Abstract
The influence of qnrA1 on the development of quinolone resistance in Enterobacteriaceae was evaluated by using the mutant prevention concentration parameter. The expression of qnrA1 considerably increased the mutant prevention concentration compared to strains without this gene. In the presence of qnrA1, mutations in gyrA and parC genes were easily selected to produce high levels of quinolone resistance.
Fluoroquinolone resistance in Enterobacteriaceae is usually the result of mutations in chromosomal genes for type II topoisomerases or in genes coding for or regulating efflux pumps and/or porins (6). Recent reports indicate that quinolone resistance may also be plasmid mediated by the qnrA, qnrS, qnrB, or aac(6′)-Ib-cr genes (12, 16). Although qnrA (and, presumably, related) genes confer low-level resistance on their own, when found in the presence of other mechanisms, they are able to help select for mutants with increased levels of fluoroquinolone resistance (6, 11, 16).
A major goal of antimicrobial therapy is to achieve sufficient drug exposure in relation to MIC for optimal efficacy at the site of infection. Activity can be assessed by using the mutant prevention concentration (MPC). This is the drug concentration that prevents the growth of the least-susceptible single-step mutant present in a large bacterial population, taking into account resistant mutant subpopulations present prior to antimicrobial treatment (1, 5). The mutant selection window, the antibiotic concentration found between MIC and MPC, is where single-step mutants can be enriched (5).
The aim of the present study was to determine the influence of qnrA1 on the development of quinolone resistance in Enterobacteriaceae using the MPC parameter. For this purpose, 14 strains were evaluated. These included: (i) four Klebsiella pneumoniae clinical strains containing qnrA1 with different levels of fluoroquinolone susceptibility (Tables 1 and 2) (11, 17) and (ii) three groups of isogenic strains that both express and do not express qnrA1 (Table 1). The primers used to clone qnrA1 in pACYC184 (New England Biolabs, Inc., Barcelona, Spain) were 5′-CGGCAGTTAAAATTGGGGCT-3′ and 5′-GACCAGACTGCATAAGCAACAC-3′.
TABLE 1.
Quinolone susceptibility characteristics of strains included in this study
| Strain | Description | CIP susceptibilitya | Mechanisms associated with quinolone resistance
|
Source or reference | |||
|---|---|---|---|---|---|---|---|
| qnrA1b | GyrA | ParC | Porinsd | ||||
| UAB1 | K. pneumoniae clinical strain | I | + | wtc | wt | + | 11 |
| N5 | K. pneumoniae clinical strain | S | + | wt | wt | + | 17 |
| 1960 | K. pneumoniae clinical strain | S | + | Ser83Phe | wt | + | 17 |
| 1132 | K. pneumoniae clinical strain | R | + | wt | wt | + | 17 |
| DH10B | E. coli reference lab strain | S | - | wt | wt | + | |
| DH10B/pACYC184 | E. coli DH10B containing pACYC184 vector | S | - | wt | wt | + | This study |
| DH10B+qnrA1 | E. coli DH10B containing qnrA1 cloned in pACYC184 vector | S | + | wt | wt | + | This study |
| E. coli J53 | E. coli reference lab strain | S | - | wt | wt | + | |
| J53/pMG252 | E. coli J53 transconjugant derived from UAB1 | S | + | wt | wt | + | 11 |
| J53/pN5 | E. coli J53 transconjugant derived from N5 | S | + | wt | wt | + | 17 |
| J53/p1960 | E. coli J53 transconjugant derived from 1960 | S | + | wt | wt | + | 17 |
| J53/p1132− | E. coli J53 transconjugant derived from 1132 | S | - | wt | wt | + | 17 |
| C2 | K. pneumoniae clinical strain | S | - | Ser83Phe | wt | - | 11 |
| C2/pMG252 | C2 transconjugant derived from UAB1 | I | + | Ser83Phe | wt | - | This study |
S, susceptible, I, intermediate susceptibility, R, resistance according to CLSI guidelines (4).
+, Presence and expression of qnrA1; −, no expression.
wt, wild type.
+ or − indicates the presence or absence of porins, respectively.
TABLE 2.
Fluoroquinolone MPCs and MICs for clinical and isogenic strains used in this study and derived resistant mutantsa
| Strain | MIC
|
MPC
|
MPC/MIC (MSW)
|
MIC for mutantsb
|
MPC time (h) windowc
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CIP | LFX | MXF | CIP | LFX | MXF | CIP | LFX | MXF | CIP | LFX | MXF | CIP | LFX | MXF | |
| UAB1 | 2 | 2 | 4 | 16 | 16 | 32 | 8 | 8 | 8 | 16-64 | 16-32 | 32-64 | 72-96 | 96 | 72 |
| N5 | 0.25 | 0.5 | 0.5 | 4 | 4 | 16 | 16 | 8 | 32 | 8 | 8 | 4-8 | 48 | 48 | 24-48 |
| 1960 | 1 | 2 | 2 | 64 | 64 | 32 | 64 | 32 | 16 | 32-64 | 64 | 16 | 72 | 72 | 24-48 |
| 1132 | 4 | 4 | 8 | 32 | 32 | 32 | 8 | 8 | 4 | 16 | 16 | 16 | 48 | 48 | 72 |
| E. coli DH10B | 0.002 | 0.004 | 0.002 | 0.015 | 0.015 | 0.06 | 7.5 | 3.8 | 30 | 0.008 | 0.008 | 0.06 | 72 | 72 | 96 |
| E. coli DH10B/pACYC184 | 0.002 | 0.004 | 0.002 | 0.015 | 0.015 | 0.06 | 7.5 | 3.8 | 30 | 0.008 | 0.008 | 0.06 | 72 | 72 | 72 |
| E. coli DH10B+qnrA | 0.125 | 0.125 | 0.125 | 2 | 2 | 2 | 16 | 16 | 16 | 1 | 1 | 0.5 | 72-96 | 96 | 96 |
| E. coli J53 | 0.004 | 0.008 | 0.015 | 0.06 | 0.06 | 0.125 | 15 | 7.5 | 16.7 | 0.03 | 0.03 | 0.5 | 48 | 48 | 72 |
| J53/pMG252 | 0.25 | 0.25 | 0.5 | 4 | 4 | 4 | 16 | 16 | 8 | 4 | 2 | 2-4 | 96 | 96 | 72 |
| J53/pN5 | 0.125 | 0.125 | 0.125 | 4 | 4 | 4 | 32 | 32 | 32 | 2 | 4 | 2 | 72 | 96 | 72 |
| J53/p1960 | 0.125 | 0.125 | 0.125 | 4 | 4 | 4 | 32 | 32 | 32 | 1 | 2 | 4 | 72 | 96 | 72 |
| J53/p1132− | 0.004 | 0.008 | 0.015 | 0.06 | 0.06 | 0.25 | 15 | 7.5 | 16.7 | 0.03 | 0.06 | 0.25 | 72 | 48 | 72 |
| C2 | 0.25 | 0.25 | 0.125 | 8 | 8 | 4 | 32 | 32 | 32 | 4-8 | 8 | 2-8 | 24-48 | 48 | 24-48 |
| C2/pMG252 | 2 | 2 | 1 | 128 | 128 | 32 | 64 | 64 | 32 | 64-128 | 64 | 32-64 | 24-48 | 72 | 24-48 |
MICs (in μg/ml) were determined by microdilution for ciprofloxacin (CIP), levofloxacin (LFX), or moxifloxacin (MXF). MPC values (in μg/ml) were determined for ciprofloxacin, levofloxacin, or moxifloxacin on Mueller-Hinton plates as the lowest antibiotic concentration (in the range of concentration steps analyzed) at which resistant colonies do not form. The MSW is the mutant selection window, the antibiotic concentration found between the MIC and the MPC.
MIC for CIP (microdilution) for ciprofloxacin- or moxifloxacin-resistant colonies were recovered on Mueller-Hinton plates one step below the MPC value.
Earliest time (in hours) at which resistant colonies were visible one step below the MPC.
qnrA1 expression as the only quinolone resistance mechanism did not bring about fluoroquinolone resistance; however, the association of qnrA1 with other mechanisms, such as porin loss and Ser83Phe substitution in the GyrA protein of the K. pneumoniae C2 pMG252 strain, did produce a phenotype of intermediate fluoroquinolone susceptibility, which supported the additive nature of the mechanism (Table 2) (10). This could imply an important tool in the acquisition of high quinolone resistance levels in relation to plasmid-mediated quinolone resistance (14, 17, 21).
MPC assays were performed on all 14 strains, as previously described (9), permitting colony growth for up to 96 h. Most strains required between 48 and 96 h of incubation before resistant colonies were observable, except in the N5 and 1960 strains for moxifloxacin and in the C2 and C2/pMG252 strains for ciprofloxacin and moxifloxacin (Table 2). The MPC values of each fluoroquinolone for the different strains expressing the qnrA1 gene as the only known quinolone resistance mechanism (Escherichia coli DH10B+qnrA1; E. coli J53 transconjugants containing qnrA1) reached levels (2 to 4 μg/ml) approaching the peak serum concentrations attained during therapy (Table 2) (5, 7, 20). MPC values for the non-qnrA1-producing isogenic controls ranged from 0.015 to 0.125 μg/ml (Table 2). These values were much higher in strains containing additional resistance mechanisms. The MPCs of clinical qnrA1-containing K. pneumoniae strains ranged from 4 to 64 μg/ml for the various fluoroquinolones. MPC values for K. pneumoniae C2 and K. pneumoniae C2 pMG252 strains were 4 to 8 μg/ml and 32 to 128 μg/ml, respectively, showing, once again, the effect of the qnrA1 gene on the MPC parameter. A potential consequence of a suboptimal therapy selecting for resistance can be exemplified by considering the case of mutants selected one dilution step below the MPC whose MIC is equal to or greater than its MPC value (Table 2).
Plasmid-mediated quinolone resistance enables mutant bacteria with low levels of resistance to survive long enough for them to grow and emerge during fluoroquinolone treatment. Interestingly, the MICs of some mutants from transconjugants containing qnrA1 and deriving from E. coli J53 and K. pneumoniae C2/pMG252 were as high as 4 μg/ml and 32 to 128 μg/ml, respectively (Table 2). Primers used to detect mutations in gyrA, parC, and qnrA1 have been described previously (8, 19). Mutations in the quinolone resistance-determining regions of gyrA or parC, which are related to the fluoroquinolone resistance in Enterobacteriaceae, were found in qnrA1-expressing mutants (Ser83Phe and Asp87Tyr in GyrA, and Ser80Ile and Val87Asp in ParC). No mutations were found either in the qnrA1 coding sequences or in their promoter sequences (data not shown) (18). Additional mutations in qnrA1-containing strains relating to quinolone resistance and detected in the type II topoisomerase genes are a reflection of the ability of this mechanism to select for mutants with additional chromosomal quinolone resistance mechanisms. Such mechanisms, because of their additive nature, lead to therapeutic failure (10). In vivo selection of fluoroquinolone-resistant E. coli expressing the qnrA1 gene was reported recently in the case of a patient with a urinary tract infection being treated with norfloxacin for an infection due to a ciprofloxacin-susceptible isolate (13). These data indicate the role of qnrA1 in the selection and generation of quinolone-resistant mutants.
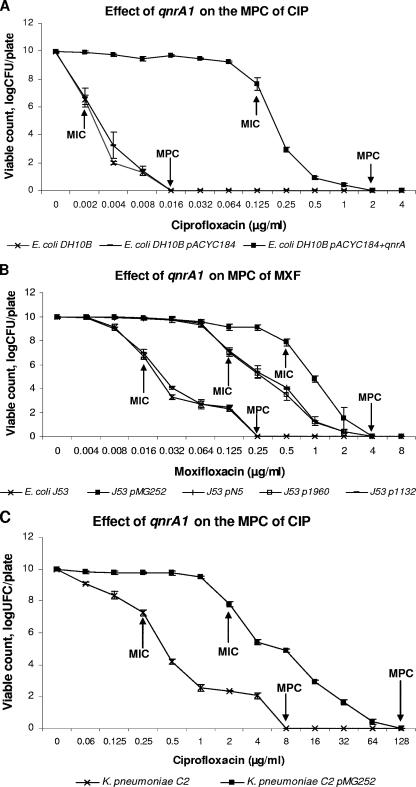
On the other hand, qnrA1 greatly eased the mutant selection (Fig. 1). For example, in the presence of qnrA1 (E. coli DH10B+qnrA1) and at 0.25 μg/ml, more than 103 resistant colonies grew from an initial population of 1010 bacteria, and colonies could still be recovered at ciprofloxacin concentrations of 1 to 2 μg/ml (Fig. 1A). Expression of qnrA1 from a natural plasmid in E. coli J53 produced over 105 resistant colonies grown at 1 μg/ml for J53 pMG252, and colonies could still be recovered at moxifloxacin concentrations of 2 to 4 μg/ml (Fig. 1B). In the case of isogenic strains of K. pneumoniae C2, the expression of qnrA1 from a natural plasmid produced more than 103 resistant colonies grown at 16 μg/ml, and colonies were still visualized at ciprofloxacin concentrations of 64 to 128 μg/ml (Fig. 1C), levels higher than peak serum concentrations attained during drug therapy. In cases of strains containing qnrA1, doses exceeding MPC in monotherapy would not be advisable with approved dosing procedures and evaluations of drug toxicity (2, 3). The increased frequency of quinolone-resistant mutants in strains which express qnrA1 at different concentrations below the MPC provides support for the role of low-level resistance mediated by the mechanism in selecting for quinolone resistance (11, 15), particularly if we take into account the MICs observed in mutants selected below the MPC and include some mutants obtained from strains whose only quinolone resistance mechanism was qnrA1 (Table 2).
FIG. 1.
MPC assays. MIC and MPC values are indicated on figures by arrows. CIP, ciprofloxacin; MXF, moxifloxacin.
Bacteria surviving at MPC were counted after 96 h, as described elsewhere (9). The MIC and MPC of these organisms (isolated at very low frequencies [data not shown]) were similar to those of the original strains. Although some bacteria survived MPC for a prolonged period, no quinolone-resistant mutants were selected, showing that the MPC parameter was working as specified. It is possible that survival is related to mutations in the hip genes (high persistence), being enhanced in the presence of β-lactams or fluoroquinolones, and producing a persistent phenotype at high antimicrobial concentrations (22).
Mechanisms, such as qnr or the recently described aac(6′)-Ib-cr gene, which are implicated in low-level fluoroquinolone resistance to plasmid-mediated determinants, may play a significant role in the generation of resistant mutants and therapeutic failure. This may be because, first, many clinical strains reported as containing genes that code for plasmid-mediated fluoroquinolone resistance are susceptible and, second, the presence of such genes in conjugative plasmid or other mobile elements may facilitate rapid dissemination in Enterobacteriaceae and other bacteria of clinical significance. Animal models will be necessary in future to perform in vivo validation of these in vitro data obtained by using the MPC parameter.
Acknowledgments
This study was supported by the Dirección General de Investigación del Ministerio de Ciencia y Tecnología of Spain (project SAF2005-04704), the Ministerio de Sanidad y Consumo, Instituto de Salud Carlos III (project PI060580), and the Spanish Network for the Research in Infectious Diseases (REIPI RD06/0008) of Spain. This study was partially supported by a grant from Bayer.
Footnotes
Published ahead of print on 2 April 2007.
REFERENCES
- 1.Baquero, F., and M. C. Negri. 1997. Strategies to minimize the development of antibiotic resistance. J. Chemother. 9(Suppl. 3):29-37. [PubMed] [Google Scholar]
- 2.Bertino, J., Jr., and D. Fish. 2000. The safety profile of the fluoroquinolones. Clin. Ther. 22:798-817. [DOI] [PubMed] [Google Scholar]
- 3.Boy, D., M. Well, M. Kinzig-Schippers, F. Sorgel, D. Nkel-Fuchs, and K. G. Naber. 2004. Urinary bactericidal activity, urinary excretion and plasma concentrations of gatifloxacin (400 mg) versus ciprofloxacin (500 mg) in healthy volunteers after a single oral dose. Int. J. Antimicrob. Agents 23(Suppl. 1):S6-S16. [DOI] [PubMed] [Google Scholar]
- 4.CLSI. 2005. Performance standards for antimicrobial susceptibility testing; 15th informational supplement. Document M100-S15. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.Drlica, K. 2003. The mutant selection window and antimicrobial resistance. J. Antimicrob. Chemother. 52:11-17. [DOI] [PubMed] [Google Scholar]
- 6.Jacoby, G. A. 2005. Mechanisms of resistance to quinolones. Clin. Infect. Dis. 41(Suppl. 2):S120-S126. [DOI] [PubMed] [Google Scholar]
- 7.Lipman, J., J. Scribante, A. G. Gous, H. Hon, S. Tshukutsoane, et al. 1998. Pharmacokinetic profiles of high-dose intravenous ciprofloxacin in severe sepsis. Antimicrob. Agents Chemother. 42:2235-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mammeri, H., L. M. Van De, L. Poirel, L. Martinez-Martinez, and P. Nordmann. 2005. Emergence of plasmid-mediated quinolone resistance in Escherichia coli in Europe. Antimicrob. Agents Chemother. 49:71-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcusson, L. L., S. K. Olofsson, L. P. Komp, O. Cars, and D. Hughes. 2005. Mutant prevention concentrations of ciprofloxacin for urinary tract infection isolates of Escherichia coli. J. Antimicrob. Chemother. 55:938-943. [DOI] [PubMed] [Google Scholar]
- 10.Martinez-Martinez, L., A. Pascual, I. Garcia, J. Tran, and G. A. Jacoby. 2003. Interaction of plasmid and host quinolone resistance. J. Antimicrob. Chemother. 51:1037-1039. [DOI] [PubMed] [Google Scholar]
- 11.Martinez-Martinez, L., A. Pascual, and G. A. Jacoby. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797-799. [DOI] [PubMed] [Google Scholar]
- 12.Nordmann, P., and L. Poirel. 2005. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J. Antimicrob. Chemother. 56:463-469. [DOI] [PubMed]
- 13.Poirel, L., J. D. Pitout, L. Calvo, J. M. Rodriguez-Martinez, D. Church, and P. Nordmann. 2006. In vivo selection of fluoroquinolone-resistant Escherichia coli isolates expressing plasmid-mediated quinolone resistance and expanded-spectrum beta-lactamase. Antimicrob. Agents Chemother. 50:1525-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poirel, L., L. M. Van De, H. Mammeri, and P. Nordmann. 2005. Association of plasmid-mediated quinolone resistance with extended-spectrum beta-lactamase VEB-1. Antimicrob. Agents Chemother. 49:3091-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robicsek, A., G. A. Jacoby, and D. C. Hooper. 2006. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis. 6:629-640. [DOI] [PubMed] [Google Scholar]
- 16.Robicsek, A., J. Strahilevitz, G. A. Jacoby, M. Macielag, D. Abbanat, C. H. Park, K. Bush, and D. C. Hooper. 2006. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 12:83-88. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez-Martinez, J. M., A. Pascual, I. Garcia, and L. Martinez-Martinez. 2003. Detection of the plasmid-mediated quinolone resistance determinant qnr among clinical isolates of Klebsiella pneumoniae producing AmpC-type beta-lactamase. J. Antimicrob. Chemother. 52:703-706. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez-Martinez, J. M., L. Poirel, R. Canton, and P. Nordmann. 2006. Common region CR1 for expression of antibiotic resistance genes. Antimicrob. Agents Chemother. 50:2544-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriguez-Martinez, J. M., C. Velasco, A. Pascual, I. Garcia, and L. Martinez-Martinez. 2006. Correlation of quinolone resistance levels and differences in basal and quinolone-induced expression from three qnrA-containing plasmids. Clin. Microbiol. Infect. 12:440-445. [DOI] [PubMed] [Google Scholar]
- 21.Turnidge, J. 1999. Pharmacokinetics and pharmacodynamics of fluoroquinolones. Drugs 58(Suppl. 2):29-36. [DOI] [PubMed] [Google Scholar]
- 22.Wang, M., J. H. Tran, G. A. Jacoby, Y. Zhang, F. Wang, and D. C. Hooper. 2003. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob. Agents Chemother. 47:2242-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolfson, J. S., D. C. Hooper, G. L. McHugh, M. A. Bozza, and M. N. Swartz. 1990. Mutants of Escherichia coli K-12 exhibiting reduced killing by both quinolone and beta-lactam antimicrobial agents. Antimicrob. Agents Chemother. 34:1938-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]