Abstract
The dissemination of AAC(6′)-I-type acetyltransferases have rendered amikacin and other aminoglycosides all but useless in some parts of the world. Antisense technologies could be an alternative to extend the life of these antibiotics. External guide sequences are short antisense oligoribonucleotides that induce RNase P-mediated cleavage of a target RNA by forming a precursor tRNA-like complex. Thirteen-nucleotide external guide sequences complementary to locations within five regions accessible for interaction with antisense oligonucleotides in the mRNA that encodes AAC(6′)-Ib were analyzed. While small variations in the location targeted by different external guide sequences resulted in big changes in efficiency of binding to native aac(6′)-Ib mRNA, most of them induced high levels of RNase P-mediated cleavage in vitro. Recombinant plasmids coding for selected external guide sequences were introduced into Escherichia coli harboring aac(6′)-Ib, and the transformant strains were tested to determine their resistance to amikacin. The two external guide sequences that showed the strongest binding efficiency to the mRNA in vitro, EGSC3 and EGSA2, interfered with expression of the resistance phenotype at different degrees. Growth curve experiments showed that E. coli cells harboring a plasmid coding for EGSC3, the external guide sequence with the highest mRNA binding affinity in vitro, did not grow for at least 300 min in the presence of 15 μg of amikacin/ml. EGSA2, which had a lower mRNA-binding affinity in vitro than EGSC3, inhibited the expression of amikacin resistance at a lesser level; growth of E. coli harboring a plasmid coding for EGSA2, in the presence of 15 μg of amikacin/ml was undetectable for 200 min but reached an optical density at 600 nm of 0.5 after 5 h of incubation. Our results indicate that the use of external guide sequences could be a viable strategy to preserve the efficacy of amikacin.
Aminoglycoside antibiotics act by interfering with the decoding process (12, 22, 25, 39) and are used to treat infections caused by aerobic gram-negative or gram-positive bacteria. In this latter case they are used in combination with other antibiotics (46). However, the rise in resistant and multiresistant strains, usually harboring aminoglycoside modifying enzymes, has limited the successful use of aminoglycosides in the treatment of serious infections (17, 37). The semisynthetic aminoglycoside amikacin (AMK) has been very useful in the treatment of multiresistant infections because a limited number of modifying enzymes, such as AAC(6′)-I-type acetyltransferases, are able to inactivate it (37). However, the spread of these enzymes has rendered AMK all but useless in some regions of the world (6, 30, 38, 45). Development of strategies to inhibit the expression of resistance genes could preserve the efficacy of AMK. Antisense oligonucleotide technologies may be an alternative for this purpose. A variety of antisense strategies have been applied before to inhibit gene expression in prokaryotic systems (4, 10, 23, 28, 34, 44).
RNase P, a ribozyme that includes an RNA component (M1) that is the catalytic subunit and a cofactor protein (C5), is responsible for generating the mature 5′ end of tRNA molecules by an endonucleolytic cleavage of the precursor tRNA (7, 15). A promising antisense strategy that takes advantage of RNase P has been recently developed (9, 10). It is based on the finding that RNase P can mediate cleavage of RNA molecules other than precursor tRNA provided that an appropriate complementary short oligoribonucleotide, known as the external guide sequence (EGS), is present and forms a duplex that results in the appropriate stem-like structure (5, 47). Oligoribonucleotides as short as 13 nucleotides efficiently mediate RNA cleavage (19). Cells are transformed with a recombinant plasmid coding for the appropriate EGS, which induces RNase P-mediated degradation of the mRNA molecule (7, 9). This strategy was successfully used to mediate conversion to susceptibility in bacteria resistant to chloramphenicol or β-lactams (10). Furthermore, EGSs targeting essential Escherichia coli genes were shown to reduce cell viability (23, 24). In the present study we designed EGSs complementary to the aac(6′)-Ib mRNA and analyzed their ability to bind native mRNA, induce RNase P-mediated degradation in vitro, and increase susceptibility to AMK in vivo.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli BL21(DE3) F− dcm ompT hsdS(rB− mB−) gal λ(DE3) (32) was used as host for pNW1 (28), an F′ derivative including the aac(6′)-Ib gene from pJHCMW1 (29) that was introduced by conjugation. E. coli TOP10 (Invitrogen) was used for regular cloning experiments. Bacterial cultures were carried out in Lennox Luria (L) broth (27). DNA fragments, including a 27-nucleotide sequence that contains a T7 promoter (GCGAAATTAATACGACTCACTATAGGG) followed by the EGS sequence (13 nucleotides, see Fig. 1), the consensus ACCA sequence, a hammerhead core as described by Guerrier-Takada et al. (9), and a T7 terminator sequence (TAGCATAACCCCTTGGGGCCTCTAAACGGGTCTTGAGGGGTTTTTTG) flanked by XbaI and BamHI sites, were cloned into pUC57 (GenBank/EMBL accession no. Y14837) to generate the recombinant clones coding for the EGSs (named pEGS, followed by the identifier letter and number, e.g., pEGSC3). Plasmid pRHC5 was generated by inserting a fragment including the rnpA gene and a T7 terminator from pBSC5 (23) into the XbaI site of the expression vector pT7-5 (33). Plasmid pJA′ includes the rnpB gene under the control of a T7 promoter (41).
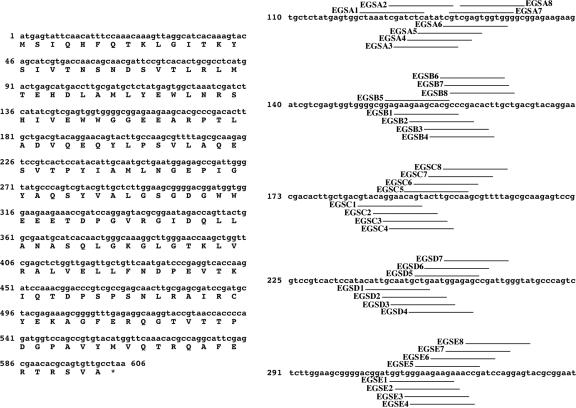
FIG. 1.
Nucleotide sequences of the EGSs. Nucleotide sequences of aac(6′)-Ib (left) and the EGSs targeting regions A to E (right). Regions A to E were identified previously (28).
General DNA and RNA procedures.
Plasmid DNA preparations were carried out by using the QIAspin miniprep kit (QIAGEN). PCRs were carried out using the HotStarTaq master mix kit (QIAGEN). All endonuclease restriction and ligase treatments were performed according to the supplier's recommendations (New England Biolabs). In vitro synthesis of RNA molecules was done using a MEGAshortscript high-yield transcription T7 kit (Ambion) according to the protocols provided. The aac(6′)-Ib mRNA was synthesized by using a linear DNA template, including aac(6′)-Ib under the control of the T7 promoter as described before (28). M1 RNA was synthesized by using pJA′ as a template. RNA labeling was carried out as described previously (28), and radioactivity was detected with a phosphorimager (Cyclone Storage Phosphor system; Packard). Denaturing polyacrylamide gel electrophoresis (PAGE) was performed as described previously (28) on 6% polyacrylamide gels using a glycerol-tolerant gel (GTG) buffer containing 16:1 (acrylamide-bis-acrylamide), 7 M urea, 89 mM Tris, 29 mM taurine, and 0.5 mM EDTA (USB Corp.). Electrophoretic mobility shift assays were carried out using 6% native (nondenaturing) PAGE (28).
Preparation of C5 protein.
The protein C5 was overexpressed and purified according to a procedure described previously (16, 41) with slight modifications. E. coli BL21(DE3)(pLysE, pRHC5) was cultured in Terrific broth (35) containing 100 μg of ampicillin (AMP)/ml and 34 μg of chloramphenicol/ml until the optical density at 600 nm (OD600) reached 0.6 to 0.8, when 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added; this was followed by incubation overnight at 37°C. Cells were harvested by centrifugation at 7,000 rpm for 10 min; resuspended in 0.02 volumes of buffer containing 50 mM Tris-HCl (pH 7.5), 60 mM NH4Cl, 10 mM magnesium acetate, 0.15% dithiothreitol, 42% urea, and 0.2 mM Pefabloc protease inhibitor (Roche); lysed by sonication; and treated with 6 μl of 2,000 U of DNase I/ml on ice for 30 min. Cell debris was removed by centrifugation at 8,000 rpm for 10 min. The cell extract was centrifuged at 30,000 × g for 30 min, and the supernatant was collected and centrifuged for 2 h at 100,000 × g. The pellet was resuspended in 10 ml of a buffer containing 50 mM Tris-HCl (pH 7.5), 1 M NH4Cl, 10 mM magnesium acetate, and 0.15% dithiothreitol and then centrifuged at 100,000 × g for 2 h. The supernatant was subjected to dialysis overnight against a buffer containing 50 mM Tris-HCl (pH 7.5), 100 mM NH4Cl, 10 mM MgCl2, and 0.15% dithiothreitol at 20°C, followed by centrifugation for 30 min at 15,500 rpm. The pellet was resuspended in 4 ml of a buffer containing 0.05 M sodium acetate (pH 7.2), 0.01 M MgCl2, 7 M urea, and 0.15% dithiothreitol. The C5 protein was then further purified by ion-exchange column chromatography using Sephadex C50 (Amersham Biosciences) and elution with a linear gradient 0 to 0.5 M NaCl. The C5 protein eluted at 0.3 M NaCl, and the procedure yielded a preparation that had C5 (13.5 kDa) and one minor larger extra band.
In vitro RNase P assays.
EGS-mediated cleavage of 5′-end-radiolabeled aac(6′)-Ib mRNA was assayed basically as described by Li et al. (19) preincubating 0.6 pmol of M1 RNA and 30 pmol of C5 protein in buffer containing 20 mM HEPES-KOH (pH 8.0), 400 mM ammonium acetate, 10 mM magnesium acetate, and 5% glycerol at 37°C for 15 min in a final volume of 7 μl. Radiolabeled aac(6′)-Ib mRNA (1.5 pmol) was preincubated with the appropriate EGS (500 pmol) at 25°C for 2 h in a volume of 3 μl. After preincubation, both solutions were combined and incubated at 37°C for 2 h. The reaction was stopped by the addition of 1 volume of gel loading buffer and analyzed by 6% denaturing GTG-PAGE.
In vivo activities of EGSs.
Overnight cultures of E. coli BL21(DE3)(pNW1) harboring the recombinant plasmid coding for the EGS to be tested were diluted to an OD600 of 0.05 in L broth containing 50 μg of AMP/ml and 15 μg of AMK/ml and then incubated at 37°C for 5 h. The growth rate of the cells was monitored by taking hourly OD600 measurements. Although the EGSs are under the control of the T7 promoter and the host E. coli BL21(DE3) includes an IPTG-inducible T7 RNA polymerase gene, we did not observe differences in growth in the presence or absence of the inducer. This effect has been described before, and it was attributed to leaky transcription of the T7 RNA polymerase gene (10).
The MICs of AMK were determined by the microdilution method (14) in the presence of 50 μg of AMP/ml to avoid loss of the plasmids coding for the EGSs.
Northern blots.
Total RNA was extracted from E. coli grown on L broth containing 50 μg of AMP/ml (27). RNA samples were electrophoresed on 2.5% formaldehyde-agarose gels and transferred to Hybond N+ membranes (Amersham Pharmacia Biotech). Equal loading and transfer of RNA were assessed by methylene blue staining of the membranes and ethidium bromide staining of duplicate gels. Probes to detect aac(6′)-Ib and 5S rRNA genes were radioactively labeled with [α-32P]dCTP by using a Prime-a-Gene labeling kit (Promega). The radioactive signals were detected and quantified by using a Storm 840 PhosphorImager (Molecular Dynamics).
RESULTS
Binding to and cleavage of aac(6′)-Ib mRNA in vitro.
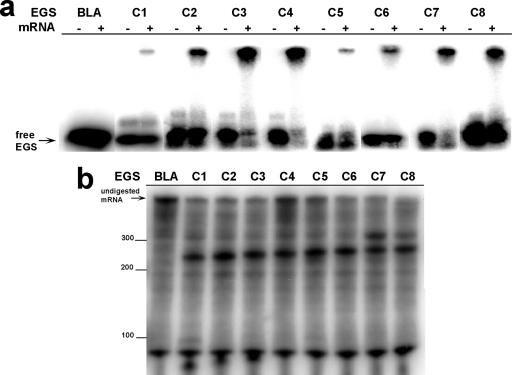
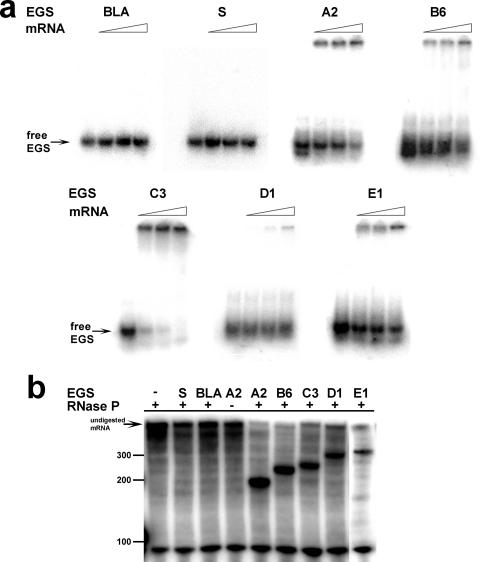
We identified aac(6′)-Ib mRNA regions accessible for interaction with complementary oligoribonucleotides by RNase H mapping (28). These regions were then used as targets to design EGSs that could induce inhibition of gene expression. We designed eight oligoribonucleotides targeting each of the regions A, B, C, and E (EGSA1 to -8, EGSB1 to -8, EGSC1 to -8, and EGSE1 to -8), and seven targeting region D (EGSD1 to -7) (Fig. 1). To select an EGS from each region for further analysis, all EGSs were analyzed to determine efficiency of binding to aac(6′)-Ib mRNA and the ability to induce RNase P-mediated degradation of aac(6′)-Ib mRNA. The selection was made based on a combination of strong binding to aac(6′)-Ib mRNA and efficient induction of RNase P-mediated cleavage of the mRNA. Figure 2a shows electrophoretic mobility shift assays obtained with EGSs targeting region C (EGSC1 through EGSC8). EGSC3, EGSC4, and EGSC8 bound the aac(6′)-Ib mRNA with the highest efficiency (Fig. 2a). However, RNase P degradation assays in vitro showed that EGSC3 and EGSC8 induced higher levels of aac(6′)-Ib mRNA cleavage than EGSC4 (Fig. 2b). These results indicate that the behavior of both EGSC3 and EGSC8 is approximately the same; we decided to select EGSC3 for further analysis. An experiment of binding in the presence of variable concentrations of aac(6′)-Ib mRNA further confirmed that binding of EGSC3 is highly efficient since the lowest concentration of mRNA assayed was sufficient to bind a high proportion of the labeled EGS (Fig. 3a). Similar experiments led us to select EGSA2 (region A), EGSB6 (region B), EGSD1 (region D), and EGSE1 (region E). All five selected EGSs were tested in binding experiments using variable concentrations of mRNA (Fig. 3a). EGSC3 bound aac(6′)-Ib mRNA with the highest affinity; EGSA2, EGSB6, and EGSE1 showed similar binding affinities but lower than that of EGSC3, and EGSD1 had the lowest affinity of all selected EGSs (Fig. 3a). As expected, neither of the two negative controls showed any aac(6′)-Ib mRNA-binding activity (Fig. 3a). One of the negative EGS controls was designed to target the β-lactamase gene, and the other carries a nucleotide sequence identical to that of the mRNA (sense). The efficiencies of all five EGSs for inducing RNase P-mediated degradation of the aac(6′)-Ib mRNA were determined by incubation in the presence of radiolabeled native mRNA and RNase P in the conditions described in the Materials and Methods section. Figure 3b shows that all five EGSs induced high levels of mRNA cleavage. As expected, in the presence of the negative EGSs controls, or the absence of RNase P or EGS, no digestion products were detected (Fig. 3b). Analysis of the results shown in Fig. 3 indicates that while all five EGSs induced high levels of mRNA cleavage by RNase P, they exhibited significant differences in binding affinities, which must be sufficient to degrade most mRNA molecules under the conditions used in these in vitro experiments.
FIG. 2.
Analysis of EGSs targeting region C (EGSC1 to EGSC8). (a) Binding to aac(6′)-Ib mRNA. EGSs (EGSC1 through EGSC8) were end labeled, mixed with aac(6′)-Ib mRNA, and analyzed by electrophoresis in 8% native polyacrylamide gels. The EGSs used in each reaction are indicated at the top of the gels. The control reaction was carried out in the presence of an EGS targeting the β-lactamase gene. A reaction with (+) or without (−) the addition of mRNA was performed for each EGS. (b) Induction of cleavage of aac(6′)-Ib mRNA. M1 RNA was preincubated with C5 protein at 37°C for 15 min, and radiolabeled aac(6′)-Ib mRNA was preincubated with the indicated EGS (shown at the top of the gel) at 25°C for 2 h. Both solutions were combined, incubated at 37°C for 2 h, and analyzed on 6% denaturing PAGE.
FIG. 3.
Binding affinities of EGSs to aac(6′)-Ib mRNA and induction of cleavage. (a) Binding of EGSs to aac(6′)-Ib mRNA. The different 5′-end-labeled EGSs and aac(6′)-Ib mRNA were incubated at EGS/mRNA ratios (from left to right) of 1:0, 1:0.5, 1:1, and 1:10 as described in Materials and Methods and then analyzed in native polyacrylamide gels. Two control reactions were carried out with an EGS designed to target the β-lactamase gene (BLA) and another with a nucleotide sequence identical to the mRNA (sense, S). (b) Induction of cleavage of aac(6′)-Ib mRNA. Cleavage reactions were carried out as described in the legend of Fig. 2. The EGSs assayed are shown at the top of the gel. Two reactions lacking RNase P or EGS (−) were carried out.
EGS-induced susceptibility to AMK.
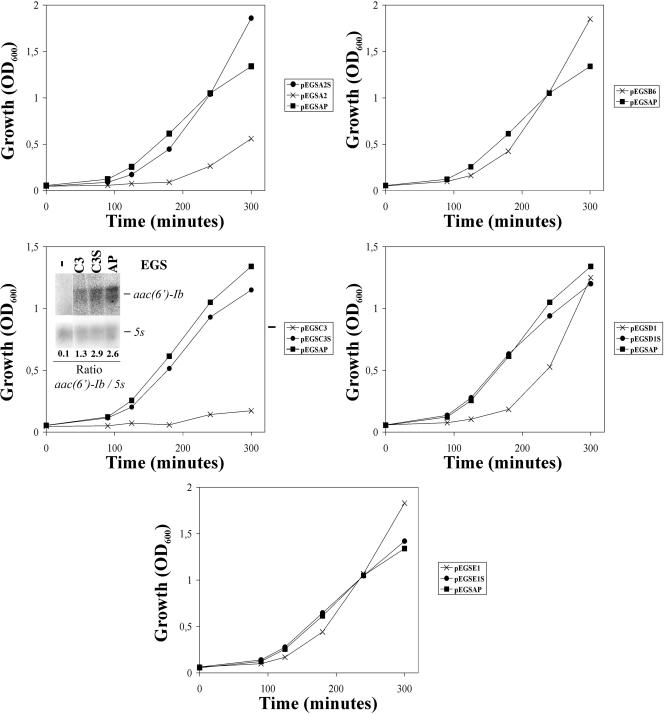
To test the ability of the selected EGSs to inhibit expression of AMK resistance in E. coli harboring pNW1, a plasmid including aac(6′)-Ib, we generated recombinant plasmids with inserts that include a T7 promoter, followed by the EGS coding region; the ACCA sequence, which enhances RNase P-substrate recognition; and the sequences required to generate a hammerhead ribozyme as described before (10). E. coli BL21(DE3)(pNW1) was transformed with plasmids pEGSA2, pEGSB6, pEGSC3, pEGSD1, or pEGSE1. As controls, the same E. coli strain was transformed with plasmids pEGSA2S, pEGSC3S, pEGSD1S, pEGSE1S, or pEGSAP. Plasmids pEGSA2S, pEGSC3S, pEGSD1S, pEGSE1S code for EGSs with sequences that are complementary to the cognate EGSs (i.e., sense oligonucleotides), and pEGSAP codes for an EGS that targets the alkaline phosphatase mRNA (10). The growth rates of all of these strains were similar when cultured in L broth containing AMP (not shown). However, upon addition of AMK the strains harboring plasmids encoding EGSA2 or EGSC3 showed growth inhibition while those harboring plasmids coding for EGSB6, EGSD1, EGSE1, or the control EGSs grew at approximately the same rate they did in the absence of the aminoglycoside (Fig. 4). Since EGSC3 was the most efficient in mediating inhibition of resistance to AMK, the E. coli strain carrying pEGSC3 was selected for comparison of its aac(6′)-Ib mRNA levels with those in E. coli harboring the controls pEGSC3S or pEGSAP. Total RNA was extracted from E. coli BL21(DE3)(pNW1) also harboring pEGSC3, pEGSC3S, or pEGSAP cultured in L broth containing AMP. Then the ratios between aac(6′)-Ib mRNA and 5S rRNA (which should be constant) were determined after quantification of the signal in Northern blot hybridization experiments. Figure 4 shows lower aac(6′)-Ib mRNA levels in E. coli BL21(DE3)(pNW1, pEGSC3) compared to those in the control strains. The MICs of AMK of E. coli BL21(DE3)(pNW1, pEGSC3), E. coli BL21(DE3)(pNW1, pEGSC3S), and E. coli BL21(DE3)(pNW1, pEGSAP) determined by using the microdilution method were 44, 96, and 96 μg/ml, respectively (average of four experiments). These results further confirmed the inhibitory effect of EGSC3 on expression of resistance to AMK. The high value could be due to the loss of expression of the EGSs due to the tendency of plasmids coding for EGSs to be lost or to suffer rearrangements. Guerrier-Takada et al. showed that liquid cultures started to lose the plasmid after 6 h (10). In addition, McKinney et al. (23) found that particularly effective EGS constructs made transformants more difficult to grow and tended to lose their plasmid vectors more rapidly and often than did the negative control EGS constructs.
FIG. 4.
Effect of EGSs on AMK-resistant E. coli. E. coli BL21(DE3)(pNW1) also harboring the recombinant plasmid coding for the indicated EGSs (indicated to the right of each panel) were cultured as described in Materials and Methods in the presence of 50 μg of AMP/ml and 15 μg of AMK/ml. Growth was monitored by measuring the OD600 at the indicated times. The controls were EGSAP, which targets the alkaline phosphatase gene, and an EGS with the corresponding sense sequence for each EGS to be tested. RNA levels in the experiment to test activity of EGSC3 were determined by Northern blot hybridization (inset). Total RNA was isolated from the cultures of E. coli BL21(DE3)(pNW1) also harboring the plasmid coding for EGSC3 (C3), EGSC3S (C3S), or EGSAP (AP) and from a culture of E. coli BL21(DE3)(pNW1) (−). The samples were run in 2.5% agarose gel, transferred to nylon membranes, and hybridized using probes to detect aac(6′)-Ib mRNA and 5S rRNA. The intensity of the signals was quantified, and the ratios aac(6′)-Ib/5S were calculated for each one of the samples (numbers below the Northern blot).
DISCUSSION
AAC(6′)-Ib has been found in a large number of gram-negative pathogenic bacteria belonging to several families and is the most prevalent among AAC(6′) type I enzymes (36, 39, and references therein). Moreover, a recently described variant of the gene, aac(6′)-Ib-cr, codes for an enzyme capable of acetylating quinolones (26). Development of viable methods to induce inhibition of expression of modifying enzymes, such as antisense technologies (18), could be a strategy to preserve the effectiveness of aminoglycosides. We have recently identified regions accessible for binding antisense oligonucleotides in the aac(6′)-Ib mRNA (28). Thirty-nine EGSs that target 13-nucleotide stretches within these regions were generated and tested to determine their mRNA binding properties, as well as their efficiency to induce RNase P-mediated degradation in vitro. We designed 39 different EGSs to be able to identify those ones that show strong aac(6′)-Ib mRNA binding affinity and are highly efficient in inducing RNase P-mediated degradation in vitro. Binding experiments showed that small changes in the locations targeted by the EGSs can result in big differences in binding affinity (illustrated for region C in Fig. 2). However, under the conditions of the experiments, there was not good correlation between binding affinity and RNase P-mediated cleavage in vitro. An EGS targeting each one of the five regions was selected using as criteria binding affinity to and cleavage of aac(6′)-Ib mRNA. Binding experiments using variable quantities of mRNA showed significantly different binding affinities, EGSC3 had the highest binding affinity of all five and EGSD1 had the lowest (Fig. 3a). However, all of them mediated high levels of mRNA cleavage (Fig. 3b). This apparent inconsistency could occur because, unlike in the binding assays, in the RNase P-mediated cleavage experiments the EGS/mRNA ratio grows as mRNA molecules are degraded. The binding affinities of all five selected EGSs must be sufficient to degrade most mRNA molecules under the conditions used in these in vitro experiments. EGSC3 showed a strong inhibitory effect on expression of resistance to AMK in vivo, and EGSA2, which was the next highest in binding efficiency, presented a lower but significant level of inhibition of AMK resistance. Interestingly, EGSs that induced similar levels of RNase P-mediated cleavage but showed a lower binding affinity to the messenger (EGSB6, EGSD1, and EGSE1) were practically unable to reverse bacterial resistance to AMK. These findings suggest that, at least in the case of the pJHCMW1 aac(6′)-Ib gene and in the conditions used to carry out these experiments, in vitro binding capacity of an EGS is the best parameter to predict its in vivo ability to inhibit gene expression. Other groups found that variations of levels of in vivo inhibition of gene expression were well correlated with RNase P activity in vitro (10). We do not know the nature of this difference; it is possible that the in vitro assays outcome is strongly dependent on the conditions of the assays or the structure of the target RNA. Further studies using a variety of RNA targets, locations, and conditions of in vitro binding and cleavage induction may help clarifying this issue.
We observed that aac(6′)-Ib mRNA levels in the presence of EGSC3 were reduced by ca. 50% with respect to those in the presence EGSC3S or AP (Fig. 4). Although the fraction of mRNA degraded was lower than one could have expected, as the goal is to completely silence the gene, it was comparable to the degradation levels achieved by others (24). Further experiments are presently being carried out to confirm that RNase P is responsible for the reduction in mRNA levels and inhibition of resistance to AMK by EGSC3.
Here we show that development of EGSs could be a way to preserve the efficacy of aminoglycosides against the rising number of resistant pathogens. However, many problems remain to be addressed. The large number of genetic environments in which the aac(6′)-Ib gene is found and the heterogeneity at the N-terminal portion of the enzyme (2, 31, 37, 40, 42) probably result in several mRNA species with different structures, not all susceptible to a given EGS. We are analyzing the length of mRNA species included in aac(6′)-Ib genes from different genetic environments, and we will perform computer analysis and mapping of these mRNA species to identify a minimum number of EGSs that can be potentially efficient at inducing the inhibition of gene expression. As presented here, although they provide proof of concept, recombinant clones coding for EGSs do not represent a realistic recourse against aminoglycoside-resistant pathogens. Therefore, an active nuclease resistance form of the EGSs and a method to ensure internalization will have to be developed. Information on nuclease-resistant oligoribonucleotides that induce cleavage of RNA by RNase P is not abundant and is mainly limited to the eukaryotic version of the enzyme (20, 21). Since the structural requirements of EGSs that induce cleavage by eukaryotic RNase P are different from those of bacterial RNase P (8), these results might not be extrapolated to EGSs to be used in prokaryotic systems. Some encouraging results have recently been reported on cell internalization of oligonucleotides and analogs using strategies such as liposome encapsulation or attachment of cell-permeabilizing peptides to peptide nucleic acids (3, 4, 11). In addition, since aac(6′)-Ib is often present as part of high-copy-number plasmids (45), there could still be synthesis of enough protein molecules to confer unacceptable levels of resistance in the presence of the EGS. This problem could be solved using inhibitors of aminoglycoside-modifying enzymes, which have been recently described (1, 13, 43). These compounds could be used in combination with EGSs to enhance susceptibility to the antibiotic. In summary, the results shown in the present study indicate that the development of viable technologies involving RNase P-mediated mRNA degradation could be an important tool to extend the useful life of existing antibiotics to which bacteria are becoming increasingly resistant.
Acknowledgments
We thank S. Altman and C. Guerrier-Takada for the gift of plasmids pBSC5 and pJA′, for their helpful guidance at various stages of this project, and for their encouragement.
This study was supported by Public Health Service grant 2R15AI047115 (to M.E.T.) from the National Institutes of Health, and grants PICT 8266 from the Agencia de Promoción Científica y Tecnológica, and X-245 from UBACyT Programación Científica, Universidad de Buenos Aires, Argentina (to A.Z.). A.Z. is a career member of CONICET. A.J.C.S.B. was supported by the Fundación Ciencias Exactas y Naturales and CONICET. M.D. was supported by LA Basin Minority Health and Health Disparities International Research Training Program (MHIRT) 5T37MD001368-09 (National Center on Minority Health and Health Disparities).
Footnotes
Published ahead of print on 26 March 2007.
REFERENCES
- 1.Boehr, D. D., K. A. Draker, K. Koteva, M. Bains, R. E. Hancock, and G. D. Wright. 2003. Broad-spectrum peptide inhibitors of aminoglycoside antibiotic resistance enzymes. Chem. Biol. 10:189-196. [DOI] [PubMed] [Google Scholar]
- 2.Canton, R., and T. M. Coque. 2006. The CTX-M beta-lactamase pandemic. Curr. Opin. Microbiol. 9:466-475. [DOI] [PubMed] [Google Scholar]
- 3.Eriksson, M., P. E. Nielsen, and L. Good. 2002. Cell permeabilization and uptake of antisense peptide-peptide nucleic acid (PNA) into Escherichia coli. J. Biol. Chem. 277:7144-7147. [DOI] [PubMed] [Google Scholar]
- 4.Fillion, P., A. Desjardins, K. Sayasith, and J. Lagace. 2001. Encapsulation of DNA in negatively charged liposomes and inhibition of bacterial gene expression with fluid liposome-encapsulated antisense oligonucleotides. Biochim. Biophys. Acta 1515:44-54. [DOI] [PubMed] [Google Scholar]
- 5.Forster, A. C., and S. Altman. 1990. External guide sequences for an RNA enzyme. Science 249:783-786. [DOI] [PubMed] [Google Scholar]
- 6.Galani, I., E. Xirouchaki, K. Kanellakopoulou, G. Petrikkos, and H. Giamarellou. 2002. Transferable plasmid mediating resistance to multiple antimicrobial agents in Klebsiella pneumoniae isolates in Greece. Clin. Microbiol. Infect. 8:579-588. [DOI] [PubMed] [Google Scholar]
- 7.Gopalan, V., A. Vioque, and S. Altman. 2002. RNase P: variations and uses. J. Biol. Chem. 277:6759-6762. [DOI] [PubMed] [Google Scholar]
- 8.Guerrier-Takada, C., and S. Altman. 2000. Inactivation of gene expression using ribonuclease P and external guide sequences. Methods Enzymol. 313:442-456. [DOI] [PubMed] [Google Scholar]
- 9.Guerrier-Takada, C., Y. Li, and S. Altman. 1995. Artificial regulation of gene expression in Escherichia coli by RNase P. Proc. Natl. Acad. Sci. USA 92:11115-11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guerrier-Takada, C., R. Salavati, and S. Altman. 1997. Phenotypic conversion of drug-resistant bacteria to drug sensitivity. Proc. Natl. Acad. Sci. USA 94:8468-8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harth, G., P. C. Zamecnik, J. Y. Tang, D. Tabatadze, and M. A. Horwitz. 2000. Treatment of Mycobacterium tuberculosis with antisense oligonucleotides to glutamine synthetase mRNA inhibits glutamine synthetase activity, formation of the poly-l-glutamate/glutamine cell wall structure, and bacterial replication. Proc. Natl. Acad. Sci. USA 97:418-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hobbie, S. N., P. Pfister, C. Bruell, P. Sander, B. Francois, E. Westhof, and E. C. Bottger. 2006. Binding of neomycin-class aminoglycoside antibiotics to mutant ribosomes with alterations in the A site of 16S rRNA. Antimicrob. Agents Chemother. 50:1489-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jana, S., and J. K. Deb. 2005. Molecular targets for design of novel inhibitors to circumvent aminoglycoside resistance. Curr. Drug Targets 6:353-361. [DOI] [PubMed] [Google Scholar]
- 14.Jorgensen, J., and J. Turnidge. 2003. Susceptibility test methods: dilution and disk diffusion methods, p. 1108-1127. In P. Murray, E. Baron, J. Jorgensen, M. Pfaller, and R. Yolken (ed.), Manual of clinical microbiology, vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 15.Kazantsev, A. V., and N. R. Pace. 2006. Bacterial RNase P: a new view of an ancient enzyme. Nat. Rev. Microbiol. 4:729-740. [DOI] [PubMed] [Google Scholar]
- 16.Kirsebom, L. A., and A. Vioque. 1995. RNase P from bacteria. Substrate recognition and function of the protein subunit. Mol. Biol. Rep. 22:99-109. [DOI] [PubMed] [Google Scholar]
- 17.Kotra, L. P., J. Haddad, and S. Mobashery. 2000. Aminoglycosides: perspectives on mechanisms of action and resistance and strategies to counter resistance. Antimicrob. Agents Chemother. 44:3249-3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurreck, J. 2003. Antisense technologies. Improvement through novel chemical modifications. Eur. J. Biochem. 270:1628-1644. [DOI] [PubMed] [Google Scholar]
- 19.Li, Y., C. Guerrier-Takada, and S. Altman. 1992. Targeted cleavage of mRNA in vitro by RNase P from Escherichia coli. Proc. Natl. Acad. Sci. USA 89:3185-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma, M., L. Benimetskaya, I. Lebedeva, J. Dignam, G. Takle, and C. A. Stein. 2000. Intracellular mRNA cleavage induced through activation of RNase P by nuclease-resistant external guide sequences. Nat. Biotechnol. 18:58-61. [DOI] [PubMed] [Google Scholar]
- 21.Ma, M. Y., B. Jacob-Samuel, J. C. Dignam, U. Pace, A. R. Goldberg, and S. T. George. 1998. Nuclease-resistant external guide sequence-induced cleavage of target RNA by human ribonuclease P. Antisense Nucleic Acid Drug Dev. 8:415-426. [DOI] [PubMed] [Google Scholar]
- 22.Magnet, S., and J. S. Blanchard. 2005. Molecular insights into aminoglycoside action and resistance. Chem. Rev. 105:477-498. [DOI] [PubMed] [Google Scholar]
- 23.McKinney, J., C. Guerrier-Takada, D. Wesolowski, and S. Altman. 2001. Inhibition of Escherichia coli viability by external guide sequences complementary to two essential genes. Proc. Natl. Acad. Sci. USA 98:6605-6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKinney, J. S., H. Zhang, T. Kubori, J. E. Galan, and S. Altman. 2004. Disruption of type III secretion in Salmonella enterica serovar Typhimurium by external guide sequences. Nucleic Acids Res. 32:848-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Possoz, C., J. Newmark, N. Sorto, D. J. Sherratt, and M. E. Tolmasky. 2007. Sublethal concentrations of the aminoglycoside amikacin interfere with cell division without affecting chromosome dynamics. Antimicrob. Agents Chemother. 51:252-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robicsek, A., J. Strahilevitz, G. A. Jacoby, M. Macielag, D. Abbanat, C. H. Park, K. Bush, and D. C. Hooper. 2006. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 12:83-88. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY7.
- 28.Sarno, R., H. Ha, N. Weinsetel, and M. E. Tolmasky. 2003. Inhibition of aminoglycoside 6′-N-acetyltransferase type Ib-mediated amikacin resistance by antisense oligodeoxynucleotides. Antimicrob. Agents Chemother. 47:3296-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarno, R., G. McGillivary, D. J. Sherratt, L. A. Actis, and M. E. Tolmasky. 2002. Complete nucleotide sequence of Klebsiella pneumoniae multiresistance plasmid pJHCMW1. Antimicrob. Agents Chemother. 46:3422-3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sekiguchi, J., T. Asagi, T. Miyoshi-Akiyama, T. Fujino, I. Kobayashi, K. Morita, Y. Kikuchi, T. Kuratsuji, and T. Kirikae. 2005. Multidrug-resistant Pseudomonas aeruginosa strain that caused an outbreak in a neurosurgery ward and its aac(6′)-Iae gene cassette encoding a novel aminoglycoside acetyltransferase. Antimicrob. Agents Chemother. 49:3734-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soler Bistue, A. J., F. A. Martin, A. Petroni, D. Faccone, M. Galas, M. E. Tolmasky, and A. Zorreguieta. 2006. Vibrio cholerae InV117, a class 1 integron harboring aac(6′)-Ib and blaCTX-M-2, is linked to transposition genes. Antimicrob. Agents Chemother. 50:1903-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 33.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan, X. X., J. K. Actor, and Y. Chen. 2005. Peptide nucleic acid antisense oligomer as a therapeutic strategy against bacterial infection: proof of principle using mouse intraperitoneal infection. Antimicrob. Agents Chemother. 49:3203-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tartof, K., and C. Hobbs. 1987. Improved media for rowing plasmid and cosmid clones. Bethesda Res. Lab Focus 9:12. [Google Scholar]
- 36.Tolmasky, M. E. 2007. Aminoglycoside-modifying enzymes: characteristics, localization, and dissemination, p. 35-52. In R. Bonomo and M. E. Tolmasky (ed.), Enzyme-mediated resistance to antibiotics: mechanisms, dissemination, and prospects for inhibition. ASM Press, Washington, DC.
- 37.Tolmasky, M. E. 2000. Bacterial resistance to aminoglycosides and beta-lactams: the Tn1331 transposon paradigm. Front. Biosci. 5:D20-D29. [DOI] [PubMed] [Google Scholar]
- 38.Tolmasky, M. E., R. M. Chamorro, J. H. Crosa, and P. M. Marini. 1988. Transposon-mediated amikacin resistance in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 32:1416-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vakulenko, S. B., and S. Mobashery. 2003. Versatility of aminoglycosides and prospects for their future. Clin. Microbiol. Rev. 16:430-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valverde, A., R. Canton, J. C. Galan, P. Nordmann, F. Baquero, and T. M. Coque. 2006. In117, an unusual In0-like class 1 integron containing CR1 and blaCTX-M-2 and associated with a Tn21-like element. Antimicrob. Agents Chemother. 50:799-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vioque, A., J. Arnez, and S. Altman. 1988. Protein-RNA interactions in the RNase P holoenzyme from Escherichia coli. J. Mol. Biol. 202:835-848. [DOI] [PubMed] [Google Scholar]
- 42.Walsh, T. R. 2006. Combinatorial genetic evolution of multiresistance. Curr. Opin. Microbiol. 9:476-482. [DOI] [PubMed] [Google Scholar]
- 43.Welch, K. T., K. G. Virga, N. A. Whittemore, C. Ozen, E. Wright, C. L. Brown, R. E. Lee, and E. H. Serpersu. 2005. Discovery of non-carbohydrate inhibitors of aminoglycoside-modifying enzymes. Bioorg. Med. Chem. 13:6252-6263. [DOI] [PubMed] [Google Scholar]
- 44.White, D. G., K. Maneewannakul, E. von Hofe, M. Zillman, W. Eisenberg, A. K. Field, and S. B. Levy. 1997. Inhibition of the multiple antibiotic resistance (mar) operon in Escherichia coli by antisense DNA analogs. Antimicrob. Agents Chemother. 41:2699-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woloj, M., M. E. Tolmasky, M. C. Roberts, and J. H. Crosa. 1986. Plasmid-encoded amikacin resistance in multiresistant strains of Klebsiella pneumoniae isolated from neonates with meningitis. Antimicrob. Agents Chemother. 29:315-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yao, J., and R. Moellering. 2003. Antibacterial agents, p. 1031-1073. In P. Murray, E. Baron, J. Jorgensen, M. Pfaller, and R. Yolken (ed.), Manual of clinical microbiology, 8th ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 47.Yuan, Y., E. S. Hwang, and S. Altman. 1992. Targeted cleavage of mRNA by human RNase P. Proc. Natl. Acad. Sci. USA 89:8006-8010. [DOI] [PMC free article] [PubMed] [Google Scholar]