (1).
Hydrolysis rates of penicillin G, ampicillin, carbenicillin, oxacillin, cephalothin, cephaloridine, cefotaxime, and ceftazidime by SCO-1 were determined by UV spectrophotometry, as described previously (10). Cell extracts from E. coli K-12 laboratory strains producing CARB-1 (PSE-4) and TEM-1 were also used for comparison. The substrate profile of SCO-1 corresponded to those of the carbenicillinases of the functional group 2c (3). The enzyme was effective against ampicillin and carbenicillin, while the relative hydrolysis rates of cephalothin and cephaloridine were low. Also, as observed for most CARB β-lactamases, hydrolysis of oxacillin by SCO-1 was relatively slow. The inhibitory activities of clavulanic acid, tazobactam, and sulbactam were assessed using penicillin G as the reporter substrate (10). Tazobactam was the most potent inhibitor of SCO-1, followed by clavulanic acid and sulbactam (Table 2).
TABLE 2.
Substrate and inhibitor profiles of SCO-1 β-lactamase and comparison with CARB-1 (PSE-4) and TEM-1
| β-Lactam | β-Lactamasesa
|
|||||
|---|---|---|---|---|---|---|
| SCO-1
|
CARB-1
|
TEM-1
|
||||
| Relative Vmax | IC50 (μM) | Relative Vmax | IC50 (μM) | Relative Vmax | IC50 (μM) | |
| Substrate | ||||||
| Penicillin G | 100 | 100 | 100 | |||
| Ampicillin | 84 | 100 | 108 | |||
| Carbenicillin | 78 | 146 | 15 | |||
| Oxacillin | 4 | 12 | ND | |||
| Cephaloridine | 21 | 15 | 154 | |||
| Cephalothin | 3 | 6 | 27 | |||
| Cefotaxime | <1 | <1 | <1 | |||
| Ceftazidime | <1 | <1 | <1 | |||
| Inhibitor | ||||||
| Clavulanic acid | 0.07 | 0.10 | 0.17 | |||
| Tazobactam | 0.01 | 0.08 | 0.09 | |||
| Sulbactam | 6.2 | 5.0 | 7.3 | |||
Hydrolysis rates are relative to that of penicillin G, which was set at 100. Values are the means of three independent measurements differing by ≤10%. IC50, 50% inhibitory concentration). ND, not determined.
Conclusions.
The data presented here show that SCO-1 is a novel class A β-lactamase functionally related to the group 2c enzymes. Also, a phylogenetic relationship of SCO-1 with the CARB β-lactamases, particularly those of the RTG subgroup, was evident. However, the relevant amino acid sequence identities were limited, suggesting a distinct origin of SCO-1. Unlike the bla genes of the RTG subgroup, which are chromosomal, blaSCO-1 was carried by a self-transferable plasmid. On the other hand, blaSCO-1 occurred as part of an apparently contiguous chromosomal sequence, as indicated by its association with a glycosidase-like gene. Diverse carbenicillinase-like β-lactamases with limited amino acid sequence homologies with CARB enzymes have been described in V. harveyi (23), Moritella marina (22), and Fulvimarina pelagi (GenBank accession no. EAU43273) from aquatic environments. Therefore, blaSCO-1 might have been derived from an unidentified environmental microorganism. The association of the described sequence with IS26, an element that is frequently involved in the mobilization of bla and other resistance genes (9, 16), could play a role in the acquisition of blaSCO-1 by pR3521. Irrespective of its origin, SCO-1 is the first RTG-type carbenicillinase known to be encoded by a plasmid.
A retrospective examination of our oxyimino-cephalosporin-resistant E. coli collection by PCR did not reveal additional isolates positive for blaSCO-1. This collection, however, represents a small fraction of the penicillin-resistant isolates. Therefore, the extent of spread of blaSCO-1 cannot be estimated. Isolation of other enterobacteria that also harbor blaSCO-1-carrying plasmids from other European countries (8) may be of epidemiological importance. Although the sequences bracketing blaSCO-1 were different from those described here, these plasmids also carried blaACC-1 and blaTEM-1 and therefore were probably related to pR3521. Despite its seemingly low prevalence among clinical enterobacteria, this plasmid type may have achieved a geographically wide dispersal. It would be interesting to examine additional ACC-1-encoding plasmids for blaSCO-1 carriage isolated from enterobacteria in other regions.
Nucleotide sequence accession number.
Nucleotide sequences described in this study have been assigned accession no. EF104648 in the GenBank database.
Footnotes
Published ahead of print on 12 March 2007.
REFERENCES
- 1.Ambler, R. P., A. F. Coulson, J.-M. Frère, J. M. Ghuysen, B. Joris, M. Forsman, R. C. Levesque, G. Tiraby, and S. G. Waley. 1991. A standard numbering scheme for the class A β-lactamases. Biochem. J. 276:269-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauernfeind, A., I. Schneider, R. Jungwirth, H. Sahly, and U. Ullmann. 1999. A novel type of AmpC β-lactamase, ACC-1, produced by a Klebsiella pneumoniae strain causing nosocomial pneumonia. Antimicrob. Agents Chemother. 43:1924-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choury, D., G. Aubert, M.-F. Szajnert, K. Azibi, M. Delpech, and G. Paul. 1999. Characterization and nucleotide sequence of CARB-6, a new carbenicillin-hydrolyzing β-lactamase from Vibrio cholerae. Antimicrob. Agents Chemother. 43:297-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choury, D., M.-F. Szajnert, M.-L. Joly-Guillou, K. Azibi, M. Delpech, and G. Paul. 2000. Nucleotide sequence of the blaRTG-2 (CARB-5) gene and phylogeny of a new group of carbenicillin-hydrolyzing β-lactamases. Antimicrob. Agents Chemother. 44:1070-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Champs, C., L. Poirel, R. Bonnet, D. Sirot, C. Chanal, J. Sirot, and P. Nordmann. 2002. Prospective survey of β-lactamases produced by ceftazidime-resistant Pseudomonas aeruginosa isolated in a French hospital. Antimicrob. Agents Chemother. 46:3031-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Decré, D., G. Arlet, E. Bergogne-Berezin, and A. Philippon. 1995. Identification of a carbenicillin-hydrolyzing β-lactamase in Alcaligenes denitrificans subsp. xylosoxydans. Antimicrob. Agents Chemother. 39:771-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doloy, A., C. Verdet, V. Gautier, D. Decré, E. Ronco, A. Hammami, A. Philippon, and G. Arlet. 2006. Genetic environment of acquired blaACC-1 β-lactamase gene in Enterobacteriaceae isolates. Antimicrob. Agents Chemother. 50:4177-4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ford, P. J., and M. B. Avison. 2004. Evolutionary mapping of the SHV β-lactamase and evidence for two separate IS26-dependent blaSHV mobilization events from the Klebsiella pneumoniae chromosome. J. Antimicrob. Chemother. 54:69-75. [DOI] [PubMed] [Google Scholar]
- 10.Giakkoupi, P., L. S. Tzouvelekis, A. Tsakris, V. Loukova, D. Sofianou, and E. Tzelepi. 2000. IBC-1, a novel integron-associated class A β-lactamase with extended-spectrum properties produced by an Enterobacter cloacae clinical strain. Antimicrob. Agents Chemother. 44:2247-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joly-Guillou, M.-L., E. Vallee, E. Bergogne-Berezin, and A. Philippon. 1988. Distribution of β-lactamases and phenotype analysis in clinical strains of Acinetobacter calcoaceticus. J. Antimicrob. Chemother. 22:597-604. [DOI] [PubMed] [Google Scholar]
- 12.Lim, D., F. Sanschagrin, L. Passmore, L. De Castro, R. C. Levesque, and N. C. J. Strynadka. 2001. Insights into the molecular basis for the carbenicillinase activity of PSE-4 β-lactamase from crystallographic and kinetic studies. Biochemistry 40:395-402. [DOI] [PubMed] [Google Scholar]
- 13.Mammeri, H., L. Poirel, N. Mangeney, and P. Nordmann. 2003. Chromosomal integration of a cephalosporinase gene from Acinetobacter baumannii into Oligella urethralis as a source of acquired resistance to β-lactams. Antimicrob. Agents Chemother. 47:1536-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matagne, A., J. Lamotte-Brasseur, and J.-M. Frère. 1998. Catalytic properties of class A β-lactamases: efficiency and diversity. Biochem. J. 330:581-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melano, R., A. Petroni, A. Garutti, H. A. Saka, L. Mange, F. Pasterán, M. Rapoport, A. Rossi, and M. F. Galas. 2002. New carbenicillin-hydrolyzing β-lactamase (CARB-7) from Vibrio cholerae non-O1, non-O139 strains encoded by the VCR region of the V. cholerae genome. Antimicrob. Agents Chemother. 46:2162-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miriagou, V., A. Carattoli, E. Tzelepi, L. Villa, and L. S. Tzouvelekis. 2005. IS26-associated In4-type integrons forming multiresistance loci in enterobacterial plasmids. Antimicrob. Agents Chemother. 49:3541-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petroni, A., R. G. Melano, H. A. Saka, A Garutti, L. Mange, F. Pasteran, M. Rapoport, M. Miranda, D. Faccone, A. Rossi, P. S. Hoffman, and M. F. Galas. 2004. CARB-9, a carbenicillinase encoded in the VCR region of Vibrio cholerae non-O1, non-O139 belongs to a family of cassette-encoded β-lactamases. Antimicrob. Agents Chemother. 48:4042-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Philippon, A. M., G. C. Paul, A. P. Thabaut, and G. A. Jacoby. 1986. Properties of a novel carbenicillin-hydrolyzing β-lactamase (CARB-4) specified by an IncP-2 plasmid from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 29:519-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poole, K. 2004. Resistance to β-lactam antibiotics. Cell. Mol. Life Sci. 61:2200-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakurai, Y., K. Tsukamoto, and T. Sawai. 1991. Nucleotide sequence and characterization of a carbenicillin-hydrolyzing penicillinase gene from Proteus mirabilis. J. Bacteriol. 173:7038-7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharp, P. M., and W.-H. Li. 1987. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15:1281-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka, M., H. Okuyama, and N. Morita. 2001. Characterization of the gene encoding the β-lactamase of the psychrophilic marine bacterium Moritella marina strain MP-1. Biosci. Biotechnol. Biochem. 65:666-669. [DOI] [PubMed] [Google Scholar]
- 23.Teo, J. W. P., A. Suwanto, and C. L. Poh. 2000. Novel β-lactamase genes from two environmental isolates of Vibrio harveyi. Antimicrob. Agents Chemother. 44:1309-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomson, K. S., D. A. Weber, C. C. Sanders, and W. E. Sanders, Jr. 1990. β-Lactamase production in members of the family Enterobacteriaceae and resistance to β-lactam-enzyme inhibitor combinations. Antimicrob. Agents Chemother. 34:622-627. [DOI] [PMC free article] [PubMed] [Google Scholar]