Abstract
DNA replication stress triggers the activation of Checkpoint Kinase 1 (Chk1) in a pathway that requires the independent chromatin loading of the ATRIP–ATR (ATR-interacting protein/ATM [ataxia-telangiectasia mutated]–Rad3-related kinase) complex and the Rad9–Hus1–Rad1 (9–1–1) clamp. We show that Rad9’s role in Chk1 activation is to bind TopBP1, which stimulates ATR-mediated Chk1 phosphorylation via TopBP1’s activation domain (AD), a domain that binds and activates ATR. Notably, fusion of the AD to proliferating cell nuclear antigen (PCNA) or histone H2B bypasses the requirement for the 9–1–1 clamp, indicating that the 9–1–1 clamp’s primary role in activating Chk1 is to localize the AD to a stalled replication fork.
Keywords: Checkpoint, replication, Rad9, ATR, TopBP1, Chk1
Genotoxic damage activates conserved checkpoint signaling pathways that maintain genomic stability by regulating cell cycle progression, triggering apoptosis, and influencing DNA repair (Zhou and Elledge 2000; Abraham 2001). One pathway that is potently activated by replication stress leads to activation of Checkpoint Kinase 1 (Chk1), which promotes cell survival by blocking the firing of replication origins, preventing entry into mitosis, stabilizing stalled replication forks, and facilitating DNA repair (Cimprich 2003; Chen and Sanchez 2004). This pathway is initiated when the replicative DNA polymerases stall and large tracts of single-stranded DNA are created by the uncoupling of the replicative helicase from the advancing replication fork (Byun et al. 2005). The single-stranded DNA is then coated by replication protein A (RPA) (Walter and Newport 2000; Byun et al. 2005), which signals the independent recruitment of two checkpoint complexes: the ataxia-telangiectasia mutated (ATM)–Rad3-related kinase–ATR-interacting protein (ATR–ATRIP) complex and the Rad9–Hus1–Rad1 (9–1–1) complex. The ATRIP–ATR complex is bound to DNA by a direct interaction between ATRIP and RPA (Zou and Elledge 2003; Ball et al. 2005, 2007; Kim et al. 2005; Namiki and Zou 2006). In contrast, loading of the 9–1–1 complex requires several steps. First, DNA polymerase α is recruited, which in turn recruits the clamp loader, Rad17-replication factor C (RFC) (You et al. 2002; Lee et al. 2003; Byun et al. 2005). Second, the Rad17–RFC then loads the proliferating cell nuclear antigen (PCNA)-like 9–1–1 complex onto chromatin in a reaction that is analogous to the loading of PCNA onto sites of DNA replication (Bermudez et al. 2003; Ellison and Stillman 2003). Although the binding of the ATRIP–ATR complex and the loading of the 9–1–1 complex occur independently of one another, both events are essential for optimal ATR-mediated Chk1 phosphorylation and activation (Melo and Toczyski 2002).
Despite the tremendous progress that has been made in deciphering the biochemical functions of the 9–1–1 complex and the in-depth understanding of the signals that lead to the loading of the 9–1–1 clamp, it has remained unclear how the chromatin-bound 9–1–1 complex initiates and propagates the Chk1-activating signal. Several studies have demonstrated that Rad9 orthologs in Schizosaccharomyces pombe (Furuya et al. 2004), Saccharomyces cerevisiae (Wang and Elledge 2002), and humans (Makiniemi et al. 2001; St. Onge et al. 2003) interact with their respective TopBP1 orthologs (Cut4, Dpb11, and TopBP1). However, the significance of the Rad9–TopBP1 interaction in 9–1–1 function has not been explored. Here we show that the role of the 9–1–1 clamp is to recruit TopBP1, which then triggers ATR-mediated Chk1 phosphorylation. Thus, TopBP1 is a molecular bridge that links the independently recruited 9–1–1 and ATRIP–ATR complexes, leading to checkpoint activation.
Results and Discussion
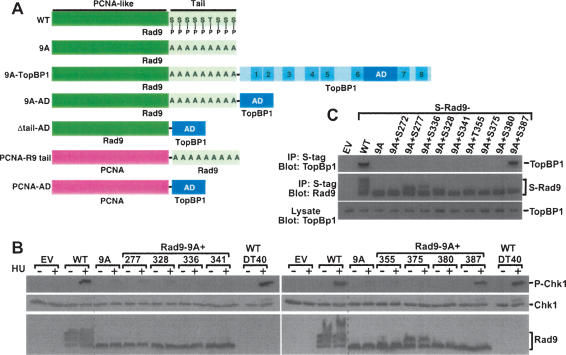
Rad9 has two predicted domains (Rauen et al. 2000; Venclovas and Thelen 2000; Roos-Mattjus et al. 2003). The N-terminal two-thirds of the protein (∼270 amino acids) forms a PCNA-like structure that associates with Hus1 and Rad1 to yield the 9–1–1 clamp complex (Fig. 1A). In contrast, the C-terminal ∼120 amino acids, dubbed the tail, are not required for formation of the 9–1–1 clamp (Rauen et al. 2000). The tail is heavily phosphorylated, with multiple sites having been identified (Chen et al. 2001; St. Onge et al. 2001, 2003; Roos-Mattjus et al. 2003). Although these sites are not required to generate the 9–1–1 clamp or for genotoxin-triggered loading of the clamp onto chromatin, a Rad9 mutant in which all the sites were simultaneously mutated could not facilitate Chk1 phosphorylation (Roos-Mattjus et al. 2003). Initial analyses of the phosphorylation sites revealed that Ser272, a site phosphorylated by ATM and ATR (Chen et al. 2001), was not essential for Chk1 phosphorylation (Roos-Mattjus et al. 2003). The individual roles of the other sites, however, were not explored. Therefore, to identify the Rad9 phosphorylation site(s) important in Chk1 activation, we created a series of mutants in which phosphorylation sites were individually added back to Rad9-9A (Fig. 1A), a mutant that lacks nine C-terminal phosphorylation sites and does not facilitate Chk1 phosphorylation (Roos-Mattjus et al. 2003). Next, we generated a series of mutants in which individual phosphorylation sites were restored. Transient expression of wild-type Rad9 in Rad9−/− DT40 cells (Kobayashi et al. 2004) restored hydroxyurea (HU)-induced Chk1 phosphorylation to levels seen in parental (wild-type) DT40 cells (Fig. 1B), whereas Rad9-9A did not enhance Chk1 phosphorylation. Analysis of the “add-back” mutants revealed that only one—Rad9-9A + Ser387—restored Chk1 phosphorylation nearly as well as wild-type Rad9, demonstrating that this site alone is sufficient for Chk1 phosphorylation (Fig. 1B).
Figure 1.
Rad9 Ser387 is involved in Chk1 phosphorylation. (A) Constructs used in this study. Rad9 and TopBP1 are not drawn to scale. Rad9 PCNA-like N-terminal domain and tail are indicated. P in tail indicates intact phosphorylation sites, whereas A indicates phosphorylation sites mutated to Ala. BRCT domains in TopBP1 are indicated; AD is the activation domain. (B) Rad9−/− DT40 cells were transiently transfected with empty vector (EV) and vectors encoding untagged wild-type Rad9 (WT); Rad9-9A (9A), the mutant lacking nine C-terminal phosphorylation sites; and Rad9-9A to which the indicated phosphorylation sites were restored (denoted as Rad9-9A + site). Following treatment with 10 mM HU for 1 h, transfected Rad9−/− DT40 cells and parental (wild-type) DT40 cells were lysed, separated by SDS-PAGE, and sequentially immunoblotted for phospho-Ser345-Chk1, Chk1, and Rad9. The multiple bands present in the Rad9 immunoblots are due to various forms of phosphorylated Rad9 (Volkmer and Karnitz 1999). The dotted line indicates the juxtaposition of nonadjacent regions of the same gel. (C) Lysates from HEK293 cells transiently transfected with empty vector (EV) or vectors encoding S-tagged wild-type Rad9 (WT), Rad-9A (9A), or the indicated Rad9-9A add-back expression plasmids were precipitated with S-protein agarose beads. Bound proteins were sequentially immunoblotted for endogenous TopBP1 (top) and Rad9 (middle). The multiple bands present in the Rad9 immunoblots are due to various forms of phosphorylated Rad9 (Volkmer and Karnitz 1999). (Bottom) A portion of the lysate was also immunoblotted to demonstrate equal TopBP1 levels in all samples.
Ser387 is constitutively phosphorylated (St. Onge et al. 2003) and conserved among vertebrate Rad9 proteins. To identify its role in checkpoint signaling, we asked whether this site was important in binding other checkpoint proteins that might participate in Chk1 activation. Rad9 was previously reported to interact with TopBP1 (Makiniemi et al. 2001; St. Onge et al. 2003), a checkpoint protein that plays a role in Chk1 activation (Parrilla-Castellar and Karnitz 2003). Intriguingly, TopBP1 also contains eight BRCA 1 C-terminal (BRCT) domains (Fig. 2A), which function as specific phosphoserine/phosphothreonine-binding domains (Manke et al. 2003; Yu et al. 2003), thus raising the possibility that phospho-Ser387 participates in TopBP1 binding. Indeed, when we examined the binding of TopBP1 to wild-type Rad9, Rad9-9A, and the panel of add-back mutants, only wild-type Rad9 and Rad9-9A + S387 bound TopBP1 (Fig. 1C).
Figure 2.
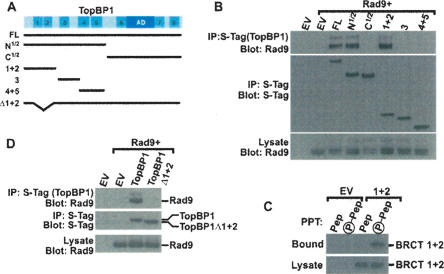
TopBP1 BRCT domains 1 and 2 bind Rad9. (A) Schematic map of TopBP1 showing the BRCT domains, the AD, and the S-tagged constructs used in this study: full-length TopBP1 (FL); the N-terminal half of TopBP1 (N1/2); the C-terminal half of TopBP1 (C1/2); fragments encoding BRCT 1 and 2 (1 + 2), BRCT 3 (3), and BRCT 4 and 5 (4 + 5); and TopBP1 lacking BRCT 1 and 2 (Δ1 + 2). (B,D) HEK293 cells were transfected with empty vector (EV) or vectors encoding AU1-tagged Rad9 and the indicated S-tagged TopBP1 proteins shown in A. Lysates were precipitated with S-protein agarose beads. Bound proteins were sequentially immunoblotted for Rad9 (top) and S-tagged TopBP1 (middle). (Bottom) A portion of the lysate was immunoblotted with Rad9 to show equal expression. The multiple bands present in the Rad9 immunoblots are due to various forms of phosphorylated Rad9 (Volkmer and Karnitz 1999). (C) HEK293 cells were transfected with empty vector or a vector expressing S-tagged TopBP1 BRCT 1 and 2 domains (1 + 2). (Top) Lysates were precipitated with nonphosphorylated (Pep) or phosphorylated (P-Pep) Ser387 Rad9 peptide covalently linked to beads. (Bottom) A portion of the lysate was immunoblotted to demonstrate equal expression of the BRCT 1 + 2 domains.
To identify the region of TopBP1 that interacts with Rad9, we created a series of TopBP1 fragments (Fig. 2A) and examined their abilities to bind Rad9. Full-length TopBP1 and the N-terminal half of TopBP1, which contains BRCT domains 1–5 interacted with Rad9, whereas the C-terminal half of TopBP1 did not interact (Fig. 2B). Because tandem BRCT domains frequently fold into compact structures that interact with phosphoserine/threonine-binding motifs, we further subdivided the N-terminal half of TopBP1 into three fragments containing the tandem BRCT domains 1 and 2, the tandem BRCT domains 4 and 5, and another fragment containing the BRCT domain 3. Coexpression studies revealed that the fragment containing BRCT domains 1 and 2 avidly interacted with Rad9, whereas fragments containing BRCT domain 3 and BRCT domains 4 and 5 did not interact with Rad9 (Fig. 2B). Likewise, the isolated BRCT 1 and 2 domains interacted with an 11-amino-acid peptide centered around phospho-Ser387 but did not bind the nonphosphorylated version of the peptide (Fig. 2C), thus demonstrating that these tandem BRCT domains bind to phosphorylated Ser387. To assess whether the BRCT domains 1 and 2 were important in the Rad9–TopBP1 interaction in the context of the full-length TopBP1, we examined the interaction of Rad9 with full-length TopBP1 or a mutant lacking BRCT domains 1 and 2. Consistent with the fragment interaction assays, deletion of BRCT domains 1 and 2 from full-length TopBP1 completely disrupted the interaction between Rad9 and TopBP1 (Fig. 2D). Collectively, these results suggest that Rad9 and TopBP1 interact via a phospho-Ser387–BRCT 1 and 2 interaction.
The interaction between Rad9 and TopBP1 raised the question of whether this interaction was indeed important for Chk1 activation. If the inability of Rad9-9A to facilitate Chk1 activation was due to a lack of interaction with TopBP1, we reasoned that constitutively tethering TopBP1 to the C-terminal tail in Rad9-9A by creating a chimeric Rad9-9A–TopBP1 fusion protein (Fig. 1A) would restore Rad9-9A’s ability to enhance Chk1 phosphorylation. Consistent with this prediction, the Rad9-9A–TopBP1 fusion facilitated Chk1 phosphorylation as well as wild-type Rad9, even when expressed at lower levels than wild-type Rad9 (Fig. 3A). Notably, expression of TopBP1 alone in Rad9−/− cells did not compensate for the Rad9 deficiency (Supplementary Fig. 1), demonstrating that TopBP1 must be linked to Rad9 to overcome a Rad9 deficiency.
Figure 3.
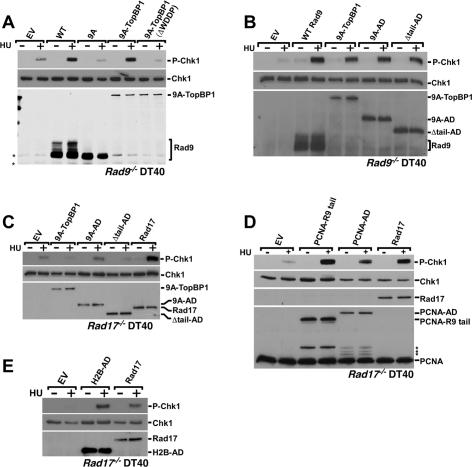
Rad9 activates Chk1 by binding TopBP1. Rad9−/− (A,B) or Rad17−/− (C–E) DT40 cells were transiently transfected with the indicated plasmids and treated with 10 mM HU for 1 h, and lysates were separated by SDS-PAGE and sequentially immunoblotted to detect phospho-Ser345-Chk1 and Chk1. To detect fusion protein and Rad17 expression, the samples were immunoblotted to detect S-tagged fusions (A–C,E), Rad17 (D), or PCNA (D). Transfections were with empty vector (EV); vectors expressing S-tagged wild-type Rad9 (WT), Rad9-9A (9A), Rad9-9A fused to full-length TopBP1 (9A–TopBP1), Rad9-9A–TopBP1 in which the WDDP motif in the AD was deleted from TopBP1 (9A–TopBP1–ΔWDDP), Rad9-9A fused to the TopBP1 AD (9A–AD), a tailless Rad9 fused to the AD (Δtail–AD), Rad17, or H2B fused to the TopBP1–AD (H2B–AD); or a vector expressing PCNA fused to the Rad9 tail (PCNA–Rad9 tail) or the TopBP1 AD (PCNA–AD). The multiple bands present in the Rad9 immunoblots are due to various forms of phosphorylated Rad9 (Volkmer and Karnitz 1999). Asterisk indicates nonspecific immunoreactive bands (A) or putative degradation products of the PCNA fusion proteins (D).
We next asked which region of TopBP1 is required for Chk1 activation in the context of a Rad9–TopBP1 fusion. In addition to BRCT domains, recent work demonstrated a region located between BRCT 6 and 7 (Fig. 2A), now dubbed the activation domain (AD), was important for ATR activation in Xenopus egg extracts (Kumagai et al. 2006). Therefore, we asked whether the TopBP1 AD participated in Chk1 phosphorylation triggered by the Rad9-9A–TopBP1 fusion. To disable the AD, we mutated the conserved tryptophan to arginine (W1145R) or deleted four highly conserved amino acids (WDDP, amino acids 1145–1148) that are essential for the AD to activate ATR (Kumagai et al. 2006). When expressed transiently in Rad9−/− DT40 cells, neither Rad9-9A–TopBP1ΔWDDP (Fig. 3A) nor Rad9-9A–TopBP1 W1145R (data not shown) restored Chk1 phosphorylation in HU-treated cells (Fig. 3A), demonstrating that the TopBP1 AD is necessary for Rad9-facilitated Chk1 phosphorylation.
To assess whether the TopBP1 AD was sufficient to restore HU-induced Chk1 phosphorylation, we fused the 30-kDa AD to the C-terminal tail of Rad9-9A (Fig. 1A). Expression of the Rad9-9A–AD fusion enhanced Chk1 phosphorylation as effectively as did Rad9-9A fused to full-length TopBP1 (Fig. 3B), thus indicating that the AD plays a critical role in Rad9-mediated Chk1 phosphorylation.
Previous work showed that the Rad9 C-terminal tail was important for Chk1 phosphorylation (Roos-Mattjus et al. 2003). To test whether the tail had another role in Chk1 phosphorylation aside from its role in associating with TopBP1, we created a “tailless” fusion between Rad9 and TopBP1 AD (Rad9 Δtail–AD). This fusion directly linked the AD to the N-terminal portion of Rad9 that is predicted to fold into a PCNA-like clamp (Fig. 1A). Like Rad9-9A–AD, Rad9 Δtail–AD also facilitated HU-induced Chk1 phosphorylation (Fig. 3B).
Rad9 is an integral part of the 9–1–1 clamp complex, which must be loaded onto chromatin by Rad17 to trigger Chk1 activation. To determine whether the Rad9–TopBP1 fusions also had to be loaded onto chromatin to enhance Chk1 phosphorylation, we expressed the Rad9–TopBP1 fusions in DT40 cells that lack Rad17 (Kobayashi et al. 2004), the clamp loader for the 9–1–1 complex (Zou et al. 2002, 2003; Bermudez et al. 2003; Ellison and Stillman 2003). As shown in Figure 3C, Rad17−/− DT40 cells have a defect in HU-induced Chk1 phosphorylation that is corrected by expression of Rad17. Notably, none of the Rad9–TopBP1 fusions stimulated Chk1 phosphorylation above that seen in cells transfected with empty vector, thus demonstrating that Rad9 fused to either full-length TopBP1 or that the AD, like wild-type Rad9, must be loaded by the Rad17 clamp loader to augment Chk1 activation.
These findings raised the possibility that the sole role for the 9–1–1 heterotrimeric complex in the Chk1 activation process is to localize the TopBP1 AD to the stalled replication fork. To address this question, we sought a way to localize the TopBP1 to stalled replication forks in the absence of a functional 9–1–1 complex. To disrupt the function of the clamp, we again used Rad17−/− DT40 cells to prevent 9–1–1 clamp loading. To localize the TopBP1 to the stalled forks in the absence of loaded 9–1–1 clamp, we fused either the Rad9 tail or the TopBP1 AD to PCNA (Fig. 1A), a homotrimeric clamp that is present at stalled forks. When expressed in Rad17−/− DT40 cells, PCNA linked to the Rad9 tail or the TopBP1 AD restored Chk1 phosphorylation to levels comparable to those seen when Rad17 was re-expressed in the Rad17-deficient cells (Fig. 3D). Similarly, tethering the AD to chromatin by fusing it to H2B also facilitated Chk1 phosphorylation (Fig. 3E). Taken together, these results demonstrate that recruitment of the TopBP1 AD to chromatin, either via the 9–1–1 complex, fusion to PCNA, or tethering to histone octamers, facilitates replication stress-induced Chk1 phosphorylation.
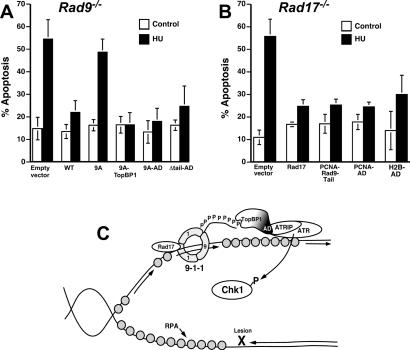
Replication stress induces apoptosis in DT40 cells lacking Rad9 or Rad17 (Fig. 4A,B), possibly because these cells have defects in Chk1 activation, a kinase that is important for cells to survive replication fork stalling (Zachos et al. 2003). To address the role of the 9–1–1 complex and TopBP1 in preventing replication stress-induced apoptosis, we transiently expressed various Rad9–TopBP1 fusions in Rad9−/− DT40 cells and treated them with HU. In the absence of Rad9 or in cells expressing Rad9-9A (the mutant lacking the tail phosphorylation sites), HU triggered robust apoptosis (Fig. 4A). In contrast, HU-induced apoptosis was blocked by the expression of wild-type Rad9, Rad9–TopBP1, Rad9–AD, or Rad9 Δtail–AD. Likewise, in cells lacking Rad17, expression of PCNA fused to the Rad9 tail or the TopBP1 AD blocked HU-induced cell death as effectively as did expression of Rad17 (Fig. 4B). Taken together, these results demonstrate that the same fusions that restore Chk1 activation also block HU-induced apoptosis, suggesting that a major role of the 9–1–1 complex in preventing HU-induced apoptosis is to activate Chk1, which in turn facilitates cell survival.
Figure 4.
Chk1 activation correlates with cell survival. Rad9−/− (A) or Rad17−/− (B) DT40 cells were cotransfected with the indicated plasmids (described in Fig. 3) and pEGFP-N1. The following day, EGFP-positive cells were purified by fluorescence-activated cell sorting, treated with HU, and stained with Hoechst 33258. For each sample, 250 cells were examined by microscopy. Apoptotic cells were identified based on nuclear morphology. Error bars indicate standard deviation of three to four independent experiments. (C) Model of Rad9’s role in Chk1 activation. See the text for details.
It has long been known that the ATRIP/ATR complex and the 9–1–1 clamp are independently loaded onto chromatin at sites of genotoxic stress, where they collaboratively participate in Chk1 phosphorylation and activation. However, the mechanism by which the 9–1–1 complex contributes to this process has remained enigmatic. A recent study shed light on the interplay between the 9–1–1 complex and ATR in S. cerevisiae (Majka et al. 2006). Using a reconstituted system of purified proteins, this work showed that the S. cerevisiae 9–1–1 complex (Ddc1, Mec3, and Rad17) directly activated the ATR homolog (Mec3) to phosphorylate multiple substrates, including the kinase Rad53.
In contrast, the results presented here suggest that higher eukaryotes have evolved a more complex mechanism for ATR-mediated Chk1 activation. In this model, TopBP1 plays a critical intermediary role by linking the 9–1–1 complex and ATR and by regulating Chk1 activation in response to replication stress (Fig. 4C). At sites of DNA polymerase stalling, MCM DNA helicase is uncoupled from the stalled replicative polymerase (Byun et al. 2005), exposing large tracts of single-stranded DNA that are then coated by RPA (Walter and Newport 2000; You et al. 2002). The DNA-bound RPA signals the binding of the ATRIP–ATR complex (Zou and Elledge 2003) and the DNA polymerase α-primase complex (You et al. 2002). DNA polymerase α-primase complex then initiates DNA synthesis on the unwound DNA (Michael et al. 2000), triggering Rad17-dependent clamp loading of the 9–1–1 complex (You et al. 2002; Zou et al. 2003; Byun et al. 2005). While ATR can phosphorylate some substrates, such as Hus1 and Rad1, in the absence of the Rad9 tail (Lupardus and Cimprich 2006), several studies have demonstrated that the Rad9 tail is critical for Chk1 phosphorylation (Roos-Mattjus et al. 2003; St. Onge et al. 2003; Lupardus and Cimprich 2006). Here we show that the role of the 9–1–1 complex in Chk1 activation is to localize TopBP1, bound to the phosphorylated Rad9 tail via TopBP1 BRCT 1 and 2, to the stalled fork. The Rad9-tethered TopBP1 AD can then interact with the ATRIP/ATR complex. Thus TopBP1 links the independently recruited 9–1–1 and ATRIP/ATR complexes, leading to ATR-mediated Chk1 phosphorylation, and cell survival.
Materials and methods
Cell culture and transfection
Human embryonic kidney (HEK) 293 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Wild-type, Rad9−/−, and Rad17−/− DT40 cells were grown as described previously (Kobayashi et al. 2004). HEK293 cells (5 × 10 to 8 × 106 per transfection) were transfected by electroporation as described previously (Volkmer and Karnitz 1999) using a total of 40 μg of DNA per transfection and a 230-V, 20-msec pulse in a 0.4-cm cuvette with a BTX T 820 electroporator. Following electroporation, the cells were replated in DMEM supplemented with 10% FBS and cultured for 14–20 h before harvest. DT40 cells (20 × 106 per transfection) were collected by centrifugation, resuspended in 0.35 mL of freshly prepared cytomix buffer (van den Hoff et al. 1992), mixed with 40 μg of DNA in 50 μL of cytomix, and electroporated with two sequential 270-V, 10-msec pulses in a 0.4-cm cuvette. Following electroporation, the cells were cultured in growth medium for 14–20 h, divided into two equal parts, treated with vehicle or 10 mM HU for 1 h, and harvested.
Western blotting
DT40 cells were lysed directly in 2× SDS-PAGE sample buffer or as described previously (Roos-Mattjus et al. 2003). Lysates were separated by SDS-PAGE (10% gel), transferred to Immobilon-P (Millipore), and immunoblotted, as indicated, with a rabbit monoclonal antibody recognizing P-Ser345-Chk1 (133D3; Cell Signaling Technology); mouse monoclonal antibodies recognizing S-peptide (Hackbarth et al. 2004), Chk1 (G-4; Santa Cruz Biotechnology), or PCNA (PC10; Santa Cruz Biotechnology); or rabbit polyclonal antisera recognizing Rad17 (Volkmer and Karnitz 1999), Rad9 (Volkmer and Karnitz 1999), or TopBP1 (BL893; Bethyl Laboratories).
Interaction experiments
HEK293 cells transiently transfected with the indicated plasmids were lysed in 50 mM Tris (pH 7.4), 150 mM NaCl, 5 mM MgCl2, and 1% Triton X-100, and were freshly supplemented with Complete protease inhibitor (Roche Applied Science), 0.4 mM phenylmethylsufonyl fluoride, 20 μM phosphoramidon, 20 μM E64, 20 nM microcystin-LR, 1 mM sodium orthovanadate, and 10 mM β-glycerol phosphate for 5 min on ice. Post-nuclear lysates were incubated with 10 μL of packed S protein-agarose (Novagen) to capture the S-peptide-tagged proteins for 1 h at 4°C, and the precipitates were washed three times with lysis buffer containing only 1 mM sodium orthovanadate and 10 mM β-glycerol phosphate. The peptide-binding experiments were performed with nonphosphorylated peptide (CPVLAEDSEGEG) and phosphorylated Ser387 peptide (CPVLAED-p-SEGEG) covalently linked via the N-terminal Cys residue to SulfoLink beads (Pierce Biotechnology) using lysates from cells transfected with empty vector or the S-tagged TopBP1–BRCT1 + 2 expression vector. For all binding experiments, bead-bound proteins were released by heating in 2× SDS-PAGE sample buffer and were separated by SDS-PAGE.
Apoptosis assays
Rad9−/− or Rad17−/− DT40 cells were electroporated as described above with pEGFP-N1 (Clontech) and the indicated plasmids and were cultured for 14–18 h. EGFP-expressing cells were purified by fluorescence-activated cell sorting and were treated with 10 mM HU for 6 h. Cells were then processed, and apoptosis was quantitated as described (Karnitz et al. 2005).
Acknowledgments
We thank the Mayo Flow Cytometry/Optical Morphology Resource facility and the Mayo DNA Sequencing and Synthesis facility. This work was supported by NIH R01 grant CA84321 and the Mayo Foundation.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1547007
References
- Abraham R.T. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes & Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- Ball H.L., Myers J.S., Cortez D., Myers J.S., Cortez D., Cortez D. ATRIP binding to replication protein A-single-stranded DNA promotes ATR–ATRIP localization but is dispensable for Chk1 phosphorylation. Mol. Biol. Cell. 2005;16:2372–2381. doi: 10.1091/mbc.E04-11-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball H.L., Ehrhardt M.R., Mordes D.A., Glick G.G., Chazin W.J., Cortez D., Ehrhardt M.R., Mordes D.A., Glick G.G., Chazin W.J., Cortez D., Mordes D.A., Glick G.G., Chazin W.J., Cortez D., Glick G.G., Chazin W.J., Cortez D., Chazin W.J., Cortez D., Cortez D. Function of a conserved checkpoint recruitment domain in ATRIP proteins. Mol. Cell. Biol. 2007;27:3367–3377. doi: 10.1128/MCB.02238-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez V.P., Lindsey-Boltz L.A., Cesare A.J., Maniwa Y., Griffith J.D., Hurwitz J., Sancar A., Lindsey-Boltz L.A., Cesare A.J., Maniwa Y., Griffith J.D., Hurwitz J., Sancar A., Cesare A.J., Maniwa Y., Griffith J.D., Hurwitz J., Sancar A., Maniwa Y., Griffith J.D., Hurwitz J., Sancar A., Griffith J.D., Hurwitz J., Sancar A., Hurwitz J., Sancar A., Sancar A. Loading of the human 9–1–1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17–replication factor C complex in vitro. Proc. Natl. Acad. Sci. 2003;100:1633–1638. doi: 10.1073/pnas.0437927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun T.S., Pacek M., Yee M.C., Walter J.C., Cimprich K.A., Pacek M., Yee M.C., Walter J.C., Cimprich K.A., Yee M.C., Walter J.C., Cimprich K.A., Walter J.C., Cimprich K.A., Cimprich K.A. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes & Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Sanchez Y., Sanchez Y. Chk1 in the DNA damage response: Conserved roles from yeasts to mammals. DNA Repair (Amst.) 2004;3:1025–1032. doi: 10.1016/j.dnarep.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Chen M.J., Lin Y.T., Lieberman H.B., Chen G., Lee E.Y., Lin Y.T., Lieberman H.B., Chen G., Lee E.Y., Lieberman H.B., Chen G., Lee E.Y., Chen G., Lee E.Y., Lee E.Y. ATM-dependent phosphorylation of human Rad9 is required for ionizing radiation-induced checkpoint activation. J. Biol. Chem. 2001;276:16580–16586. doi: 10.1074/jbc.M008871200. [DOI] [PubMed] [Google Scholar]
- Cimprich K.A. Fragile sites: Breaking up over a slowdown. Curr. Biol. 2003;13:R231–R233. doi: 10.1016/s0960-9822(03)00158-1. [DOI] [PubMed] [Google Scholar]
- Ellison V., Stillman B., Stillman B. Biochemical characterization of DNA damage checkpoint complexes: Clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol. 2003;1:231–243. doi: 10.1371/journal.pbio.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya K., Poitelea M., Guo L., Caspari T., Carr A.M., Poitelea M., Guo L., Caspari T., Carr A.M., Guo L., Caspari T., Carr A.M., Caspari T., Carr A.M., Carr A.M. Chk1 activation requires Rad9 S/TQ-site phosphorylation to promote association with C-terminal BRCT domains of Rad4TOPBP1. Genes & Dev. 2004;18:1154–1164. doi: 10.1101/gad.291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackbarth J.S., Lee S.H., Meng X.W., Vroman B.T., Kaufmann S.H., Karnitz L.M., Lee S.H., Meng X.W., Vroman B.T., Kaufmann S.H., Karnitz L.M., Meng X.W., Vroman B.T., Kaufmann S.H., Karnitz L.M., Vroman B.T., Kaufmann S.H., Karnitz L.M., Kaufmann S.H., Karnitz L.M., Karnitz L.M. S-peptide epitope tagging for protein purification, expression monitoring, and localization in mammalian cells. Biotechniques. 2004;37:835–839. [PubMed] [Google Scholar]
- Karnitz L.M., Flatten K.S., Wagner J.M., Loegering D., Hackbarth J.S., Arlander S.J., Vroman B.T., Thomas M.B., Baek Y.U., Hopkins K.M., Flatten K.S., Wagner J.M., Loegering D., Hackbarth J.S., Arlander S.J., Vroman B.T., Thomas M.B., Baek Y.U., Hopkins K.M., Wagner J.M., Loegering D., Hackbarth J.S., Arlander S.J., Vroman B.T., Thomas M.B., Baek Y.U., Hopkins K.M., Loegering D., Hackbarth J.S., Arlander S.J., Vroman B.T., Thomas M.B., Baek Y.U., Hopkins K.M., Hackbarth J.S., Arlander S.J., Vroman B.T., Thomas M.B., Baek Y.U., Hopkins K.M., Arlander S.J., Vroman B.T., Thomas M.B., Baek Y.U., Hopkins K.M., Vroman B.T., Thomas M.B., Baek Y.U., Hopkins K.M., Thomas M.B., Baek Y.U., Hopkins K.M., Baek Y.U., Hopkins K.M., Hopkins K.M., et al. Gemcitabine-induced activation of checkpoint signaling pathways that affect tumor cell survival. Mol. Pharmacol. 2005;68:1636–1644. doi: 10.1124/mol.105.012716. [DOI] [PubMed] [Google Scholar]
- Kim S.M., Kumagai A., Lee J., Dunphy W.G., Kumagai A., Lee J., Dunphy W.G., Lee J., Dunphy W.G., Dunphy W.G. Phosphorylation of Chk1 by ATM- and Rad3-related (ATR) in Xenopus egg extracts requires binding of ATRIP to ATR but not the stable DNA-binding or coiled-coil domains of ATRIP. J. Biol. Chem. 2005;280:38355–38364. doi: 10.1074/jbc.M508673200. [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Hirano A., Kumano T., Xiang S.L., Mihara K., Haseda Y., Matsui O., Shimizu H., Yamamoto K., Hirano A., Kumano T., Xiang S.L., Mihara K., Haseda Y., Matsui O., Shimizu H., Yamamoto K., Kumano T., Xiang S.L., Mihara K., Haseda Y., Matsui O., Shimizu H., Yamamoto K., Xiang S.L., Mihara K., Haseda Y., Matsui O., Shimizu H., Yamamoto K., Mihara K., Haseda Y., Matsui O., Shimizu H., Yamamoto K., Haseda Y., Matsui O., Shimizu H., Yamamoto K., Matsui O., Shimizu H., Yamamoto K., Shimizu H., Yamamoto K., Yamamoto K. Critical role for chicken Rad17 and Rad9 in the cellular response to DNA damage and stalled DNA replication. Genes Cells. 2004;9:291–303. doi: 10.1111/j.1356-9597.2004.00728.x. [DOI] [PubMed] [Google Scholar]
- Kumagai A., Lee J., Yoo H.Y., Dunphy W.G., Lee J., Yoo H.Y., Dunphy W.G., Yoo H.Y., Dunphy W.G., Dunphy W.G. TopBP1 activates the ATR–ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- Lee J., Kumagai A., Dunphy W.G., Kumagai A., Dunphy W.G., Dunphy W.G. Claspin, a Chk1-regulatory protein, monitors DNA replication on chromatin independently of RPA, ATR, and Rad17. Mol. Cell. 2003;11:329–340. doi: 10.1016/s1097-2765(03)00045-5. [DOI] [PubMed] [Google Scholar]
- Lupardus P.J., Cimprich K.A., Cimprich K.A. Phosphorylation of Xenopus Rad1 and Hus1 defines a readout for ATR activation that is independent of Claspin and the Rad9 carboxy terminus. Mol. Biol. Cell. 2006;17:1559–1569. doi: 10.1091/mbc.E05-09-0865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majka J., Niedziela-Majka A., Burgers P.M., Niedziela-Majka A., Burgers P.M., Burgers P.M. The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol. Cell. 2006;24:891–901. doi: 10.1016/j.molcel.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makiniemi M., Hillukkala T., Tuusa J., Reini K., Vaara M., Huang D., Pospiech H., Majuri I., Westerling T., Makela T.P., Hillukkala T., Tuusa J., Reini K., Vaara M., Huang D., Pospiech H., Majuri I., Westerling T., Makela T.P., Tuusa J., Reini K., Vaara M., Huang D., Pospiech H., Majuri I., Westerling T., Makela T.P., Reini K., Vaara M., Huang D., Pospiech H., Majuri I., Westerling T., Makela T.P., Vaara M., Huang D., Pospiech H., Majuri I., Westerling T., Makela T.P., Huang D., Pospiech H., Majuri I., Westerling T., Makela T.P., Pospiech H., Majuri I., Westerling T., Makela T.P., Majuri I., Westerling T., Makela T.P., Westerling T., Makela T.P., Makela T.P., et al. BRCT domain-containing protein TopBP1 functions in DNA replication and damage response. J. Biol. Chem. 2001;276:30399–30406. doi: 10.1074/jbc.M102245200. [DOI] [PubMed] [Google Scholar]
- Manke I.A., Lowery D.M., Nguyen A., Yaffe M.B., Lowery D.M., Nguyen A., Yaffe M.B., Nguyen A., Yaffe M.B., Yaffe M.B. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- Melo J., Toczyski D., Toczyski D. A unified view of the DNA-damage checkpoint. Curr. Opin. Cell Biol. 2002;14:237–245. doi: 10.1016/s0955-0674(02)00312-5. [DOI] [PubMed] [Google Scholar]
- Michael W.M., Ott R., Fanning E., Newport J., Ott R., Fanning E., Newport J., Fanning E., Newport J., Newport J. Activation of the DNA replication checkpoint through RNA synthesis by primase. Science. 2000;289:2133–2137. doi: 10.1126/science.289.5487.2133. [DOI] [PubMed] [Google Scholar]
- Namiki Y., Zou L., Zou L. ATRIP associates with replication protein A-coated ssDNA through multiple interactions. Proc. Natl. Acad. Sci. 2006;103:580–585. doi: 10.1073/pnas.0510223103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrilla-Castellar E.R., Karnitz L.M., Karnitz L.M. Cut5 is required for the binding of Atr and DNA polymerase α to genotoxin-damaged chromatin. J. Biol. Chem. 2003;278:45507–45511. doi: 10.1074/jbc.C300418200. [DOI] [PubMed] [Google Scholar]
- Rauen M., Burtelow M.A., Dufault V.M., Karnitz L.M., Burtelow M.A., Dufault V.M., Karnitz L.M., Dufault V.M., Karnitz L.M., Karnitz L.M. The human checkpoint protein hRad17 interacts with the PCNA-like protein hRad1, hHus1, and hRad9. J. Biol. Chem. 2000;275:29767–29771. doi: 10.1074/jbc.M005782200. [DOI] [PubMed] [Google Scholar]
- Roos-Mattjus P., Hopkins K.M., Oestreich A.J., Vroman B.T., Johnson K.L., Naylor S., Lieberman H.B., Karnitz L.M., Hopkins K.M., Oestreich A.J., Vroman B.T., Johnson K.L., Naylor S., Lieberman H.B., Karnitz L.M., Oestreich A.J., Vroman B.T., Johnson K.L., Naylor S., Lieberman H.B., Karnitz L.M., Vroman B.T., Johnson K.L., Naylor S., Lieberman H.B., Karnitz L.M., Johnson K.L., Naylor S., Lieberman H.B., Karnitz L.M., Naylor S., Lieberman H.B., Karnitz L.M., Lieberman H.B., Karnitz L.M., Karnitz L.M. Phosphorylation of human Rad9 is required for genotoxin-activated checkpoint signaling. J. Biol. Chem. 2003;278:24428–24437. doi: 10.1074/jbc.M301544200. [DOI] [PubMed] [Google Scholar]
- St Onge R.P., Besley B.D., Park M., Casselman R., Davey S., Besley B.D., Park M., Casselman R., Davey S., Park M., Casselman R., Davey S., Casselman R., Davey S., Davey S. DNA damage-dependent and -independent phosphorylation of the hRad9 checkpoint protein. J. Biol. Chem. 2001;276:41898–41905. doi: 10.1074/jbc.M105152200. [DOI] [PubMed] [Google Scholar]
- St Onge R.P., Besley B.D., Pelley J.L., Davey S., Besley B.D., Pelley J.L., Davey S., Pelley J.L., Davey S., Davey S. A role for the phosphorylation of Rad9 in checkpoint signaling. J. Biol. Chem. 2003;278:26620–26628. doi: 10.1074/jbc.M303134200. [DOI] [PubMed] [Google Scholar]
- van den Hoff M.J., Moorman A.F., Lamers W.H., Moorman A.F., Lamers W.H., Lamers W.H. Electroporation in “intracellular” buffer increases cell survival. Nucleic Acids Res. 1992;20:2902. doi: 10.1093/nar/20.11.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venclovas C., Thelen M.P., Thelen M.P. Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes. Nucleic Acids Res. 2000;28:2481–2493. doi: 10.1093/nar/28.13.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmer E., Karnitz L.M., Karnitz L.M. Human homologs of Schizosaccharomyces pombe Rad1, Hus1, and Rad9 form a DNA damage-responsive protein complex. J. Biol. Chem. 1999;274:567–570. doi: 10.1074/jbc.274.2.567. [DOI] [PubMed] [Google Scholar]
- Walter J., Newport J., Newport J. Initiation of eukaryotic DNA replication: Origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase α. Mol. Cell. 2000;5:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- Wang H., Elledge S.J., Elledge S.J. Genetic and physical interactions between DPB11 and DDC1 in the yeast DNA damage response pathway. Genetics. 2002;160:1295–1304. doi: 10.1093/genetics/160.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z., Kong L., Newport J., Kong L., Newport J., Newport J. The role of single-stranded DNA and polymerase α in establishing the ATR, Hus1 DNA replication checkpoint. J. Biol. Chem. 2002;277:27088–27093. doi: 10.1074/jbc.M204120200. [DOI] [PubMed] [Google Scholar]
- Yu X., Chini C.C., He M., Mer G., Chen J., Chini C.C., He M., Mer G., Chen J., He M., Mer G., Chen J., Mer G., Chen J., Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- Zachos G., Rainey M.D., Gillespie D.A., Rainey M.D., Gillespie D.A., Gillespie D.A. Chk1-deficient tumour cells are viable but exhibit multiple checkpoint and survival defects. EMBO J. 2003;22:713–723. doi: 10.1093/emboj/cdg060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B.B., Elledge S.J., Elledge S.J. The DNA damage response: Putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- Zou L., Elledge S.J., Elledge S.J. Sensing DNA damage through ATRIP recognition of RPA–ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- Zou L., Cortez D., Elledge S.J., Cortez D., Elledge S.J., Elledge S.J. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes & Dev. 2002;16:198–208. doi: 10.1101/gad.950302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Liu D., Elledge S.J., Liu D., Elledge S.J., Elledge S.J. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc. Natl. Acad. Sci. 2003;24:13827–13832. doi: 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]