Abstract
band, an allele enabling clear visualization of circadianly regulated spore formation (conidial banding), has remained an integral tool in the study of circadian rhythms for 40 years. bd was mapped using single-nucleotide polymorphisms (SNPs), cloned, and determined to be a T79I point mutation in ras-1. Alterations in light-regulated gene expression in the ras-1bd mutant suggests that the Neurospora photoreceptor WHITE COLLAR-1 is a target of RAS signaling, and increases in transcription of both wc-1 and fluffy show that regulators of conidiation are elevated in ras-1bd. Comparison of ras-1bd with dominant active and dominant-negative ras-1 mutants and biochemical assays of RAS function indicate that RAS-1bd displays a modest enhancement of GDP/GTP exchange and no change in GTPase activity. Because the circadian clock in ras-1bd appears to be normal, ras-1bd apparently acts to amplify a subtle endogenous clock output signal under standard assay conditions. Reactive oxygen species (ROS), which can affect and be affected by RAS signaling, increase conidiation, suggesting a link between generation of ROS and RAS-1 signaling; surprisingly, however, ROS levels are not elevated in ras-1bd. The data suggest that interconnected RAS- and ROS-responsive signaling pathways regulate the amplitude of circadian- and light-regulated gene expression in Neurospora.
Keywords: Circadian, output, band, ras-1, ROS
Neurospora crassa has served for several decades as a model organism for the study of diverse biological processes ranging from classic metabolism to circadian rhythms or RNA interfering (RNAi) (quelling) to name a few (Borkovich et al. 2004). Forty years ago a strain of Neurospora was isolated that exhibited robust and rhythmic formation of asexual spores (conidia), which fulfilled the characteristics of a circadian-regulated process (Sargent et al. 1966). This strain, originally called Timex, contained mutations in two unlinked loci; one caused an invertase deficiency that did not have any effect on circadian output, and the second, called band (bd), that was responsible for the augmented conidiation (Sargent and Woodward 1969). Since then, bd has been integrated into the genetic background of essentially every strain used for circadian biology research in Neurospora.
The bd mutation has been integral to the study of circadian biology in Neurospora because of the ease with which rhythmic conidia formation in this strain can be scored on glass growth tubes (race tubes) containing agar media (Fig. 1A); this has allowed numerous clock genes to be identified (for review, see Dunlap and Loros 2005; Liu and Bell-Pedersen 2006). The Neurospora circadian clock includes interconnected transcriptional-translational positive and negative feedback loops where the frequency (frq) gene is positively regulated by the WHITE COLLAR-1 (WC-1) and WHITE COLLAR-2 (WC-2) complex and FRQ protein feeds back to inhibit its own expression (Dunlap 1999; Bell-Pedersen et al. 2005; Brunner and Schafmeier 2006). WC-1 and WC-2 are regulators of light- and circadian-regulated gene expression and are thought to be responsive to the intracellular redox state (Crosthwaite et al. 1997; Linden and Macino 1997; Yoshida and Hasunuma 2004).
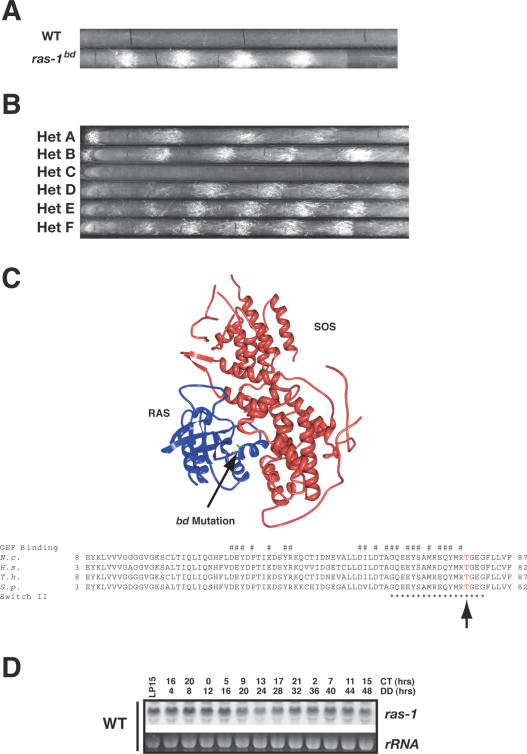
Figure 1.
Cloning and identification of the bd mutation. (A) Germinated spores isolated from cross 402 between wild-type Mauriceville and Oak Ridge bd strains were scored on race tubes. The panel shows representative strains with and without the bd mutation grown on standard race tube media. (B) Forced heterokaryon analysis was used to establish the dominance of ras-1bd. Heterokaryons were generated and banding was scored on race tubes. The corresponding genotype of each heterokaryon is Het A, 901-64 (inl, bd, A) and 418-11 (pan-2, A); Het B, 418-11 (pan-2, A) and 901-64 (inl, bd, A); Het C, 418-11 (pan-2, A) and 418-24 (inl, A); Het D, 901-62 (pan-2, bd, A) and 418-18 (inl, A); Het E, 418-18 (inl, A) and 901-62 (pan-2, bd, A); and Het F, 901-62 (pan-2, bd, A) and 418-16 (inl, bd, A). (C) Structure and sequence alignment indicating the location of the bd mutation in RAS-1. The structure of RAS and SOS (Boriack-Sjodin et al. 1998) was modified using PDB (PDB:1BKD) (http://www.rcsb.org/pdb/explore/pubmed.do?structureid=1BKD). The number sign (#) marks the region comprising the GEF-binding domain, and the asterisk (*) marks the switch II loop. The homology alignment compares RAS-1 between Neurospora (N.c.), Homo sapien (H.s.), Trametes hirsuta (T.h.), and Schizosaccharomyces pombe (S.p.). The T mutated to I in RAS-1bd is highlighted with an arrow. (D) Northern blots showing the regulation of ras-1 in a wild-type strain where RNA was extracted at the times shown after standard circadian entrainment.
A growing body of evidence suggests that cell signaling is fundamentally linked to circadian oscillations. For example, a circadian rhythm in tyrosine phosphorylation of the chicken mitogen-activated protein kinase (MAPK) coincides with changes in its activity (Sanada et al. 2000). Plus, null mutants of Neurofibromatosis-1 (Nf1), a gene known to be upstream of RAS/MAPK, have defects in circadianly regulated locomotor activity in Drosophila (Williams et al. 2001). BMAL1 is a direct downstream target of MAPK phosphorylation (Sanada et al. 2002), and CLOCK/CYCLE-dependent transcription is regulated in part by signaling through protein kinase A (PKA), calcium/calmodulin-dependent kinase (CaMK) II, and MAPK pathways (Weber et al. 2006).
The small membrane-anchored G-protein RAS is a central component in numerous signaling cascades that link extracellular stimuli to gene expression (Marshall 1996). RAS is activated by exchange of GDP with GTP that is mediated by GEFs (guanine nucleotide exchange factors), and these function as regulators, effectors, and integrators in multiple signaling cascades (Mitin et al. 2005). In Neurospora, there are two ras genes, ras-1 (Altschuler et al. 1990) and ras-2 (Kana-uchi et al. 1997), and at least three genes are predicted to encode RAS–GEFs (Borkovich et al. 2004); however, the dynamics of RAS signaling have yet to be determined.
A variety of studies indicate that activated RAS can cause elevations in cellular levels of reactive oxygen species (ROS). For example, mitogenic RAS leads to increased superoxide formation in transformed fibroblasts, and ROS itself is becoming an important and widely recognized second messenger in cell signaling (Hansberg and Aguirre 1990; Irani et al. 1997). In addition, different reports have highlighted the direct effect of ROS on RAS activity (e.g., Heo and Campbell 2006). Excessive amounts of ROS, which incidentally are also a byproduct of respiration, accompany apoptosis, and its removal is catalyzed by superoxide dismutases (SODs) and catalases (CATs) (Aguirre et al. 2005). In Neurospora, elevated ROS levels lead to increased differentiation, asexual spore development, and carotenoid synthesis and are thought to activate WC-mediated transcription (Yoshida and Hasunuma 2004; Aguirre et al. 2005).
Although the circadian clock in strains containing bd is normal, the bd allele exerts profound effects on circadian output and on growth and development in general: bd acts to turn a subtle and generally invisible circadianly regulated aspect of physiology or metabolism into a robust and readily identifiable phenotype under the conditions typically used to monitor the circadian clock (constant darkness at 25°C in a tube with restricted airflow) (Fig. 1A). The identity of the bd mutation has remained elusive due to difficulties in genetic mapping and cloning; however, aided by the completion of the Neurospora genome (Galagan et al. 2003), we used SNPs to map and identify bd. The mutation, in ras-1, causes an increase in transcripts whose gene products control asexual development, resulting in the “banding” phenotype on race tubes. In searching for the nature of a cue upon which bd could be keying its regulation, we found that manipulation of ROS levels can influence the appearance of circadian banding, strongly suggesting a link between RAS signaling and oxidative stress (ROS levels) in promotion of asexual sporulation and development. The likely involvement of multiple interconnected signaling pathways influenced by RAS and ROS has many ramifications for the study of circadian output in this and other organisms.
Results
Mapping the bd mutation
The bd mutation was initially mapped to the right arm of Linkage Group IV (LGIV) distal to pan-1 by 1.5 map units (MU) (Sargent and Woodward 1969). The mapping was further refined using three point crosses and determined to be between cot-1 (16.6 MU) and nit-3 (18.8 MU), and distal to his-4 (18.3 MU) and met-5 (8.9 MU) (Supplementary Fig. 1). At ∼20 kb per map unit, these distances still posed a daunting challenge for a chromosomal walk; therefore we developed a local single-nucleotide polymorphism (SNP) map of LG IV to more precisely map the location of bd. SNPs are a powerful tool in mapping loci, and Neurospora crassa is an ideal organism for this technique; Neurospora is haploid, independent natural isolates contain SNPs, a single cross can generate thousands of progeny for scoring purposes, and a physical map is available consisting of contigs anchored to the existing genetic map (Galagan et al. 2003).
To better map the bd mutation, 96 independent progeny were obtained from a single cross between an Oak Ridge strain containing bd and a wild-type Mauriceville strain, and these were scored for the banding phenotype (Fig. 1A). SNPs between Oak Ridge and Mauriceville strains were identified by sequencing regions from Mauriceville and comparing these to published sequence data obtained from an Oak Ridge strain (Galagan et al. 2003), and recombination frequencies were calculated between bd and each SNP (Supplementary Fig. 1). We identified one SNP (SNP12) that always segregated with the bd locus (96 out of 96 progeny cosegregated), thus localizing bd to a defined region on the physical map. The boundaries of the bd mutation were determined by identifying additional SNPs on both sides of SNP12. SNP11 and SNP13, each ∼35 kb from SNP12, had ∼1% recombination, respectively (one of 96 recombinants), and were the closest SNPs to show recombination with bd, indicating that bd was localized to within a 70-kb region on LGIV.
bd is a point mutation in ras-1
Using forced heterokaryon analysis, we determined that bd is a dominant mutation: All heterokaryons containing the bd allele exhibited banding on race tubes regardless of the nuclear ratio (Fig. 1B). To identify the bd allele, we generated 10 individual overlapping plasmids containing ∼10-kb segments of DNA that spanned the 70-kb region from a bd strain. DNA from these constructs was transformed into wild type, and transformants were scored for banding on race tubes. A single plasmid (pBC6), when transformed into wild type, caused circadian conidiation on race tubes, and this plasmid was sequenced, identifying a single C-to-T point mutation in ras-1 (NCU08823.2) that changed Thr 79 to isoleucine. The ras-1 transcript contains three introns that are spliced, resulting in an ∼1650-base-pair (bp) transcript (with an unusually long 3′ untranslated region [UTR]) that gives rise to a 213-amino-acid protein (Supplementary Fig. 1). The mutation occurs in a highly conserved region that includes the switch II surface loop whose conformation is known to change upon GTP binding and is adjacent to the GEF-binding domain (Fig. 1C). We surmise that the mutation may alter RAS-1 enough that its association with one or more GEFs, or any one of a number of effector molecules, is altered, because it seems unlikely that the mutation would solely alter the conformational change brought on by GTP binding. Because RAS-1 is a well-known signaling molecule, we use ras-1bd to refer to this allele as a means of connecting with existing nomenclature.
The ras-1bd allele causes heightened circadian output under standard laboratory conditions, so we examined whether the ras-1 transcript was circadianly regulated. RNA was extracted from wild-type and ras-1bd cultures grown under a standard 48-h time course in constant darkness, and one culture was subjected to a 15-min light pulse to examine light-induced transcription. Northern blot analysis using a ras-1 specific probe indicated that ras-1 is neither circadianly nor light-regulated, nor is there a substantial difference in the amount of transcript between wild-type and the bd strains (Fig. 1D; data not shown). The lack of circadian regulation and the similarity in transcript levels between the two strains suggest that the phenotype is due to an altered function of the protein.
ras-1bd has a subtle affect on activity
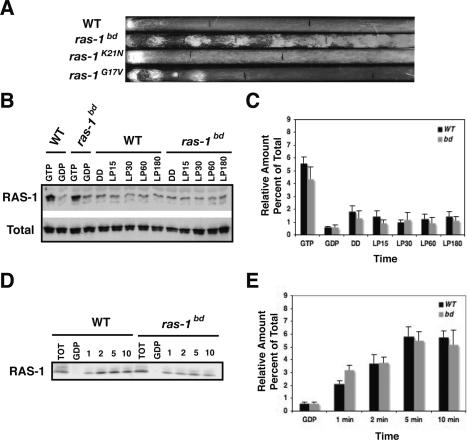
To understand how ras-1bd produces such profound effects on development and clock expression, we needed to understand the biochemical consequences of the mutation. However, ras-1bd differs from previously reported oncogenic ras mutations, so we compared its phenotype with dominant active and dominant-negative point mutants in RAS. We obtained site-directed mutants that caused RAS-1 to be in a GTP-bound active form (Gly 17 to valine) or GDP-bound negative form (Lys 21 to asparagine) and examined their phenotypes. Both the dominant active and dominant-negative strains had a severe growth defect when grown on minimal media that is not observed in ras-1bd, indicating that the T79I mutation causes a subtle effect. Because of the growth defect in the G17V and K21N strains, we were not able to examine growth on race tubes using our standard media but instead used media supplemented with yeast and malt extracts to improve growth. When we compared wild type, ras-1bd, ras-1G17V, and ras-1K21N on race tubes, we found that the dominant active G17V strain produced circadianly regulated banding that was rapidly damped after 2 d, whereas the dominant-negative K21N strain produced no conidia (Fig. 2A). This indicates that circadianly regulated conidiation requires the GTP-bound active form of RAS-1, and presumably hydrolysis of GTP to GDP is required for sustained banding. It is important to note that strains bearing ras-1bd produced substantially more conidia than those having dominant active ras-1G17V, and after 4 d constant conidiation was observed.
Figure 2.
Analysis of RAS-1bd activity. (A) Comparison of wild type and ras-1bd with dominant-negative GDP-bound (K21N) and dominant active GTP-bound (G17V) forms of RAS-1. Strains were grown on Vogel’s media supplemented with 0.1% glucose, 0.17% arginine, yeast extract, and malt extract. (B) The amount of activated RAS-1 was measured in wild-type and ras-1bd strains in the dark and following exposure to light (LP) for 30, 60, or 180 min. GTP-bound RAS was assessed by its ability to bind GST–RAF–RBD and was detected with a monoclonal antibody that specifically recognizes RAS-1. Lanes marked GTP represent lysate preincubated with GTPγS, and lanes marked GDP represent those incubated with GDP. (C) Densitometric analysis of GTP-bound RAS-1 obtained from five independent experiments; error bars represent ±SEM. (D) GDP/GTP exchange was measured by incubating lysates in the presence of GTPγS, and then GST–RBD-bound RAS-1 was detected by immunoblot analysis. TOT represents 10% of the total lysate used, and lanes marked GDP show lysate incubated with GDP; see Materials and Methods. (E). Graphical representation of GTP exchange in wild type compared with ras-1bd: Error bars represent ±SEM; n = 6.
These data suggested that RAS-1bd, like RASG17V, might be more active than wild type so we evaluated the relative levels of RAS–GTP in wild type and ras-1bd using a RAS-activation assay. This assay measures the amount of RAS-1 that is in the GTP-bound form based on binding of RAS-1–GTP to the RAS-binding domain (RBD) of RAF (Taylor et al. 2001). In both wild type and ras-1bd, we found that the vast majority of RAS-1 was in the GDP-bound form and did not bind to GST–RAF–RBD; the levels of GTP-bound RAS were comparable to lysates incubated with GDP (Fig. 2B,C), demonstrating that cellular pools of RAS-1 are mostly in the GDP-bound form. There was no reproducible difference in RAS–GTP levels between wild type and ras-1bd. The GDP exchange rate of RAS-1bd was compared with wild-type RAS-1 and was determined by measuring the rate at which GDP was exchanged with GTPγS, a nonhydrolyzable analog of GTP, using the RAS-activation assay. In this assay, RAS-1bd displays a small but consistent and reproducible increase in the amount of GTP bound at the 1-min time point (Fig. 2D,E), suggesting a small increase in the GDP exchange rate. This observation is consistent with the idea that the mutation affects the interaction of RAS-1 with one or more GEFs, and presumably downstream effectors—that is, although not inherently more active, RAS-1bd might respond more acutely or robustly to endogenous signals. We conclude that although ras-1bd causes a dramatic developmental phenotype, the mutation has, at best, only a minor effect on activity. It is important to note that ras-1 appears to be essential for Neurospora viability because we were unable to generate a homokaryotic knockout of this gene.
To get further insight into the nature of the ras-1bd mutation and its behavior in comparison with dominant active and dominant-negative RAS, we measured the ability of ras-1bd to germinate after a heat shock. It has been reported that yeast strains harboring dominant active RAS mutations show extreme sensitivity to heat shock (Engelberg et al. 1994). As expected, we saw that ras-1G17V was severely compromised in its ability to germinate after a heat shock, while in contrast, ras-1bd displayed germination levels comparable with wild type (data not shown), reinforcing the idea that the ras-1bd mutation differs from a GTP-bound dominant active RAS. We also evaluated the ability of ras-1bd to germinate in the presence of oxidative stress, and observed a decrease in the germination rate compared with wild type; in contrast, ras-1G17V did not show a dramatic phenotype. In the aggregate then, while ras-1G17V has a defect in responding to thermal stress, ras-1bd shows an alteration in its oxidative stress-response, supporting the different nature of the two mutations.
wc-1-associated transcription is elevated in ras-1bd
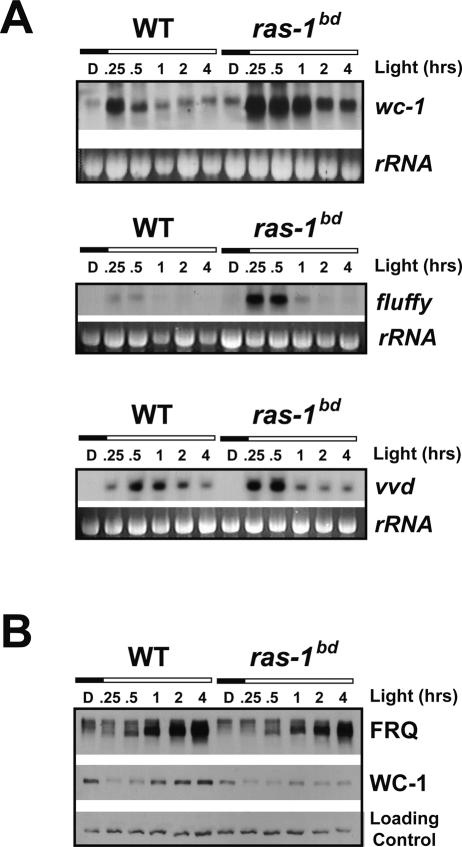
Light responses are heightened in the bd strain. For instance, in a ras-1bd background the light-activated genes eas (ccg-2), con-6, and con-10 have higher expression and remain elevated even after 4 h of light treatment, a time when expression is reduced in wild type (Arpaia et al. 1993; Lauter and Yanofsky 1993). Because WC-1 is the photoreceptor, VVD mediates photoadaptation, and FLUFFY regulates development, we examined regulation of wc-1, vvd, and fluffy in response to light in ras-1bd and wild type (Fig. 3A). All of these genes show moderately to sharply enhanced expression shortly after exposure to light in the ras-1bd background (see also Supplementary Fig. 2). However, analysis of WC-1 protein obtained from the same samples showed lower amounts in the ras-1bd strain. It has been demonstrated that WC-1 is turned over in response to light-activated transcription (Talora et al. 1999) so the lower levels of WC-1 seen in ras-1bd may reflect more light-activated turnover due to increased WC-1 activity. The elevated light response in ras-1bd, as well as the associated increase in WC-1-dependent transcription, pinpoint WC-1 as a downstream component in RAS signaling.
Figure 3.
Light-activated gene expression is elevated in ras-1bd. (A) Following treatment with saturating amounts of light for the indicated times, total RNA from wild-type and ras-1bd strains was isolated and analyzed by Northern blotting using probes specific for wc-1, fluffy, and vvd. rRNA represents the loading control. (B) Protein lysates were isolated from the same cultures used in A, and immunoblot analysis was performed using antibodies specific for FRQ and WC-1. Blots are representative of three replicates. (See also Supplementary Fig. 2 for densitometric analyses.)
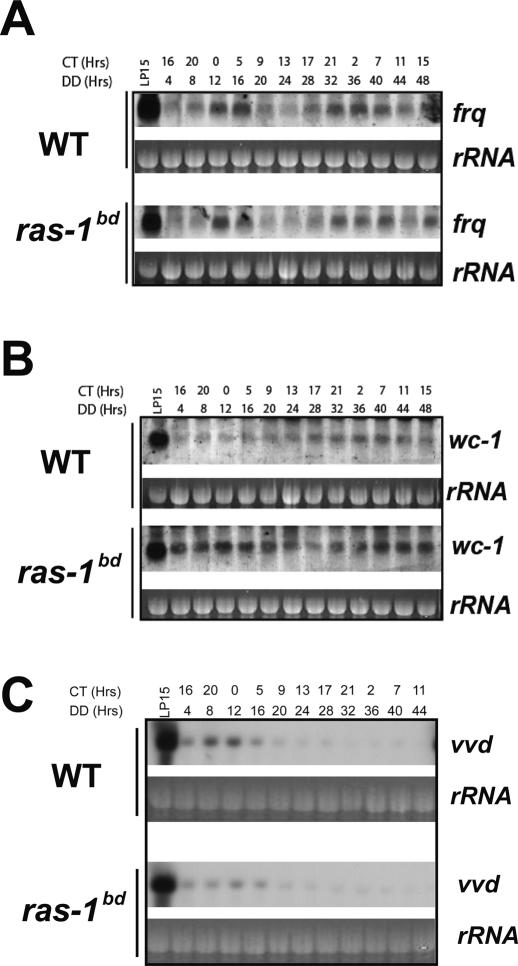
Light-activated gene expression in Neurospora is closely linked to circadian regulation with many of the components playing dual roles in these processes. Because of this, and to confirm the frequent assertion that ras-1bd strains have normal clocks, we examined circadian regulation of frq, a core component in the Neurospora circadian system, in wild type versus ras-1bd from liquid cultures expressing a circadian rhythm (Fig. 4A). Similar oscillations were observed with no discernable difference in period length, consistent with the assertion that the ras-1bd mutation does not appear to affect the clock but only enhances an output (see also Supplementary Fig. 3A). We also examined wc-1 expression and found that the amount of transcript was elevated in ras-1bd, and because WC-1 is autoregulated, we conclude once again that WC-1 is more active (Fig. 4B; Supplementary Fig. 3B). Interestingly, in extended darkness, vvd levels are initially reduced about twofold in ras-1bd (Fig. 4C; Supplementary Fig. 3C, confirmed by RT–PCR; data not shown), a fact that could contribute to the sustained WC-1 activity seen in ras-1bd.
Figure 4.
Circadian-regulated transcription in wild type versus ras-1bd. Following growth in liquid culture after standard circadian entrainment, total RNA from wild-type and ras-1bd strains was isolated at the indicated times in constant darkness (or after a 15-min light pulse given at DD24) and analyzed by Northern blotting with probes specific for frq (A), wc-1 (B), or vvd (C). Blots are representative of three to four replicates. (See also Supplementary Fig. 3 for densitometric analyses.)
Elevated ROS causes circadian banding
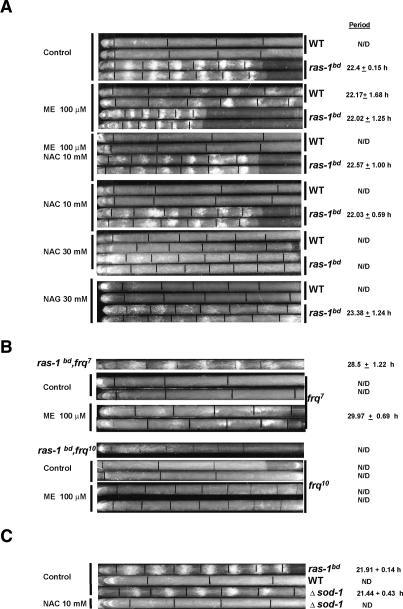
The observation that ras-1bd strains have a normal clock but profoundly enhanced light- and clock-regulated gene expression suggests that an effect of ras-1bd is amplification of an endogenous signal leading to an unnatural effect—that is, ras-1bd exposes a robust visible rhythm in growth and development that is absent in wild type under identical growth conditions. This suggests that the signal amplified in ras-1bd is perhaps an aspect of the normal primary regulation of development, so we looked for effects that would phenocopy ras-1bd. Recently it has been reported that mutations in catalase-3 (cat-3) (Michan et al. 2003) or superoxide dismutase-1 (sod-1) (Yoshida and Hasunuma 2004) show increased asexual spore development, higher carotenoid levels, and defects in photoadaptation. Because both CAT-3 and SOD-1 are involved in removal of ROS and because of the extensive literature indicating that activated ras-1 mutations cause an increase in cellular ROS levels, we asked whether we could manipulate ROS levels and affect circadian banding—i.e., would pro-oxidant small molecule ROS generators such as menadione (a ROS-generating quinone) phenocopy ras-1bd and, if so, would antioxidants such as N-acetyl-cysteine (NAC) block this effect and also phenotypically revert ras-1bd? Also, could mutations in enzymes such as CAT-3 and SOD-1 phenocopy ras-1bd? All four suppositions proved true (Fig. 5).
Figure 5.
Increased ROS causes circadianly regulated banding. (A) The wild-type strain exhibits circadian banding when evaluated in race tube media containing the ROS-generating compound menadione (ME). The effect can be reverted by the addition of the antixodant NAC, while higher concentrations of NAC abrogate banding in the ras-1bd strain. Race tube assays were conducted under conditions of constant darkness (DD) and temperature (25°C). Standard race tube media was supplemented with the indicated compounds at the specified concentrations. NAG was used as a control for NAC addition. Two out of six replicate tubes are shown for each treatment, and period lengths are reported as mean ± SD for n = 6 race tubes. (B) To confirm the circadian nature of the rhythm observed for wild type, the long-period mutant frq7 and the arrhythmic frq10 were also evaluated in menadione-race tubes and compared with ras-1bd, frq7 and ras-1bd, frq10. (C) The Δsod-1 strain displays circadian banding when assayed in race tubes, and the banding phenotype can be reverted by addition of 10 mM NAC. Where possible, circadian period lengths were measured and are reported as mean ± SD (n = 6 race tubes).
When wild type is grown in the presence of menadione, there is a clear banding pattern with periods similar to the ones observed in the ras-1bd strain (Fig. 5A; Supplementary Fig. 4). To confirm that the effect of menadione was mediated by changes in ROS levels, we added the antioxidant NAC to menadione supplemented media, and the wild-type strains were restored to their normal untreated phenotypes; treatment with higher levels of antioxidant NAC (30 mM) abrogated banding in ras-1bd, whereas the treatment with N-acetyl glycine (NAG), which is structurally similar but has no effect on oxidation, has no effect and serves as a control. To confirm that the rhythm exposed by manipulation of ROS levels was the same as that seen in ras-1bd, we tested a wild-type ras-1 strain carrying alleles of the clock gene frq, the long period clock mutant frq7, and the arrhythmic frq10, on race tubes with menadione, and observed an appropriate period of ∼29 h in frq7 and arrhythmic conidiation in frq10, similar to what is seen in a strain with both frq10 and ras-1bd (Fig. 5B).
Next we reasoned that manipulation of ROS levels through genetic means should work as well as chemicals, so genes responsible for the removal of intracellular reactive oxygen (sod-1, cat-1, cat-3, and mt-sod) were disrupted to create knockouts (Colot et al. 2006); of these, Δsod-1 showed the most striking phenotype (Fig. 5C; Supplementary Fig. 4). Circadian banding was observed in this strain, with a period appropriately increased to 29 h if combined with frq7 allele (data not shown), and addition of the antioxidant NAC eliminated banding (Fig. 5C; Supplementary Fig. 4). Addition of 100 μM of menadione to Δsod-1 resulted in oxidative stress that prevented growth, and this was ameliorated by addition of NAC (data not shown). A last set of experiments drew on the observation (Fig. 2A) that strains bearing dominant active (G17V) ras-1 banded briefly on race tubes. Menadione treatment of these dominant active ras-1 strains intensified and extended circadian banding to more than five cycles, but similar treatment failed to stimulate conidiation in the dominant-negative (K21N) allele (Supplementary Fig. 5); this indicates that active RAS is needed for conidiation and suggests that ROS may not act only through RAS. Taken together, these results confirm that artificial manipulation of ROS levels can phenocopy ras-1bd and unveil an expression of the underlying FRQ-based circadian oscillator present in a wild-type strain.
ras-1bd leads to unexpected effects on ROS levels
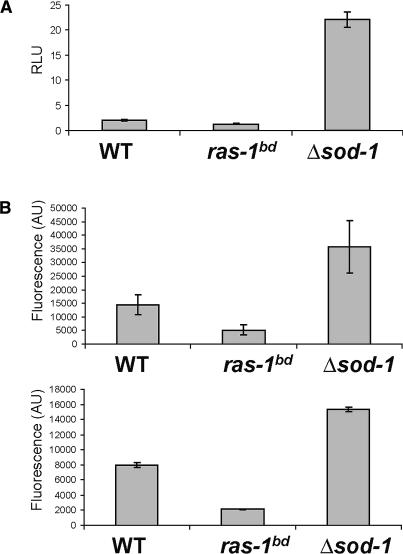
Based of the ability of elevated ROS to elicit banding, a straightforward explanation for the ras-1bd phenotype would have been that the T79I mutation in RAS-1 resulted in elevated ROS levels. To test this prediction, three different types of assays were conducted to evaluate the relative ROS levels in wild type, ras-1bd, and Δsod-1. First, superoxide was measured by lucigenin-enhanced chemiluminescence (e.g., Bai et al. 2001). Young mycelial cultures were grown for 16–20 h prior to measuring ROS levels by chemiluminescence (normalized to protein) as indicated in Materials and Methods. Δsod-1 showed levels of superoxide significantly higher than wild type or ras-1bd, and levels of superoxide appeared marginally lower in ras-1bd than in wild type (Fig. 6A). Comparable results were obtained from mycelia harvested from a culture grown on a Petri dish or race tube media topped with cellophane as described in Materials and Methods. (data not shown). To corroborate this result, we used 2′,7′-dichorodihydrofluorescein diacetate (H2DCFDA), which detects a broader spectrum of ROS with a higher specificity for H2O2. When oxidation of H2DCFDA (normalized by dry weight) was evaluated in static mycelial mats as described in Materials and Methods, the levels of fluorescence showed the same trend detected in the previous experiments (Fig. 6B). Interestingly after 1 h of incubation considerably higher levels of oxidized H2DCFDA could be detected in the extracellular media. Because intracellular oxidation of H2DCF tends to be accompanied by leakage of the product, 2′,7′-dichlorofluorescein (Ubezio and Civoli 1994), we also examined the H2DCFDA oxidation-derived fluorescence in the extracellular media of liquid cultures. After 1 h of incubation in darkness, high levels of 2′,7′-dichlorofluorescein were detected in Δsod-1 followed by wild type, while in ras-1bd they were consistently lower.
Figure 6.
Measurement of ROS levels in wild type, ras-1bd, and Δsod-1. (A) Superoxide was detected by lucigenin-enhanced chemiluminescence in mycelial layers grown for 20 h at 30°C in 0.1% liquid culture medium as described in Materials and Methods. (RLU) Relative light units. (B) ROS levels were measured following H2DCFDA diacetate oxidation in mycelia (top panel) or extracellular media (bottom panel) as indicated in Material and Methods. Values are averages of six samples and corrected by protein (A) or dry weight (B). Error bars represent the standard deviation of the mean.
Lastly, the generation of superoxide was also examined using nitroblue tetrazolium (NBT), which upon reduction forms a blue insoluble precipitate (formazan; see Materials and Methods). Consistent with the other assays, Δsod-1 presented the highest levels of formazan, reflecting higher levels of superoxide, followed by wild type and ras-1bd. The growth fronts as well as the intermediate regions of the plates were examined, and areas with low tissue density were selected in order to account for biomass differences. NBT reduction was overall lower in ras-1bd than in wild type, although some fields showed uneven reactivity toward NBT. In both strains, formazan deposition appeared as defined and punctuated areas along the hypha, while in Δsod-1 it adopted a stronger coloration spreading extensively along the cells (Supplementary Fig. 6).
Taken together, these results indicate that under the experimental conditions tested, which include those leading to conidial banding, ras-1bd displays slightly reduced or quasinormal ROS levels, while Δsod-1 presents levels significantly higher than those seen in wild-type strains. We also saw no evidence for rhythmic ROS production (data not shown). Thus it appears that, although elevated ROS are sufficient to induce banding, they are, perhaps surprisingly, not necessary for banding in a ras-1bd background: ras-1bd enhances clock-driven conidiation through a mechanism other than increased ROS, suggesting that interconnected signaling pathways influenced by both RAS-1 and ROS can regulate conidiation.
The observation that ras-1bd had altered ROS levels led us to examine the expression of genes known to be involved in ROS scavenging and production. We examined the expression of sod-1, cat-1, cat-3, nox-1, and nox-2 in both wild type and ras-1bd and noted minor reductions of sod-1 and cat-3 expression in the ras-1bd strain (data not shown). The reason for the decrease remains an enigma; it is not known whether these are reduced because ras-1bd affects their transcription or because they are down-regulated due to reduced ROS levels.
Increased fluffy expression accounts for banding
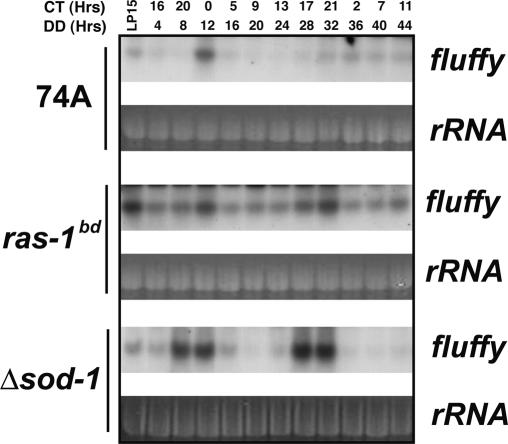
Conidiation in Neurospora is a complex developmental process that is regulated by light, the circadian clock, and oxidative stress among other factors. Genetic analyses have revealed that fluffy, which encodes a binuclear zinc cluster transcription factor, acts to integrate many environmental cues and to trigger the developmental process leading to conidiation. Because increased expression of fluffy in a wild-type background is necessary and sufficient to induce conidiation (Bailey-Shrode and Ebbole 2004), and fluffy is clock-regulated in a ras-1bd background (Correa and Bell-Pedersen 2002), we characterized the effects of alterations in RAS-1 activity and ROS levels on fluffy expression by comparing ras-1bd and Δsod-1 strains with wild type. The Δsod-1 strain has elevated ROS levels and should by all accounts mimic the treatment of wild type with menadione.
In wild-type liquid cultures, fluffy is weakly expressed, with a clear peak present only on the first day in darkness; this is consistent with race tube data (e.g., Fig. 5) that show, at best, a weak band only on the first day in darkness and no rhythmic conidiation thereafter. In contrast, fluffy expression is distinctly elevated in both ras-1bd and Δsod-1 (Fig. 7; Supplementary Fig. 3D) compared with wild type, although the mode of regulation differs slightly between the two strains. In ras-1bd there is a sharp increase in the response to light and an overall increase in the basal and peak level of transcription, with the peaks and troughs both higher than in wild type. Alternatively, in Δsod-1, light produces only a modest increase, whereas the amplitude of the rhythm in fluffy expression is greatly increased with troughs similar to wild type but peaks distinctly higher than in wild type or ras-1bd (Supplementary Fig. 3D). These differences in fluffy transcription among wild-type, ras-1bd, and Δsod-1 strains are consistent with multiple modes of regulation where both RAS and ROS can affect expression through interconnected signaling pathways. Because fluffy is a major regulator whose increased expression leads conidiation, it seems likely that the increase in banding seen on race tubes in both ras-1bd and Δsod-1 is the result of this enhanced fluffy expression.
Figure 7.
RAS-1bd and ROS increase fluffy trancription. Following growth in liquid culture after standard circadian entrainment, total RNA from wild-type, ras-1bd, and Δsod-1 strains was isolated at the indicated times in constant darkness (or after a 15-min light pulse given at DD24) and analyzed by Northern blotting with a probes specific for fluffy. Blots are representative of three to four replicates. (See also Supplementary Fig. 3D for densitometric analyses.)
Discussion
The bd mutation, long known and universally used as a key tool in the study of Neurospora circadian biology, is a dominant mutation in ras-1. ras-1bd results in increased expression of clock controlled genes (ccgs), including fluffy, a major regulator of conidiation. The identity of ras-1bd serves to highlight an aspect of circadian-regulated gene expression that, in hindsight, is not unexpected: Although the clock controls timing, other pathways, including the MAPK pathway, are known to affect circadian regulated gene expression as well as development. Furthermore, it has long been known that certain media can heighten circadianly regulated conidiation (Sargent and Kaltenborn 1972), and we show here that oxidative stress also contributes to an increase in ccg expression and circadian output.
The ras-1bd mutation (T79I, equivalent to T74 in H-Ras coordinates) resides in the distal switch II region, a solvent-exposed domain that has been established as vital in the productive interaction of Ras with GEFs (Milburn et al. 1990). This region of the protein is highly conserved and could presumably affect its interaction with one or more of the GEFs or one of the numerous downstream effectors or both. While mutations in residues such as D69 or R73 of H-Ras have an impact on the association with GEFs, CDC25 (Segal et al. 1995), SOS, or GRP (Nielsen et al. 2001), the changes T74I and T74A have been shown not to be critical for interaction with any of these exchange factors (Segal et al. 1995; Nielsen et al. 2001). Interestingly genetic data suggest that mutations in T74 may be important for proper activation of adenylyl cyclase in yeast (Segal et al. 1995), although this seems to vary with the assay conditions (Nielsen et al. 2001). In addition, a single recessive substitution in the position N81 of yeast Ras2 (equivalent to H-Ras T74) does not significantly change the Ras–GEF interaction, although it affects adenylyl cyclase activity as inferred from complementation assays (Hermann-Le Denmat and Jacquet 1997).
In addition, other mutations in the switch II domain give similarly mixed phenotypes; for example, one of these, yeast RAS2-Y78F, is capable of constitutively activating downstream effectors through a mechanism independent of elevated RAS2–GTP levels (Dalley and Cannon 1996). We found that circadian banding requires the GTP found form of RAS-1 and that the dominant T79I mutation in ras-1bd results in slightly increased GTP exchange. ras-1bd may result in a slightly more active protein that has a higher affinity for downstream effectors and/or slightly increased GTP exchange, perhaps enhancing downstream responses that would otherwise require defined and/or additional stimuli to be triggered.
The interdependent link between RAS and ROS, whereby dominant active RAS mutants cause increased ROS levels and ROS promotes the exchange of RAS-GDP for GTP, highlights ROS not only as a by-product of oxidative respiration but also as an important second messenger in signaling (Irani et al. 1997; Heo and Campbell 2006). The identification of bd as an allele of ras-1 and the observation that ROS levels are increased during cell differentiation (Hansberg and Aguirre 1990; Aguirre et al. 2005), in conjunction with the results presented here, provide a clear link between RAS and ROS in circadianly regulated conidiation. An initial trivial explanation for the data was that the T79I mutation in RAS-1bd simply made RAS-1 more active/responsive to ambient levels of ROS, thereby amplifying an endogenous signal and triggering circadian development. However, the results showing that intracellular ROS levels are lower in the ras-1bd strain compared with wild type came as a surprise and led to a refined model in which, although RAS-1 and ROS can still affect each other, the two can work through separate pathways. This model of separate action toward a common endpoint is supported by several observations. First, increased ROS resulting from menadione treatment of strains bearing the dominant active ras-1 further enhanced circadian banding (Supplementary Fig. 4), consistent with independent and additive action. Second, both ras-1bd and Δsod-1 resulted in elevated levels of fluffy, but they have opposite effects on ROS: RAS-1bd results in low levels of ROS but higher than normal RAS activity, and in Δsod-1, elevated ROS levels bypass RAS signaling. Consistent with other studies on fungal growth and development (Chen and Dickman 2004; Boyce et al. 2005), RAS and ROS are seen as acting in an interconnected signaling cascade that includes RAC and RAS–GEF, both of which can regulate conidiation; the pathways may be interconnected through the GEF, SOS, that interconnects RAS and RAC signaling (Nimnual et al. 1998), so that an increase in either RAS activity or ROS levels can trigger a developmental switch.
Regardless of the mechanisms through which RAS and ROS act and interact, it is clear that WC-1 is a downstream target, and this indicates another level of complexity in WC-related activity. It has been proposed that ROS may have a direct effect on WC-1, whereby the LOV domain is oxidized directly by superoxide increasing its activity (Yoshida and Hasunuma 2004). In contrast, the data presented here indicate that RAS-1bd stimulates WC-1 even though ROS levels are not elevated. What remains cryptic is whether WC-1-specific transcripts are elevated in ras-1bd strains because WC-1-associated transcription is activated or derepressed, but of these the former seems more likely—namely, that RAS-1 and ROS positively influence WC-1 activity. Support for this comes from two observations: First, the dominant active GTP-bound RAS displays a certain level of circadian banding, whereas the GDP-bound form is devoid of conidiation altogether (Fig. 3), indicating that activated RAS leads to banding. Second, oxygen is required for WC-1-mediated transcription, and in cultures lacking oxygen, there is no light-induced transcription until oxygen is supplied (Iigusa et al. 2005). These data suggest that RAS and ROS contribute positively to WC-1 activation and that multiple pathways feed into the biological clock and affect the amplitude of clock-regulated expression.
A remaining point not addressed in this study is the inability of CO2 to suppress conidiation in ras-1bd. Low levels of conidiation and weakly rhythmic circadian banding can be observed in a wild-type strain if air is slowly passed through a race tube; this banding is normally suppressed by increasing CO2 levels, yet in ras-1bd conidiation persists at CO2 concentrations up to 20%–25% (Sargent and Kaltenborn 1972). Recently it has been described in Candida and Cryptococcus that high CO2 levels can induce different cellular processes through the activation of adenylate cyclase and the concomitant production of cAMP (Bahn et al. 2005; for review, see Bahn and Muhlschlegel 2006; Bahn et al. 2007). In different systems, it has been shown that high cAMP levels can down-regulate the RAS/MAPK signaling pathway (e.g., Dumaz and Marais 2005), so it is plausible that the ras-1bd mutation renders a MAPK cascade less sensitive to the effects of cAMP, or that RAS-1bd in turn might down-regulate cAMP levels. Either hypothesis could account for the peculiarities of the ras-1bd strain, and indeed it has been reported that cAMP levels in ras-1bd are lower than in wild type (Hasunuma and Shinohara 1985). In addition, cpd-1 and cpd-2 (mutants in an orthophosphate cyclic phosphodiesterase) exhibit rhythmic conidiation in solid media while also showing decreased cAMP levels (Hasunuma and Shinohara 1985).
As noted in the Introduction, because of its utility for visualizing the overt expression of the circadian clock, the band allele of ras has been incorporated into the genetic background of essentially every strain used for chronobiology in Neurospora. As a result, and with very few exceptions, every study done on circadian rhythms in this organism has been carried out in the context of this altered signaling molecule that may act by affecting the production or response to intracellular ROS levels and that clearly amplifies endogenous signals to an unnatural level. There are several significant practical and conceptual implications of these results. First, a number of studies in Neurospora over the past decades have focused on circadian control of gene expression, aiming at identifying the spectrum of clock-controlled genes (ccgs) and the pathways connecting the core circadian oscillator with the target genes (for review, see Dunlap and Loros 2004, 2006; Liu and Bell-Pedersen 2006; Vitalini et al. 2006). The realization that RAS and ROS responses are affected by ras-1bd strongly suggests that a subset of the genes and pathways previously identified as primarily clock controlled may instead simply be RAS-responsive or downstream from RAS-induced developmental transitions that are muted or absent in wild type. A careful reanalysis of the meaning of clock control and the extent of this regulation in the genome is clearly warranted. Second, there is an important practical implication of the data presented in Figure 5, namely, that with the simple manipulation of intracellular ROS levels circadianly regulated banding can now be visualized in any wild-type or mutant strain of Neurospora so long as it shows adequate growth on race tubes. Specifically, there is no longer the explicit requirement that the ras-1bd allele be incorporated into the genetic background of any strain targeted for circadian rhythm studies. For instance, as gene knockouts are generated through the ongoing high-throughput systematic deletion component (http://www.dartmouth.edu/~neurosporagenome) of the Neurospora Genome Project, the effects of these gene deletions on the circadian system can be quickly assessed. Likewise, elimination of the requirement for ras-1bd will greatly facilitate the examination of natural variation of phase and period length of the circadian system in wild collected strains and should pave the way for QTL analyses.
Materials and methods
Strains, growth conditions, and race-tube assay
The Oak Ridge parent strain used for cross #402 to SNP map bd was 87-3 (bd; a), and the Mauriceville parent was FGSC 2225 (bd+; A). General conditions for growth, maintenance, and crossing have been described (Davis and deSerres 1970; Davis 2000). Forced heterokaryons were generated as previously described (Loros et al. 1986). Race tube medium consisted of 1× Vogel’s salts, 0.1% glucose, 0.17% arginine, 50 ng/mL biotin, and 1.5% bacto-agar, and liquid culture medium (LCM) is 1× Vogel’s, 0.5% arginine, and 50 ng/mL biotin with glucose at 0.1% or 2% as a carbon source. Race tubes were inoculated and incubated in constant light for ∼24 h at 25°C before transfer to constant darkness at 25°C. For SNP mapping and heterokaryon analysis, the ends of the tubes were individually wrapped with parafilm to prevent gas exchange. Race tubes were then kept in constant darkness at 25°C and the growth fronts were marked every 24 h. Rhythmic changes in conidial density as a function of time were quantified by digitally scanning race tube replicates (n = 6 to 12) with an Epson V750Pro scanner. The digital image was analyzed using Chrono (Roenneberg and Taylor 2000) as previously described (Dunlap and Loros 2005) in order to determine period values. Knockout strains were constructed as described (Colot et al. 2006)
SNP identification, SNAP PCR, and primer design
SNPs between Oak Ridge and Mauriceville were identified by (1) PCR-amplifying ∼700- to 800-bp regions of Mauriceville genomic DNA using the Roche Expand High-Fidelity PCR system according to the manufacturer’s instructions, (2) gel-purifying and sequencing the fragments from both ends using the PCR primers, and (3) comparing this sequence to the Neurospora genome sequence of an Oak Ridge strain, assembly version 3 (Galagan et al. 2003). SNPs were then scored in individual strains using SNAP PCR as previously described (Drenkard et al. 2000). Briefly, SNAP uses a standard PCR reaction with primers containing a single base pair mismatch within 3 nucleotides of the 3′ end of one allele (the specific allele) and in addition having a 3′ mismatch with the nonspecific allele. Two PCR reactions are performed for each SNP for each strain; one reaction has a Mauriceville-specific primer and a common reverse primer, and a second reaction has an Oak Ridge-specific primer and the same common reverse primer. Only one of the two reactions will yield a product for each strain, indicating which allele of a given SNP a strain possesses. SNAP PCR primers were designed using the SNAPPER program (Drenkard et al. 2000; http://ausubellab.mgh.harvard.edu).
Sequencing of genomic DNA and identification of band
To identify the bd mutation, SNP markers were used to localize bd to a 70-kb region. While ∼5-kb regions of genomic DNA from a bd strain (PCR-amplified using Takara LA Taq polymerase following the manufacturer’s instructions) were made and sequenced using primers spaced approximately every 500 bp, overlapping ∼10-kb PCR fragments spanning this 70-kb region from a bd mutant strain were transformed into Neurospora (Mauriceville strain) (Supplementary Fig. 1); one yielded transformants that banded, reflecting the dominance of the bd allele. The sequencing revealed only one variation, a C to G within the ras-1 gene that was contained in the 10-kb fragment that conferred banding. The mutation was confirmed by sequencing the region containing the mutation in three separate strains: 402-12 bd, 402-11 bd, and FGSC 6103 bd+; his-3.
Northern blots and immunoblot analysis
Standard Northern blots were performed using digoxigenin-labeled DNA probes following Roche guidelines. RNA was isolated from cells using a hot phenol extraction, and 15 or 25 μg/μL of RNA were fractionated on a 1.3% agarose formaldehyde gel (Aronson et al. 1994). DNA was isolated using the Puragene Kit following the manufacturer’s protocol. Immunoblot analysis was as described (Garceau et al. 1997). RAS activity was determined by the ability of RAS-1 GTP to associate with GST-RBD (Taylor et al. 2001) using a RAS activation kit (Pierce) following the manufacturer’s protocol.
Measurement of ROS levels
For superoxide detection, Petri dishes (35 × 10 mm) containing race tube media were point-inoculated with 5 μL of 1 × 105 conidiospores per milliliter. After 16–20 h at constant light (25°C), plates were flooded with 1 mL of NBT 0.6 mM in MOPS 5 mM (pH 7.6) and further incubated with gentle agitation (75 rpm). After 30 min, the reaction was stopped by addition of HCl to a final concentration of 0.05 N. The reduction of NBT (as formazan production) was assessed under a Nikon SMZ1500 microscope. The experiment was repeated five independent times.
Alternatively, superoxide production was detected using lucigenin-enhanced chemiluminescence. Petri dishes (35 × 10 mm) containing race tube media, covered with cellophane, were point-inoculated with 5 μL of 1 × 105 conidiospores per milliliter. After 16–20 h at constant light (25°C), 5 × 5-mm regions were cut and submerged in a 2-mL microfuge tube containing 0.5 mL of 0.2 mM lucigenin. Luminescence was immediately measured in a Turner 20e luminometer (Turner Designs) using an integration time of 15 sec. In addition, the assay was conducted using young mycelia derived from liquid cultures in 24-well plates. For this, wells containing 0.5 mL of LCM (0.1% glucose) were inoculated with 1 × 106 spores and incubated for 16–20 h at 30°C. The plate was then equilibrated for 10 min at room temperature, and the thin mycelial layer was transferred to a 2-mL microfuge tube containing 0.5 mL of 0.2 mM lucigenin and assayed as described. To correct for biomass differences, a quick protein extraction was performed for each of the samples, and the luminescence values were normalized by the total amount of proteins estimated (by the Bradford method) in the sample. Six to eight wells were analyzed for each strain.
Oxidative stress was also measured using H2DCFDA. For this, 1 × 106 spores were inoculated in a Petri dish plate containing 20 mL of LCM (2% glucose) media. After 48 h at 30°C, 6-mm-diameter plugs were cut and washed in LCM (0.1% glucose). Plugs where then transferred to a 96-well plate, and 150 μL of LCM (0.1% glucose) containing H2DCFDA (10 μg/mL) were added per well. Incubation proceeded under darkness for 1 h, after which total fluorescence was evaluated for each well at 485ex/530em nm. Twelve plugs were evaluated for each strain, and values were corrected by dry weight. Alternatively, 48-h LCM (0.1% glucose) liquid cultures (25°C LL) were complemented with H2DCFDA (10 μg/mL final) and transferred to 25°C DD (125 rpm). After 1 h, 1-mL aliquots were taken and fluorescence was measured in 150 μL fractions (in sixtuplicate)
Acknowledgments
We thank H. Inoue for kindly supplying strains bearing the dominant active and dominant-negative ras-1 alleles. This work was supported by grants from the National Institutes of Health to J.C.D. (GM34985 and GM68087) and to J.C.D. and J.J.L. (MH44651). W.J.B. is funded in part by a Ruth L. Kirschstein Post-doctoral Fellowship (GM071223) from the NIH, and L.F.L. is Pew Latin American Fellow.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1551707
References
- Aguirre J., Rios-Momberg M., Hewitt D., Hansberg W., Rios-Momberg M., Hewitt D., Hansberg W., Hewitt D., Hansberg W., Hansberg W. Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 2005;13:111–118. doi: 10.1016/j.tim.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Altschuler D.L., Muro A., Schijman A., Almonacid F.B., Torres H.N., Muro A., Schijman A., Almonacid F.B., Torres H.N., Schijman A., Almonacid F.B., Torres H.N., Almonacid F.B., Torres H.N., Torres H.N. Neurospora crassa cDNA clones coding for a new member of the ras protein family. FEBS Lett. 1990;273:103–106. doi: 10.1016/0014-5793(90)81061-r. [DOI] [PubMed] [Google Scholar]
- Aronson B.D., Johnson K.A., Loros J.J., Dunlap J.C., Johnson K.A., Loros J.J., Dunlap J.C., Loros J.J., Dunlap J.C., Dunlap J.C. Negative feedback defining a circadian clock: Autoregulation of the clock gene frequency. Science. 1994;263:1578–1584. doi: 10.1126/science.8128244. [DOI] [PubMed] [Google Scholar]
- Arpaia G., Loros J.J., Dunlap J.C., Morelli G., Macino G., Loros J.J., Dunlap J.C., Morelli G., Macino G., Dunlap J.C., Morelli G., Macino G., Morelli G., Macino G., Macino G. The interplay of light and the circadian clock. Independent dual regulation of clock-controlled gene ccg-2(eas) Plant Physiol. 1993;102:1299–1305. doi: 10.1104/pp.102.4.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn Y.S., Muhlschlegel F.A., Muhlschlegel F.A. CO2 sensing in fungi and beyond. Curr. Opin. Microbiol. 2006;9:572–578. doi: 10.1016/j.mib.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Bahn Y.S., Cox G.M., Perfect J.R., Heitman J., Cox G.M., Perfect J.R., Heitman J., Perfect J.R., Heitman J., Heitman J. Carbonic anhydrase and CO2 sensing during Cryptococcus neoformans growth, differentiation, and virulence. Curr. Biol. 2005;15:2013–2020. doi: 10.1016/j.cub.2005.09.047. [DOI] [PubMed] [Google Scholar]
- Bahn Y.S., Xue C., Idnurm A., Rutherford J.C., Heitman J., Cardenas M.E., Xue C., Idnurm A., Rutherford J.C., Heitman J., Cardenas M.E., Idnurm A., Rutherford J.C., Heitman J., Cardenas M.E., Rutherford J.C., Heitman J., Cardenas M.E., Heitman J., Cardenas M.E., Cardenas M.E. Sensing the environment: Lessons from fungi. Nat. Rev. Microbiol. 2007;5:57–69. doi: 10.1038/nrmicro1578. [DOI] [PubMed] [Google Scholar]
- Bai Z., Harvey L.M., McNeil B., Harvey L.M., McNeil B., McNeil B. Use of the chemiluminescent probe lucigenin to monitor the production of the superoxide anion radical in a recombinant Aspergillus niger (B1-D) Biotechnol. Bioeng. 2001;75:204–211. doi: 10.1002/bit.1180. [DOI] [PubMed] [Google Scholar]
- Bailey-Shrode L., Ebbole D.J., Ebbole D.J. The fluffy gene of Neurospora crassa is necessary and sufficient to induce conidiophore development. Genetics. 2004;166:1741–1749. doi: 10.1534/genetics.166.4.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell-Pedersen D., Cassone V.M., Earnest D.J., Golden S.S., Hardin P.E., Thomas T.L., Zoran M.J., Cassone V.M., Earnest D.J., Golden S.S., Hardin P.E., Thomas T.L., Zoran M.J., Earnest D.J., Golden S.S., Hardin P.E., Thomas T.L., Zoran M.J., Golden S.S., Hardin P.E., Thomas T.L., Zoran M.J., Hardin P.E., Thomas T.L., Zoran M.J., Thomas T.L., Zoran M.J., Zoran M.J. Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat. Rev. Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boriack-Sjodin P.A., Margarit S.M., Bar-Sagi D., Kuriyan J., Margarit S.M., Bar-Sagi D., Kuriyan J., Bar-Sagi D., Kuriyan J., Kuriyan J. The structural basis of the activation of Ras by Sos. Nature. 1998;394:337–343. doi: 10.1038/28548. [DOI] [PubMed] [Google Scholar]
- Borkovich K.A., Alex L.A., Yarden O., Freitag M., Turner G.E., Read N.D., Seiler S., Bell-Pedersen D., Paietta J., Plesofsky N., Alex L.A., Yarden O., Freitag M., Turner G.E., Read N.D., Seiler S., Bell-Pedersen D., Paietta J., Plesofsky N., Yarden O., Freitag M., Turner G.E., Read N.D., Seiler S., Bell-Pedersen D., Paietta J., Plesofsky N., Freitag M., Turner G.E., Read N.D., Seiler S., Bell-Pedersen D., Paietta J., Plesofsky N., Turner G.E., Read N.D., Seiler S., Bell-Pedersen D., Paietta J., Plesofsky N., Read N.D., Seiler S., Bell-Pedersen D., Paietta J., Plesofsky N., Seiler S., Bell-Pedersen D., Paietta J., Plesofsky N., Bell-Pedersen D., Paietta J., Plesofsky N., Paietta J., Plesofsky N., Plesofsky N., et al. Lessons from the genome sequence of Neurospora crassa: Tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 2004;68:1–108. doi: 10.1128/MMBR.68.1.1-108.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce K.J., Hynes M.J., Andrianopoulos A., Hynes M.J., Andrianopoulos A., Andrianopoulos A. The Ras and Rho GTPases genetically interact to co-ordinately regulate cell polarity during development in Penicillium marneffei. Mol. Microbiol. 2005;55:1487–1501. doi: 10.1111/j.1365-2958.2005.04485.x. [DOI] [PubMed] [Google Scholar]
- Brunner M., Schafmeier T., Schafmeier T. Transcriptional and post-transcriptional regulation of the circadian clock of cyanobacteria and Neurospora. Genes & Dev. 2006;20:1061–1074. doi: 10.1101/gad.1410406. [DOI] [PubMed] [Google Scholar]
- Chen C., Dickman M.B., Dickman M.B. Dominant active Rac and dominant negative Rac revert the dominant active Ras phenotype in Colletotrichum trifolii by distinct signalling pathways. Mol. Microbiol. 2004;51:1493–1507. doi: 10.1111/j.1365-2958.2003.03932.x. [DOI] [PubMed] [Google Scholar]
- Colot H.V., Park G., Turner G.E., Ringelberg C., Crew C.M., Litvinkova L., Weiss R.L., Borkovich K.A., Dunlap J.C., Park G., Turner G.E., Ringelberg C., Crew C.M., Litvinkova L., Weiss R.L., Borkovich K.A., Dunlap J.C., Turner G.E., Ringelberg C., Crew C.M., Litvinkova L., Weiss R.L., Borkovich K.A., Dunlap J.C., Ringelberg C., Crew C.M., Litvinkova L., Weiss R.L., Borkovich K.A., Dunlap J.C., Crew C.M., Litvinkova L., Weiss R.L., Borkovich K.A., Dunlap J.C., Litvinkova L., Weiss R.L., Borkovich K.A., Dunlap J.C., Weiss R.L., Borkovich K.A., Dunlap J.C., Borkovich K.A., Dunlap J.C., Dunlap J.C. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. 2006;103:10352–10357. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa A., Bell-Pedersen D., Bell-Pedersen D. Distinct signaling pathways from the circadian clock participate in regulation of rhythmic conidiospore development in Neurospora crassa. Eukaryot. Cell. 2002;1:273–280. doi: 10.1128/EC.1.2.273-280.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosthwaite S.K., Dunlap J.C., Loros J.J., Dunlap J.C., Loros J.J., Loros J.J. Neurospora wc-1 and wc-2: Transcription, photoresponses, and the origins of circadian rhythmicity. Science. 1997;276:763–769. doi: 10.1126/science.276.5313.763. [DOI] [PubMed] [Google Scholar]
- Dalley B.K., Cannon J.F., Cannon J.F. Novel, activated RAS mutations alter protein-protein interactions. Oncogene. 1996;13:1209–1220. [PubMed] [Google Scholar]
- Davis R.H. Neurospora: Contributions of a model organism. Oxford University Press; Oxford, UK: 2000. [Google Scholar]
- Davis R.L., deSerres D., deSerres D. Genetic and microbial research techniques for Neurospora crassa. Methods Enzymol. 1970;27A:79–143. [Google Scholar]
- Drenkard E., Richter B.G., Rozen S., Stutius L.M., Angell N.A., Mindrinos M., Cho R.J., Oefner P.J., Davis R.W., Ausubel F.M., Richter B.G., Rozen S., Stutius L.M., Angell N.A., Mindrinos M., Cho R.J., Oefner P.J., Davis R.W., Ausubel F.M., Rozen S., Stutius L.M., Angell N.A., Mindrinos M., Cho R.J., Oefner P.J., Davis R.W., Ausubel F.M., Stutius L.M., Angell N.A., Mindrinos M., Cho R.J., Oefner P.J., Davis R.W., Ausubel F.M., Angell N.A., Mindrinos M., Cho R.J., Oefner P.J., Davis R.W., Ausubel F.M., Mindrinos M., Cho R.J., Oefner P.J., Davis R.W., Ausubel F.M., Cho R.J., Oefner P.J., Davis R.W., Ausubel F.M., Oefner P.J., Davis R.W., Ausubel F.M., Davis R.W., Ausubel F.M., Ausubel F.M. A simple procedure for the analysis of single nucleotide polymorphisms facilitates map-based cloning in Arabidopsis. Plant Physiol. 2000;124:1483–1492. doi: 10.1104/pp.124.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumaz N., Marais R., Marais R. Integrating signals between cAMP and the RAS/RAF/MEK/ERK signalling pathways. Based on the anniversary prize of the Gesellschaft fur Biochemie und Molekularbiologie Lecture delivered on 5 July 2003 at the Special FEBS Meeting in Brussels. FEBS J. 2005;272:3491–3504. doi: 10.1111/j.1742-4658.2005.04763.x. [DOI] [PubMed] [Google Scholar]
- Dunlap J.C. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Dunlap J.C., Loros J.J., Loros J.J. The Neurospora circadian system. J. Biol. Rhythms. 2004;19:414–424. doi: 10.1177/0748730404269116. [DOI] [PubMed] [Google Scholar]
- Dunlap J.C., Loros J.J., Loros J.J. Analysis of circadian rhythms in neurospora: Overview of assays and genetic and molecular biological manipulation. Methods Enzymol. 2005;393:3–22. doi: 10.1016/S0076-6879(05)93001-2. [DOI] [PubMed] [Google Scholar]
- Dunlap J.C., Loros J.J., Loros J.J. How fungi keep time: Circadian system in Neurospora and other fungi. Curr. Opin. Microbiol. 2006;9:579–587. doi: 10.1016/j.mib.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Engelberg D., Zandi E., Parker C.S., Karin M., Zandi E., Parker C.S., Karin M., Parker C.S., Karin M., Karin M. The yeast and mammalian Ras pathways control transcription of heat shock genes independently of heat shock transcription factor. Mol. Cell. Biol. 1994;14:4929–4937. doi: 10.1128/mcb.14.7.4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan J.E., Calvo S.E., Borkovich K.A., Selker E.U., Read N.D., Jaffe D., FitzHugh W., Ma L.J., Smirnov S., Purcell S., Calvo S.E., Borkovich K.A., Selker E.U., Read N.D., Jaffe D., FitzHugh W., Ma L.J., Smirnov S., Purcell S., Borkovich K.A., Selker E.U., Read N.D., Jaffe D., FitzHugh W., Ma L.J., Smirnov S., Purcell S., Selker E.U., Read N.D., Jaffe D., FitzHugh W., Ma L.J., Smirnov S., Purcell S., Read N.D., Jaffe D., FitzHugh W., Ma L.J., Smirnov S., Purcell S., Jaffe D., FitzHugh W., Ma L.J., Smirnov S., Purcell S., FitzHugh W., Ma L.J., Smirnov S., Purcell S., Ma L.J., Smirnov S., Purcell S., Smirnov S., Purcell S., Purcell S., et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- Garceau N.Y., Liu Y., Loros J.J., Dunlap J.C., Liu Y., Loros J.J., Dunlap J.C., Loros J.J., Dunlap J.C., Dunlap J.C. Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell. 1997;89:469–476. doi: 10.1016/s0092-8674(00)80227-5. [DOI] [PubMed] [Google Scholar]
- Hansberg W., Aguirre J., Aguirre J. Hyperoxidant states cause microbial cell differentiation by cell isolation from dioxygen. J. Theor. Biol. 1990;142:201–221. doi: 10.1016/s0022-5193(05)80222-x. [DOI] [PubMed] [Google Scholar]
- Hasunuma K., Shinohara Y., Shinohara Y. Characterization of cpd-1 and cpd-2 mutants which affect the activity of orthophosphate regulated cyclic phosphodiesterase in Neurospora. Curr. Genet. 1985;10:197–203. doi: 10.1007/BF00798749. [DOI] [PubMed] [Google Scholar]
- Heo J., Campbell S.L., Campbell S.L. Ras regulation by reactive oxygen and nitrogen species. Biochemistry. 2006;45:2200–2210. doi: 10.1021/bi051872m. [DOI] [PubMed] [Google Scholar]
- Hermann-Le Denmat S., Jacquet M., Jacquet M. Yeast RAS2 mutations modulating the ras-guanine exchange factor interaction. FEBS Lett. 1997;403:95–99. doi: 10.1016/s0014-5793(97)00030-6. [DOI] [PubMed] [Google Scholar]
- Iigusa H., Yoshida Y., Hasunuma K., Yoshida Y., Hasunuma K., Hasunuma K. Oxygen and hydrogen peroxide enhance light-induced carotenoid synthesis in Neurospora crassa. FEBS Lett. 2005;579:4012–4016. doi: 10.1016/j.febslet.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Irani K., Xia Y., Zweier J.L., Sollott S.J., Der C.J., Fearon E.R., Sundaresan M., Finkel T., Goldschmidt-Clermont P.J., Xia Y., Zweier J.L., Sollott S.J., Der C.J., Fearon E.R., Sundaresan M., Finkel T., Goldschmidt-Clermont P.J., Zweier J.L., Sollott S.J., Der C.J., Fearon E.R., Sundaresan M., Finkel T., Goldschmidt-Clermont P.J., Sollott S.J., Der C.J., Fearon E.R., Sundaresan M., Finkel T., Goldschmidt-Clermont P.J., Der C.J., Fearon E.R., Sundaresan M., Finkel T., Goldschmidt-Clermont P.J., Fearon E.R., Sundaresan M., Finkel T., Goldschmidt-Clermont P.J., Sundaresan M., Finkel T., Goldschmidt-Clermont P.J., Finkel T., Goldschmidt-Clermont P.J., Goldschmidt-Clermont P.J. Mitogenic signaling mediated by oxidants in Ras-transformed fibroblasts. Science. 1997;275:1649–1652. doi: 10.1126/science.275.5306.1649. [DOI] [PubMed] [Google Scholar]
- Kana-uchi A., Yamashiro C.T., Tanabe S., Murayama T., Yamashiro C.T., Tanabe S., Murayama T., Tanabe S., Murayama T., Murayama T. A ras homologue of Neurospora crassa regulates morphology. Mol. Gen. Genet. 1997;254:427–432. doi: 10.1007/s004380050435. [DOI] [PubMed] [Google Scholar]
- Lauter F.R., Yanofsky C., Yanofsky C. Day/night and circadian rhythm control of con gene expression in Neurospora. Proc. Natl. Acad. Sci. 1993;90:8249–8253. doi: 10.1073/pnas.90.17.8249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden H., Macino G., Macino G. White collar 2, a partner in blue-light signal transduction, controlling expression of light-regulated genes in Neurospora crassa. EMBO J. 1997;16:98–109. doi: 10.1093/emboj/16.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Bell-Pedersen D., Bell-Pedersen D. Circadian rhythms in Neurospora crassa and other filamentous fungi. Eukaryot. Cell. 2006;5:1184–1193. doi: 10.1128/EC.00133-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loros J.J., Richman A., Feldman J.F., Richman A., Feldman J.F., Feldman J.F. A recessive circadian clock mutation at the frq locus of Neurospora crassa. Genetics. 1986;114:1095–1110. doi: 10.1093/genetics/114.4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall C.J. Ras effectors. Curr. Opin. Cell Biol. 1996;8:197–204. doi: 10.1016/s0955-0674(96)80066-4. [DOI] [PubMed] [Google Scholar]
- Michan S., Lledias F., Hansberg W., Lledias F., Hansberg W., Hansberg W. Asexual development is increased in Neurospora crassa cat-3-null mutant strains. Eukaryot. Cell. 2003;2:798–808. doi: 10.1128/EC.2.4.798-808.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milburn M.V., Tong L., deVos A.M., Brunger A., Yamaizumi Z., Nishimura S., Kim S.H., Tong L., deVos A.M., Brunger A., Yamaizumi Z., Nishimura S., Kim S.H., deVos A.M., Brunger A., Yamaizumi Z., Nishimura S., Kim S.H., Brunger A., Yamaizumi Z., Nishimura S., Kim S.H., Yamaizumi Z., Nishimura S., Kim S.H., Nishimura S., Kim S.H., Kim S.H. Molecular switch for signal transduction: Structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990;247:939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- Mitin N., Rossman K.L., Der C.J., Rossman K.L., Der C.J., Der C.J. Signaling interplay in Ras superfamily function. Curr. Biol. 2005;15:R563–R574. doi: 10.1016/j.cub.2005.07.010. [DOI] [PubMed] [Google Scholar]
- Nielsen K.H., Gredsted L., Broach J.R., Willumsen B.M., Gredsted L., Broach J.R., Willumsen B.M., Broach J.R., Willumsen B.M., Willumsen B.M. Sensitivity of wild type and mutant ras alleles to Ras specific exchange factors: Identification of factor specific requirements. Oncogene. 2001;20:2091–2100. doi: 10.1038/sj.onc.1204306. [DOI] [PubMed] [Google Scholar]
- Nimnual A.S., Yatsula B.A., Bar-Sagi D., Yatsula B.A., Bar-Sagi D., Bar-Sagi D. Coupling of Ras and Rac guanosine triphosphatases through the Ras exchanger Sos. Science. 1998;279:560–563. doi: 10.1126/science.279.5350.560. [DOI] [PubMed] [Google Scholar]
- Roenneberg T., Taylor W., Taylor W. Automated recordings of bioluminescence with special reference to the analysis of circadian rhythms. Methods Enzymol. 2000;305:104–119. doi: 10.1016/s0076-6879(00)05481-1. [DOI] [PubMed] [Google Scholar]
- Sanada K., Hayashi Y., Harada Y., Okano T., Fukada Y., Hayashi Y., Harada Y., Okano T., Fukada Y., Harada Y., Okano T., Fukada Y., Okano T., Fukada Y., Fukada Y. Role of circadian activation of mitogen-activated protein kinase in chick pineal clock oscillation. J. Neurosci. 2000;20:986–991. doi: 10.1523/JNEUROSCI.20-03-00986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanada K., Okano T., Fukada Y., Okano T., Fukada Y., Fukada Y. Mitogen-activated protein kinase phosphorylates and negatively regulates basic helix–loop–helix–PAS transcription factor BMAL1. J. Biol. Chem. 2002;277:267–271. doi: 10.1074/jbc.M107850200. [DOI] [PubMed] [Google Scholar]
- Sargent M.L., Kaltenborn S.H., Kaltenborn S.H. Effects of medium composition and carbon dioxide on circadian conidiation in Neurospora. Plant Physiol. 1972;50:171–175. doi: 10.1104/pp.50.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent M.L., Woodward D.O., Woodward D.O. Genetic determinants of circadian rhythmicity in Neurospora. J. Bacteriol. 1969;97:861–866. doi: 10.1128/jb.97.2.861-866.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent M.L., Briggs W.R., Woodward D.O., Briggs W.R., Woodward D.O., Woodward D.O. Circadian nature of a rhythm expressed by an invertaseless strain of Neurospora crassa. Plant Physiol. 1966;41:1343–1349. doi: 10.1104/pp.41.8.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal M., Marbach I., Willumsen B.M., Levitzki A., Marbach I., Willumsen B.M., Levitzki A., Willumsen B.M., Levitzki A., Levitzki A. Two distinct regions of Ras participate in functional interaction with GDP–GTP exchangers. Eur. J. Biochem. 1995;228:96–101. doi: 10.1111/j.1432-1033.1995.0096o.x. [DOI] [PubMed] [Google Scholar]
- Talora C., Franchi L., Linden H., Ballario P., Macino G., Franchi L., Linden H., Ballario P., Macino G., Linden H., Ballario P., Macino G., Ballario P., Macino G., Macino G. Role of a white collar-1–white collar-2 complex in blue-light signal transduction. EMBO J. 1999;18:4961–4968. doi: 10.1093/emboj/18.18.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.J., Resnick R.J., Shalloway D., Resnick R.J., Shalloway D., Shalloway D. Nonradioactive determination of Ras–GTP levels using activated ras interaction assay. Methods Enzymol. 2001;333:333–342. doi: 10.1016/s0076-6879(01)33067-7. [DOI] [PubMed] [Google Scholar]
- Ubezio P., Civoli F., Civoli F. Flow cytometric detection of hydrogen peroxide production induced by doxorubicin in cancer cells. Free Radic. Biol. Med. 1994;16:509–516. doi: 10.1016/0891-5849(94)90129-5. [DOI] [PubMed] [Google Scholar]
- Vitalini M.W., de Paula R.M., Park W.D., Bell-Pedersen D., de Paula R.M., Park W.D., Bell-Pedersen D., Park W.D., Bell-Pedersen D., Bell-Pedersen D. The rhythms of life: Circadian output pathways in Neurospora. J. Biol. Rhythms. 2006;21:432–444. doi: 10.1177/0748730406294396. [DOI] [PubMed] [Google Scholar]
- Weber F., Hung H.C., Maurer C., Kay S.A., Hung H.C., Maurer C., Kay S.A., Maurer C., Kay S.A., Kay S.A. Second messenger and Ras/MAPK signalling pathways regulate CLOCK/CYCLE-dependent transcription. J. Neurochem. 2006;98:248–257. doi: 10.1111/j.1471-4159.2006.03865.x. [DOI] [PubMed] [Google Scholar]
- Williams J.A., Su H.S., Bernards A., Field J., Sehgal A., Su H.S., Bernards A., Field J., Sehgal A., Bernards A., Field J., Sehgal A., Field J., Sehgal A., Sehgal A. A circadian output in Drosophila mediated by neurofibromatosis-1 and Ras/MAPK. Science. 2001;293:2251–2256. doi: 10.1126/science.1063097. [DOI] [PubMed] [Google Scholar]
- Yoshida Y., Hasunuma K., Hasunuma K. Reactive oxygen species affect photomorphogenesis in Neurospora crassa. J. Biol. Chem. 2004;279:6986–6993. doi: 10.1074/jbc.M310060200. [DOI] [PubMed] [Google Scholar]