Abstract
Phosphorylation is an important timekeeping mechanism in the circadian clock that has been closely studied at the level of the kinases involved but may also be tightly controlled by phosphatase action. Here we demonstrate a role for protein phosphatase 1 (PP1) in the regulation of the major timekeeping molecules in the Drosophila clock, TIMELESS (TIM) and PERIOD (PER). Flies with reduced PP1 activity exhibit a lengthened circadian period, reduced amplitude of behavioral rhythms, and an altered response to light that suggests a defect in the rising phase of clock protein expression. On a molecular level, PP1 directly dephosphorylates TIM and stabilizes it in both S2R+ cells and clock neurons. However, PP1 does not act in a simple antagonistic manner to SHAGGY (SGG), the kinase that phosphorylates TIM, because the behavioral phenotypes produced by inhibiting PP1 in flies are different from those achieved by overexpressing SGG. PP1 also acts on PER, and TIM regulates the control of PER by PP1, although it does not affect PP2A action on PER. We propose a modified model for post-translational regulation of the Drosophila clock, in which PP1 is critical for the rhythmic abundance of TIM/PER while PP2A also regulates the nuclear translocation of TIM/PER.
Keywords: Circadian rhythms, TIM, PER, phosphorylation, protein phosphatase
Circadian rhythms in Drosophila require cycling of the protein products of two major clock genes: period (per) and timeless (tim) (Scully and Kay 2000; Yang and Sehgal 2001). Cyclic expression of per and tim is executed by a feedback loop, in which PER and TIM accumulate in the cytoplasm during the early night, and subsequently enter the nucleus to inhibit their own transcription by repressing transcriptional activators CLOCK (CLK) (Allada et al. 1998) and CYCLE (CYC) (Rutila et al. 1998). In the late night/early morning, timely degradation of PER and TIM relieves the repression and allows the transcription of per and tim to start over again (Harms et al. 2004).
In the absence of rhythmic transcription, PER and TIM protein levels can still oscillate and drive behavioral rhythms (Yang and Sehgal 2001). This suggests that post-translational regulation, such as cyclic phosphorylation of PER and TIM, plays an important role in the timekeeping mechanism of the clock (Harms et al. 2004). Phosphorylation has been shown to regulate the stability of PER and the timing of nuclear expression of PER and TIM in the lateral neurons (LNs), the Drosophila central pacemaker cells (Harms et al. 2004). PER is phosphorylated by the casein kinase Iε (CKIε) homolog DOUBLETIME (DBT) (Price et al. 1998; Kloss et al. 2001) and by CK2 (Lin et al. 2002; Akten et al. 2003). Phosphorylated PER is a substrate of the E3 ligase SLMB, which targets PER for degradation (Grima et al. 2002; Ko et al. 2002). Rhythmic abundance and phosphorylation of PER also rely on its partner TIM as suggested by the following data: (1) PER and TIM form heterodimers in fly heads (Zeng et al. 1996), (2) PER protein levels are constitutively low and the circadian oscillation of PER phosphorylation is suppressed in tim0 flies (Price et al. 1995), and (3) acute expression of TIM using a heat-shock promoter rescues the accumulation and phosphorylation pattern of PER in the tim01 mutant (Suri et al. 1999). TIM abundance is controlled through light-dependent and light-independent mechanisms, both of which involve the ubiquitin-mediated proteasome pathway (Naidoo et al. 1999; Grima et al. 2002; Koh et al. 2006). Phosphorylation is implicated in mediating light-triggered TIM degradation (Zeng et al. 1996; Naidoo et al. 1999) and appears to also play a role in light-independent degradation driven by SLMB, based on the accumulation of hyperphosphorylated forms of TIM in slmb mutants (Grima et al. 2002). The only specific kinase thus far known to phosphorylate TIM is the Drosophila glycogen synthase kinase-3β (GSK-3β) ortholog SHAGGY (SGG) (Martinek et al. 2001). Overexpression of SGG promotes TIM nuclear translocation and shortens the period of behavioral rhythms in flies. However, phosphorylation by SGG does not have a major effect on TIM stability, suggesting that other kinases and/or phosphatases are involved (Martinek et al. 2001).
In contrast to the well-characterized clock kinases, little is known about the potential action of protein phosphatases in the clock. Unlike the large number of protein kinases (∼400) in eukaryotes, there are only ∼25 protein phosphatases (Honkanen and Golden 2002). Of these, protein phosphatase 2A (PP2A) and PP1 together contribute ∼90% of the total serine/threonine phosphatase activity in mammalian cells (Oliver and Shenolikar 1998). Previously, we demonstrated that PP2A dephosphorylates PER, thereby stabilizing PER and promoting its nuclear translocation (Sathyanarayanan et al. 2004). In this study, we examined whether PP1 also has a circadian function. PP1 is a ubiquitous eukaryotic enzyme and plays an important role in many cellular processes, including metabolism, cell cycle, muscle relaxation, and synaptic plasticity (Ceulemans and Bollen 2004). In Drosophila, four genes encode a catalytic subunit of PP1 (PP1c) and are named according to their chromosomal loci: 9C (also called flapwing, flw), 13C, 87B, and 96A. PP1c is highly conserved across species, and the four Drosophila PP1c isoforms are ∼90% identical to each other at the amino acid level with indistinguishable activities in vitro. Most PP1 targets and associated proteins contain a conserved PP1c-binding motif, [R/K]–X0–1–[V/I]–{P}–[F/W] (where X denotes any residue and {P} any residue except proline, so-called RVxF motif) (Egloff et al. 1997; Wakula et al. 2003), which is also found in TIM (RAIGF, amino acids 77–81). This prompted us to test TIM as a possible PP1 target. Here we show that PP1 dephosphorylates and stabilizes TIM, which is a prerequisite for the rhythmic abundance of TIM/PER, and thus plays an essential role in the post-translational regulation of the Drosophila clock.
Results
PP1 regulates TIM protein levels in S2R+ cells
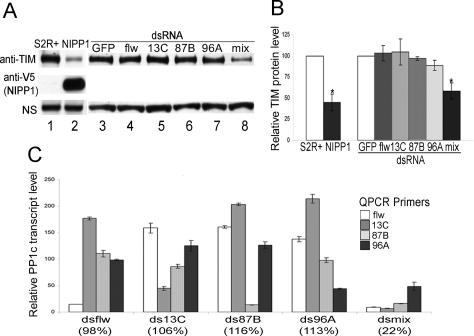
Sequence analysis of the core clock proteins in Drosophila revealed the presence of a PP1-binding motif, RVxF, in TIM, suggesting that TIM may be a target of PP1. To determine if this is the case, we examined the effect of PP1 inhibition by overexpressing an endogenous PP1 inhibitor, nuclear inhibitor of PP1 (NIPP1) in Drosophila S2R+ cells. NIPP1 is a potent and specific inhibitor of PP1 with an IC50 value of <1 pM, and it does not inhibit PP2A or other phosphatases (Sheppeck et al. 1997; McCluskey et al. 2002). In S2R+ cells, transfected TIM was stably expressed; however, when NIPP1 was coexpressed, TIM levels were decreased by ∼55% (Fig. 1A [lanes 1,2], B [left panel]).
Figure 1.
Inhibition of PP1 decreases TIM protein levels in S2R+ cells. (A) Representative Western blots show levels of TIM in cells transfected with pAct-tim alone (lane 1), or along with pAct-nipp1-V5 (lane 2), or incubated with dsRNA against the indicated proteins (lanes 3–8). Cell lysates were separated on 3%–8% tris-acetate gels and membranes were probed sequentially with anti-TIM and anti-V5 (NIPP1) antibodies. A nonspecific band (NS) that appeared while probing with the anti-TIM antibody is shown as a loading control. (B) Quantification of TIM levels from three independent experiments. Normalized anti-TIM signals are shown as the average percentages ± SEM relative to TIM levels in cells transfected with TIM alone (for NIPP1) or cells incubated with dsRNA against GFP (for dsRNA of PP1c). (*) P < 0.05. (C) Quantitative RT–PCR shows the specificity and efficiency of the dsRNA against each PP1c isoform. actin was used as an internal control to normalize PP1c transcript levels. Transcript levels of each PP1c in cells treated with the indicated dsRNA are shown as the average percentages ± SEM of their levels in control cells (dsRNA against GFP). Only the targeted PP1c isoform was efficiently knocked down and nontargeted PP1c isoforms often showed a compensatory increase in expression. The total PP1c transcript levels, relative to those in control cells, are indicated in parentheses and were calculated as described in Supplementary Figure 1.
In spite of the high sequence identity, isoform-specific function of PP1c has been reported (Rahgavan et al. 2000). To determine if the PP1 effect on TIM was isoform specific, we knocked down the expression of each PP1c isoform in S2R+ cells by RNA-mediated interference (RNAi). Knocking down each PP1c using isoform-specific PP1c double-stranded RNA (dsRNA) did not significantly change TIM levels; however, a PP1c dsRNA mix decreased TIM levels by ∼42% compared with levels in the control cells that were treated with dsRNA against green fluorescent protein (GFP) (Fig. 1A [lanes 3–8], B [right panel]). Careful inspection of PP1c dsRNA efficiency and specificity by quantitative PCR (qPCR) revealed an interesting phenomenon (Fig. 1C): Knocking down a single PP1c resulted in an increase in the expression of other isoforms, presumably to keep the total PP1 mRNA at normal levels; only when levels of all PP1c transcripts were decreased by the mix of dsRNAs was the TIM level significantly decreased. We conclude that stable expression of transfected TIM in S2R+ cells does not require any specific PP1c isoform, but rather relies on total PP1 activity. Overexpression of any single PP1c did not change TIM levels significantly, probably because PP1 activity is already saturated in S2R+ cells (Fig. 5B, the input control [below]; data not shown).
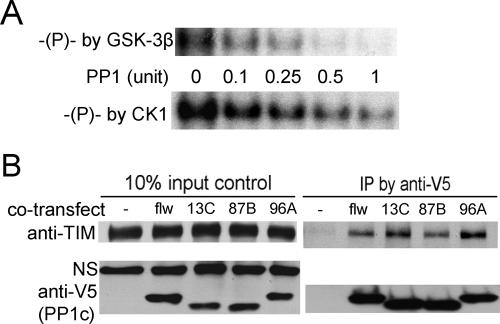
Figure 5.
PP1 directly dephosphorylates TIM in vitro and interacts with TIM in cultured cells. (A) Representative autoradiograph showing direct dephosphorylation of TIM by PP1. Immunoprecipitated TIM was in vitro phosphorylated with GSK-3β (top panel) or CK1δ (bottom panel) in the presence of γ-P32 ATP. Equal amounts of phosphorylated TIM were incubated with varying amounts (0.1–1 U) of purified PP1 in the presence of kinase inhibitor 5-IT. The blots shown are representative of two independent experiments. (B) Co-IP of TIM and PP1 from S2R+ cells. S2R+ cells were transfected with TIM alone or along with V5-tagged PP1c isoforms as indicated. Cell lysates were immunoprecipitated with anti-V5 antibody (against PP1c-V5). The presence of PP1c (34∼38 kDa) and TIM was determined by Western blots of the cell lysates (left panels) and the pellets (right panels) with anti-V5 and anti-TIM antibodies. A nonspecific band (NS) detected by the anti-TIM antibody is shown as a loading control. Three independent experiments yielded the same result.
Inhibition of PP1 alters behavioral rhythms in flies
The presence of multiple PP1c loci in Drosophila made it difficult to conduct genetic analysis using loss-of-function mutants. In fact, flies carrying hypomorphic or amorphic mutations in genes encoding PP1c-flw, 13C, and 96A did not exhibit circadian behavioral phenotypes, nor did flies heterozygous for the recessive lethal mutation in 87B (data not shown). Therefore, in order to test the physiological function of PP1 in the Drosophila clock, we used the GAL4-UAS system (Brand and Perrimon 1993) to express the endogenous PP1 inhibitor NIPP1 in clock neurons. Transgenic expression of NIPP1 was shown to specifically reduce PP1 activity in vivo and produce phenotypes similar to those of PP1c mutants (Parker et al. 2002; Bennett et al. 2003).
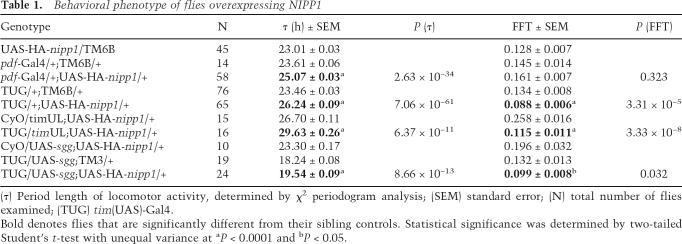
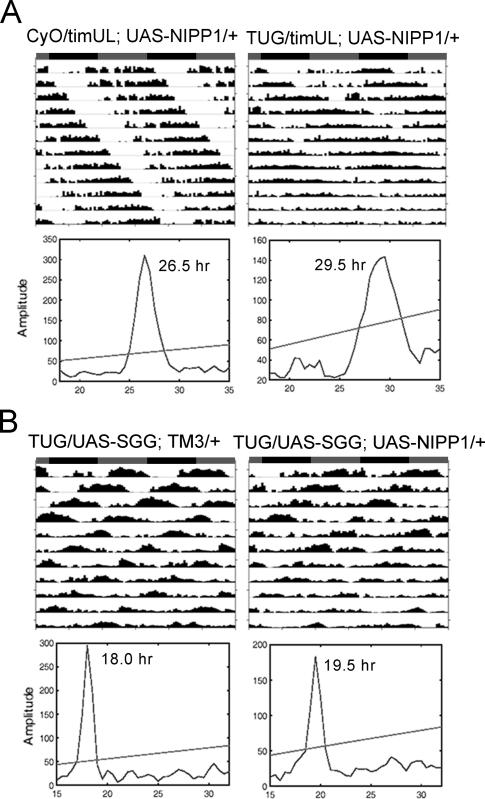
Ubiquitous expression of NIPP1, as achieved with widespread drivers such as actin-Gal4 and elav-Gal4, causes lethality (data not shown). However, flies carrying the UAS-HA-nipp1 transgene under control of the Pdf-Gal4 driver displayed a ∼1.5 h longer period than their sibling controls (Table 1). Use of the tim(UAS)-Gal4 (TUG) driver, which expresses in additional cells and perhaps also at higher levels, lengthened the period by ∼2.8 h (Fig. 2A; Table 1). Coexpression of one of the PP1 catalytic subunits, PP1-87B, rescued the long period phenotype of NIPP1 flies (Supplementary Fig. S2), suggesting that the effect of NIPP1 on circadian period is due to its inhibition of PP1. In addition to the lengthened period, elevated expression of NIPP1 by TUG significantly reduced the amplitude of the circadian rhythm (P = 3.31 × 10−5). As shown in Figure 2A, these flies displayed long period rhythms that degenerated into arrhythmia after 4–6 d in constant darkness (DD).
Table 1.
Behavioral phenotype of flies overexpressing NIPP1
(τ) Period length of locomotor activity, determined by χ2 periodogram analysis; (SEM) standard error; (N) total number of flies examined; (TUG) tim(UAS)-Gal4.
Bold denotes flies that are significantly different from their sibling controls. Statistical significance was determined by two-tailed Student’s t-test with unequal variance at aP < 0.0001 and bP < 0.05.
Figure 2.
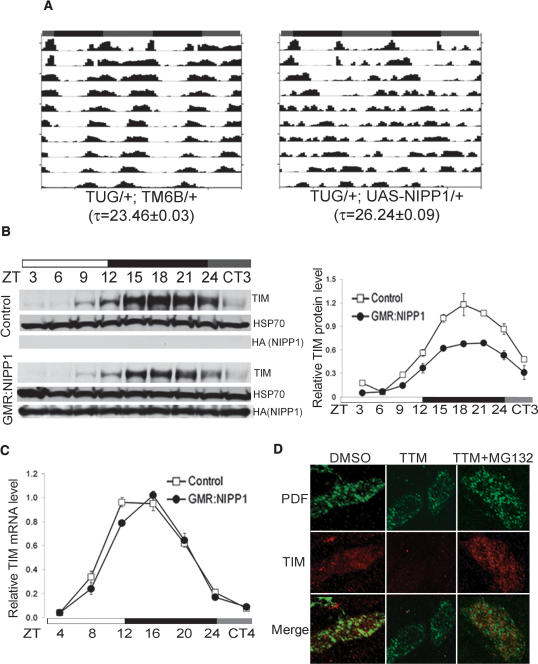
Inhibition of PP1 alters behavioral rhythms and TIM oscillation in flies. (A) Representative locomotor activity records of individual flies kept in DD for 10 d after LD entrainment. The subjective LD phases (gray:black bars), the genotype, and the circadian period (τ ± SEM) as determined by χ2 periodogram analysis are indicated. (B) Representative Western blots show decreased steady-state levels of TIM in adult fly heads overexpressing NIPP1 using a GMR-Gal4 driver. After being entrained to LD cycles, flies were collected at the indicated times in LD and on the first day in DD. (ZT0) Lights on; (ZT12) lights off; (ZT24/CT0) subjective lights on. Quantification of TIM levels in NIPP1-overexpressing flies is shown on the right. TIM signals were normalized to HSP70 (loading control) in three independent experiments and plotted as averages ± SEM over time. (C) tim mRNA levels are not reduced in NIPP1-overexpressing flies. The transcript levels of tim were quantified using actin as an internal control and then normalized to peak levels of control flies, which were set as 1. Data from two independent experiments were pooled, and averages ± SEM are shown. (D) Acute inhibition of PP1 reduces TIM stability in per01 larval LNs. TIM (red) and PDF (green) expression was visualized essentially as described in Materials and Methods. Compared with the DMSO vehicle control, samples treated with TTM had significantly decreased TIM levels, and this effect was abolished by the addition of the proteasome inhibitor MG132 (100 μM). Quantification of TIM immunointensity is shown in Supplementary Figure 2.
Flies overexpressing NIPP1 have altered TIM oscillations
To determine the molecular basis of the behavioral phenotype in flies overexpressing NIPP1, TIM abundance was examined by Western blots of fly head extracts. Since TIM expression in photoreceptor cells of the compound eye constitutes the majority of the TIM signal seen on Western blots of adult heads, we crossed flies containing the UAS-HA-nipp1 transgene to flies carrying an eye-specific driver, glass multimer reporter (GMR)-Gal4. In both the control and NIPP1-overexpressing flies, TIM protein levels oscillated with a 24-h rhythm in light:dark 12:12 (LD) cycles (Fig. 2B). However, TIM abundance was decreased in NIPP1-overexpressing flies and the amplitude of the TIM oscillation was blunted relative to the control (Fig. 2C). We noticed that, although NIPP1 locates predominantly to the nucleus (Parker et al. 2002), a decrease in TIM abundance was also observed at times when TIM is believed to be in the cytoplasm. Since cytoplasmic TIM is thought to represent TIM actively exported from the nucleus (Ashmore et al. 2003; Meyer et al. 2006), it may be exposed to NIPP1 prior to its visible nuclear expression. Alternatively, PP1 activity in the cytoplasm of LNs may be down-regulated by overexpression of NIPP1 as well, since the nuclear pools of PP1 are dynamic and in equilibrium with the cytoplasmic pools, and overexpression of NIPP1 retargets and retains cytoplasmic PP1 in the nucleus (Trinkle-Mulcahy et al. 2001; Lesage et al. 2004).
We then examined whether inhibition of PP1 affects tim mRNA cycling (Fig. 2C). In contrast to TIM protein levels, which were significantly decreased at almost all times, tim mRNA levels were similar to those of the control at all time points examined except one. Thus, although we cannot exclude the possibility that the slight difference in tim mRNA levels contributes to the overall change in the clock, it is unlikely that the robust decrease in TIM abundance is due to this subtle change in tim mRNA. More likely, the change in mRNA, which is basically a minor delay in the peak, results from the longer period of NIPP1 flies.
PP1 is required for TIM stability in LNs
To further exclude the possibility that the decreased TIM abundance in NIPP1-overexpressing flies was largely an indirect effect caused by an altered feedback loop, we inhibited PP1 in per01 flies that do not have a functional endogenous clock. PP1 was inhibited pharmacologically in dissected brains maintained in DD for 4 h at 18°C. To minimize light-triggered TIM degradation during the dissection, third instar larval brains were examined because adult brains cannot be dissected rapidly in the presence of dim red light (such light conditions are equivalent to dark for flies). The brains were incubated in Schneider’s culture medium containing a PP1 selective inhibitor Tautomycin (TTM, 4 μM). TTM is a potent cell-permeable PP1 inhibitor with ∼10-fold greater potency for PP1 as compared with PP2A (Ubukata et al. 1990); it has been used in cell culture experiments to completely inhibit PP1 activity without affecting PP2A at concentrations of up to 10 μM (Favre et al. 1997; Chen et al. 2005).
The larval brains were fixed and double-stained for TIM and Pigment Dispersing Factor (PDF). PDF is secreted by LNs and was used here to locate LNs and to define their cytoplasm (Kaneko et al. 1997). Representative samples of TIM expression (red) are shown with their corresponding PDF staining (green) in Figure 2D. Compared with the vehicle (DMSO) control, the group treated with TTM showed significantly decreased TIM intensity, as quantified with densitometry (Supplementary Fig. S3). The addition of MG132 abolished the TTM-induced decrease in TIM levels, indicating that the decrease in TIM levels induced by PP1 inhibition is due to proteasome-mediated degradation. Thus, PP1 regulates TIM stability in LNs. Together with the largely unchanged tim mRNA levels shown in Figure 2C, these data suggest that inhibition of PP1 in flies reduces TIM stability independent of the feedback loop. Since PER phosphorylation and abundance cycle in a TIM-dependent manner (see above), we also examined PER oscillations by Western blots. We found that PER protein levels were also reduced by NIPP1 (Supplementary Fig. S4). However, there was no detectable change in phosphorylation-induced mobility shifts for either PER or TIM.
Inhibition of PP1 slows down the nuclear translocation of TIM in ventrolateral neurons (LNvs)
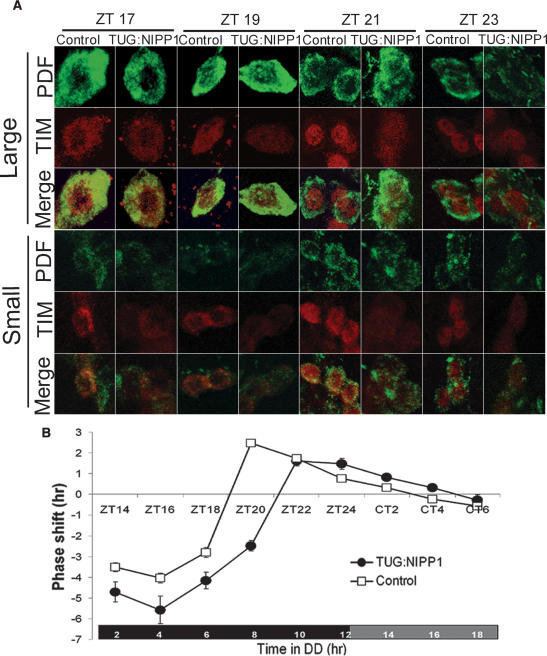
The timing of TIM/PER nuclear entry in LNs is believed to constitute an important determinant of circadian period (Harms et al. 2004). To determine whether the lengthened period of NIPP1-overexpressing flies was due to delayed TIM nuclear entry, we examined the subcellular location of TIM in both large and small LNvs of TUG:NIPP1 flies by immunostaining. After entraining flies to an LD cycle, adult brains were collected and dissected at the indicated zeitgeber times (ZT; ZT0, lights on; ZT12, lights off), and were then double-labeled for PDF and TIM (Fig. 3A).
Figure 3.
Delayed TIM accumulation causes an early- to mid-night defect in NIPP1-overexpressing flies. (A) TIM accumulation in nuclei is diminished and delayed in NIPP1-overexpressing flies. Adult fly heads were collected at the indicated times in an LD cycle after 3 d of LD entrainment. TIM expression was assayed in large LNvs (top) and small LNvs (bottom) at various times of the night. The cytoplasm of LNvs is defined by PDF staining (green). TIM staining (red) is lower in TUG:NIPP1 flies than in control flies at all time points. TIM starts entering the nucleus (uniform staining in the cytoplasm and nucleus) at the same time in TUG:NIPP1 flies and control flies (ZT19 in large-LNvs and ZT21 in small-LNvS), but the peak of TIM nuclear expression is delayed, suggesting a slower rate of nuclear accumulation of TIM in NIPP1-overexpressing flies. (B) Altered PRC of NIPP1-overexpressing flies. The phase shift (<0, phase delay; >0, phase advance) in response to a 5-min light pulse is plotted as a function of the time when the pulse was delivered. For TUG:NIPP1 flies, the time domain of phase-delaying shifts is expanded (cf. ∼11 h for TUG:NIPP1 flies and ∼9 h for the control flies) and the transition point (from phase delay to phase advance) is 2 h later (approximately ZT21 vs. approximately ZT19), while the phase-advancing domain is about the same (∼8 h) as that of the control.
In the large LNvs, TIM was predominantly cytoplasmic at ZT17 in both the control and the TUG:NIPP1 flies. At ZT19, TIM staining was distributed uniformly in the cytoplasm and the nucleus in both groups of flies; however, the intensity was lower in the TUG:NIPP1 flies. At ZT21, TIM displayed very strong nuclear staining in control flies but was much less condensed in the nuclei of the TUG:NIPP1 flies, probably because of dramatically decreased TIM levels. At ZT23, TIM in both groups was predominantly nuclear.
As reported by Shafer et al. (2002) TIM nuclear translocation differed significantly between the large and the small LNvs. In the small LNvs, TIM remained exclusively cytoplasmic through ZT19. At ZT21, uniform staining of TIM was seen in small LNvs of both the control and the TUG:NIPP1 flies, although at this time point, TIM in large neurons was already restricted to the nucleus. At ZT23, TIM was predominantly nuclear in most small LNvs of the control flies, while in TUG: NIPP1 flies only a few small LNvs displayed exclusively nuclear TIM and most were uniformly stained.
Thus, despite the difference in TIM nuclear translocation between the large and small LNvs, we found that the onset of TIM nuclear expression in both subsets of LNvs occurs at about the same time in the control and TUG: NIPP1 flies, but the peak of TIM nuclear staining is delayed in NIPP1-overexpressing flies. This is consistent with cell culture experiments indicating that the rate, but not the onset, of nuclear accumulation of TIM is positively correlated with the level of TIM (Meyer et al. 2006). Therefore, we speculate that the long period and reduced rhythmicity phenotype of NIPP1-overexpressing flies is due to reduced TIM abundance, especially the diminished and delayed nuclear accumulation of TIM.
Early- to mid-night defect in NIPP1-overexpressing flies
A change in periodicity caused by altered TIM stability was also observed in timUL flies, in which a point mutation in tim increases TIM stability and produces a ∼2.5-h longer period (Rothenfluh et al. 2000). To determine if increased stability of TIMUL could antagonize the instability caused by inhibition of PP1, we expressed NIPP1 in the timUL background. As with overexpression of NIPP1 in a wild-type background, the period of timUL flies was lengthened by ∼2.9 h (Figs. 2A, 4A; Table 1), indicating that the mutation in timUL does not make it resistant to inhibition of PP1. This completely additive effect of the timUL mutation and PP1 inhibition suggests that the two act on different aspects of the pathway. Alternatively, since timUL flies have a specific late-night defect (Rothenfluh et al. 2000), the additive effect may also occur because PP1 acts at a specific time of the circadian cycle that does not overlap with that of timUL. In fact, analysis of TIM cycling in timUL flies supported the idea that the two act at different times of the cycle (Supplementary Fig. S5).
Figure 4.
The effects of inhibition of PP1 are reduced in SGG-overexpressing flies but not in timUL flies. (A) Overexpression of NIPP1 has additive effects with the timUL mutation in lengthening the circadian period and reducing rhythmicity. (B) Inhibition of PP1 in an SGG-overexpressing background has a reduced effect on the circadian period. In a timUL background, NIPP1 reduces the amplitude of circadian rhythms and lengthens the circadian period by ∼3 h, similar to the effect it has in a wild-type background. The period-lengthening and rhythmicity-reducing effects of NIPP1 are less noticeable in an SGG-overexpressing background (see activity records), and the χ2 periodograms indicate a 1.5-h lengthened period as compared with an average of ∼2.8 h in the wild-type background. Circadian periods (hr in figure) determined by the χ2 analysis are indicated. Group averages for all circadian statistics are summarized in Table 1.
To further address when NIPP1 acts in the circadian cycle, we examined the phase response curve (PRC) for light of NIPP1-overexpressing flies. A light pulse at night triggers rapid TIM degradation, which in turn destabilizes PER (Price et al. 1995; Zeng et al. 1996; Suri et al. 1999). Depending on the time of night at which a light pulse is given, it can either slow down the accumulation of TIM/PER proteins and delay the repression of transcription, or speed up the depletion of TIM/PER proteins and advance the release of the repression, which correspondingly leads to a phase delay or a phase advance of the behavioral cycles (Young 1998). Comparing the TUG:NIPP1 and the control PRCs (Fig. 3B), we found that both the amplitude of phase shifts and the effective duration of the phase delay domain in TUG:NIPP1 flies were increased. The phase delay-to-phase advance transition point for the TUG:NIPP1 flies occurred ∼2 h later than in control flies (approximately ZT21 vs. approximately ZT19), but the phase advance domains were about the same duration (∼8 h) in both fly lines. The extension of the delay domain indicates that the accumulation of PER–TIM proteins occurs at a slower rate in NIPP1 flies. Thus, inhibition of PP1 specifically affects the early- and mid-night parts of the circadian cycle.
Taken together, NIPP1-overexpressing flies display diminished and delayed TIM nuclear accumulation (Fig. 3A) and an early- to mid-night defect (Fig. 3B), while timUL flies have prolonged nuclear expression and a late-night defect (Rothenfluh et al. 2000). These data suggest that different mechanisms for regulating TIM stability may be employed at different times of the circadian cycle.
Genetic interaction between PP1 and SGG
Both PP1 and SGG target TIM, and SGG promotes nuclear entry of TIM in the middle of the night (Martinek et al. 2001). This overlap of the substrates and of the timing of action prompted us to investigate a possible genetic interaction between SGG and PP1. It is worth noting that, although overexpression of SGG and inhibition of PP1 presumably tip the balance of TIM phosphorylation in the same direction, they display different effects on the nuclear expression of TIM (Fig. 3A; Martinek et al. 2001) and opposite effects on the circadian period: Overexpression of SGG shortens the period by ∼5 h (Table 1; Yuan et al. 2005), while overexpression of NIPP1 lengthens the period by ∼2.8 h (Table 1; Fig. 2A).
Flies coexpressing NIPP1 and SGG exhibited an average period of 19.5 h (Table 1; Fig. 4B), which is only ∼1.3 h longer than that of flies overexpressing SGG alone. Thus, the period-lengthening effect of NIPP1 was reduced by ∼50% in an SGG-overexpressing background. In addition, overexpression of NIPP1 did not reduce rhythmicity in this background as much as it did in the wild-type background (Table 1, cf. P = 0.032 and P = 3.31 × 10−5). Together, the effect of PP1 inhibition on behavioral rhythms is reduced in the SGG-overexpressing background, suggesting an interaction between SGG and PP1. It should be mentioned that cross-talk between PP1 and GSK-3β has been indicated in a few mammalian studies (DePaoli-Roach 1984; Szatmari et al. 2005). In addition, GSK-3β and other clock kinases have been shown to regulate the activity of PP1 holoenzymes (Vulsteke et al. 1997; Sakashita et al. 2003). Thus, complex interactions and feedback mechanisms bridge kinases and phosphatases, which may contribute to precise control of the intrinsic clock.
TIM is a direct target of PP1
A question raised from the above data is whether the regulation of TIM by PP1 is indirect, through its interactions with the clock kinases, or whether TIM is a direct substrate of PP1. To determine if PP1 dephosphorylates TIM directly, we immunoprecipitated TIM from transfected S2R+ cells and phosphorylated it in vitro using the active form of CK1δ and GSK-3β. The CK1δ used in these experiments is 93% similar to the corresponding region in DBT, and GSK-3β is 85% identical to SGG within the kinase domain. We found that TIM was readily phosphorylated by CK1δ, which also primed it for phosphorylation by GSK-3β.
The in vitro phosphorylation was stopped by a kinase inhibitor 5-Iodotubericidin (5-IT), and purified PP1 was then introduced into the reaction. The addition of purified PP1 resulted in a dose-dependent loss of the phosphorylation signal on TIM (Fig. 5A). Effects were seen at concentrations as low as 0.1 U (∼7.5 ng) of PP1—well within the physiological range for PP1 activity (Bennett et al. 2003). Hence, PP1 directly dephosphorylates TIM.
Given that TIM is a substrate of PP1 in vitro (Fig. 5A), we used coimmunoprecipitation (co-IP) assays to determine if PP1c actually interacts with TIM in cells. pAct-tim was cotransfected with either empty vector or an expression construct of each of the PP1c isoforms tagged with V5. As shown in Figure 5B, the anti-V5 antibody (against PP1c-V5) did not pull down TIM from the cell lysates where TIM was transfected alone, but co-IP of TIM was observed when any of the four PP1c isoforms were coexpressed. This indicates a physical interaction between PP1 and TIM.
Moderate PP1 inhibition reconstitutes the dependence of PER stability on TIM in S2R+ cells
NIPP1-expressing flies show decreased levels of PER, which we believed result from the dependence of PER stability on TIM (Price et al. 1995). However, we noticed that high concentrations of PP1 have some dephosphorylation activity toward PER in vitro (Sathyanarayanan et al. 2004). Thus, we sought to compare the effects of inhibiting PP1 on PER expression with those on TIM expression in cultured cells. Unlike what has been observed in clock neurons, PER does not require TIM for its accumulation in S2R+ cells (Fig. 6A, lane 1), indicating that the concentrations of some PER-stabilizing and/or destabilizing factors differ between these two cell types. Nevertheless, PER protein levels greatly decreased when PP1 was inhibited by TTM at concentrations of >1 μM (Fig. 6A, lanes 1–5), although a significant reduction of TIM levels required 4 μM TTM (Fig. 6A, lanes 13–17). TIM is more resistant to PP1 inhibition than PER, indicating that less PP1 activity is needed to dephosphorylate and stabilize TIM in S2R+ cells. Thus, TIM is more sensitive to PP1 action than PER.
Figure 6.
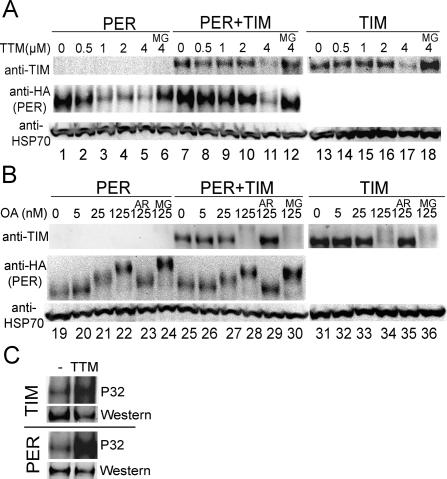
TIM protects PER from degradation induced by PP1 inhibition, but not from phosphorylation promoted by PP2A inhibition. (A,B) Inhibition of PP1 by TTM induces degradation of TIM and PER while inhibition of PP2A by OA causes phosphorylation-related mobility shifts. S2R+ cells were transfected with pAct-per-HA alone (lanes 1–6,19–24), or pAct-tim alone (lanes 13–18,31–36), or both pAct-per-HA and pAct-tim (lanes 7–12,25–30). Transfected cells were treated with varying concentrations of TTM (A) or OA (B), in some cases along with proteasome inhibitor MG132 (MG, 100 μM), or along with the GSK-3β inhibitor AR-A014418 (AR, 10 μM) as indicated (low mobility forms of TIM, produced by PP2A inhibition, are faint most likely because the antibody poorly recognizes phosphorylated TIM). The HSP70 band is shown as a loading control as well as a baseline for comparing mobility shifts between lanes across the gel. TIM is more resistant to PP1 inhibition than PER, and the presence of TIM prevents PER from degradation when PP1 is moderately inhibited. However, TIM does not block PER from hyperphosphorylation induced by PP2A inhibition. (C) Inhibition of PP1 increases TIM and PER phosphorylation. Transfected S2R+ cells were metabolically labeled with 32P (see Materials and Methods). Cells treated with 4 μM TTM (along with 100 μM MG132) display more incorporation of 32P, indicating increased phosphorylation of TIM and PER, as compared with control cells (DMSO). Western blots of one-third of the samples are shown to demonstrate protein levels. The blots shown are representative of two independent experiments.
As expected, PER levels were slightly increased when it was coexpressed with TIM in S2R+ cells (Fig. 6A, lane 7). To our surprise, PER became relatively insensitive to PP1 inhibition when TIM was present (Fig. 6A, lanes 7–11). In the presence of TIM, PER levels did not decrease until the TTM concentration reached 4 μM, at which point TIM itself was unstable. This indicates that inhibition of PP1 decreases PER levels only if TIM is decreased, suggesting that the decreased PER abundance in NIPP1-overexpressing flies is secondary to the decreased TIM protein levels (Fig. 2B; Supplementary Fig. S4). The addition of proteasome inhibitor MG132 abolished the effect of TTM (Fig. 6A, lanes 6,12,18), confirming that the decrease in protein levels was due to degradation. Thus, with moderate PP1 inhibition, which renders PER unstable but allows TIM to accumulate and to stabilize PER, we have reconstituted the dependence of PER stability on TIM in the S2 cell culture system.
TIM does not interfere with the effects of PP2A inhibition on PER in S2R+ cells
Since PP2A dephosphorylates and stabilizes PER (Sathyanarayanan et al. 2004), we were interested in determining whether TIM also protects PER from PP2A inhibition. Thus, we examined the response of TIM and PER to okadaic acid (OA), a potent PP2A inhibitor with 100-fold greater selectivity for PP2A over PP1 (Chatfield and Eastman 2004). At higher concentrations of OA (>250 nM), both TIM and PER were rendered unstable and, unlike in response to PP1 inhibition, TIM was not more resistant than PER to PP2A inhibition (Supplementary Fig. S6). At lower concentrations (≤125 nM), PER exhibited striking mobility shifts on Western blots that showed a dose dependence on OA (Fig. 6B, lanes 19–22). A mobility shift of TIM was also observed at OA concentrations of 125 nM (Fig. 6B, lanes 31–34). To confirm that the mobility shift of TIM was due to phosphorylation, a GSK-3β selective inhibitor AR-A014418 (Bhat et al. 2003) was added along with OA. At the concentration we used (10 μM), AR-A014418 specifically inhibits GSK-3β activity (Bhat et al. 2003), and it was predicted to block TIM phosphorylation. To our surprise, it blocked the mobility shifts of PER as well as TIM (Fig. 6B, lanes 23,29,35), suggesting that GSK-3β also may be involved in the phosphorylation of PER. More importantly, the presence of TIM did not block the OA-induced mobility shift of PER (Fig. 6B, lanes 25–30). Thus, although TIM was previously shown to stabilize PER in S2 cells and block its phosphorylation by DBT (Ko et al. 2002), our results indicate that not all phosphorylation of PER is affected by TIM. Taken together, TIM protects PER from degradation induced by PP1 inhibition, but it does not block hyperphosphorylation of PER caused by PP2A inhibition.
Inhibition of PP1 increases phosphorylation of TIM and PER in S2R+ cells
We noticed that, although the in vitro phosphatase and co-IP assays suggested direct dephosphorylation of TIM by PP1 (Fig. 5), inhibition of PP1 did not cause a significant change in TIM and PER electrophoretic mobility in either fly head protein extracts (Fig. 2C; Supplementary Fig. S4) or culture cells (Fig. 6A), although phosphorylation-induced mobility shifts were observed with PP2A inhibition. Since not all phosphorylation events lead to noticeable changes in protein mobility (Grasser and Konig 1992), one explanation is that PP1 only affects one or a small number of phosphorylation sites. Of note, difficulty in discerning mobility shifts upon phosphorylation of clock proteins was also reported elsewhere (Akten et al. 2003; Lin et al. 2005). Nevertheless, in order to determine whether the decreased stability of TIM and PER produced by PP1 inhibition is associated with increased phosphorylation, we metabolically labeled S2R+ cells with orthophosphates and examined the integration of 32P, a more direct indication of phosphorylation (Fig. 6C). We found that 4 μM TTM increased 32P incorporation into TIM and PER in S2R+ cells, indicating increased phosphorylation in response to PP1 inhibition. The increased phosphorylation was observed even though overall levels of protein were reduced by TTM (some degradation occurred despite the presence of MG132).
Discussion
We show here that PP1 plays a role in the Drosophila circadian clock by regulating the stability of TIM and PER. PP1 directly dephosphorylates and stabilizes TIM, which promotes accumulation of PER (Fig. 6A; Supplementary Fig. S4). The reduction in TIM/PER abundance caused by PP1 inhibition, somewhat resembling a response to continuous dim light (Konopka et al. 1989), generates a long period coupled with a reduced circadian rhythmicity phenotype.
TIM/PER accumulation, a controlled step of the timekeeping mechanism
Inhibition of PP1 in flies significantly decreases TIM abundance (Fig. 2B), especially TIM accumulation in the nucleus (Fig. 3A), but the onset of TIM nuclear entry appears intact (Fig. 2). In cell culture experiments as well we found that neither PP1c nor NIPP1 affects the subcellular localization of TIM (data not shown). Thus, although PP1 regulates TIM stability, it does not play a major role in trigging the nuclear entry of TIM, suggesting that additional regulation is required to initiate nuclear translocation. This is consistent with the finding that the onset of nuclear accumulation of TIM and PER is not correlated with their protein levels in the cytoplasm (Meyer et al. 2006). Moreover, we found that TIM protects PER from inhibition of PP1 but not of PP2A (Fig. 6), which allows TIM-stabilized PER to undergo further phosphorylation/dephosphorylation. Since dephosphorylation of PER by PP2A promotes nuclear translocation of PER (Sathyanarayanan et al. 2004) and nuclear expression of TIM appears to depend on PER (Ashmore et al. 2003; Meyer et al. 2006), we conclude that, while PP2A primarily targets PER and controls the timing of TIM/PER nuclear translocation, PP1 plays a central role in stabilizing TIM and PER and regulating their rhythmic abundance.
NIPP1-overexpressing flies have a specific early- to mid-night defect (Fig. 3B), and overexpression of NIPP1 produces an additive effect on period lengthening in a timUL background (Fig. 4A; Table 1). We infer that the destabilizing effect of NIPP1 during the accumulation (rising phase) does not affect the action of the timUL mutation, which increases TIM stability specifically in the nucleus during the late night (falling phase). The regulation of TIM stability may involve different mechanisms at different times of the cycle, which is also implied by the different mechanisms used for light-independent and light-triggered TIM degradation (Grima et al. 2002; Koh et al. 2006). These data also suggest that opposite effects on TIM stability can have the same effect on circadian period if they occur at different times of day; in this case, longer periods are produced either by decreased TIM stability during the rising phase (as produced by inhibition of PP1) or by increased TIM stability during the falling phase (produced by the timUL mutation).
Different phosphorylation sites for different regulation
Although inhibition of PP1 does not lead to a significant mobility shift, direct dephosphorylation of TIM by PP1 is suggested by the in vitro phosphatase and co-IP assays (Fig. 5) as well as by the 32P metabolic labeling in S2R+ cells (Fig. 6C). A mobility change of TIM was observed in SGG-overexpressing flies, in which TIM nuclear entry is advanced but TIM stability is not significantly decreased (Martinek et al. 2001). Hence, although PP1 dephosphorylates GSK-3β-phosphorylated TIM in vitro, their target sites may not completely overlap; in addition, the functions of PP1 and SGG in regulating the clock are not simply antagonistic (Fig. 4B). It is likely that different phosphorylation sites on TIM mediate different cellular processes and are regulated by different mechanisms. A similar idea has been proposed for the regulation of Drosophila and mammalian PER (Takano et al. 2004; Lin et al. 2005; Vanselow et al. 2006; Xu et al. 2007).
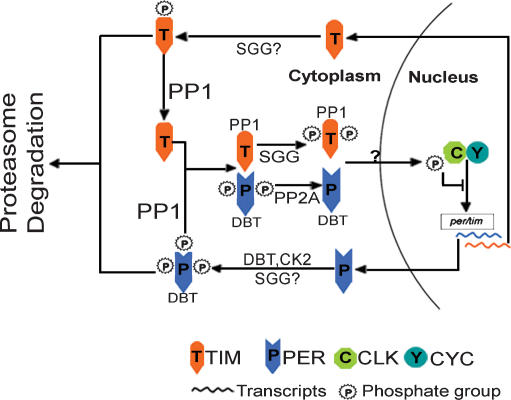
Based on these findings, we propose a modified model for the post-translational regulation of the Drosophila clock by multiple phosphorylation events (Fig. 7): Once translated, TIM and PER proteins are subject to modifications including phosphorylation, which targets them for proteasome-mediated degradation. PP1 dephosphorylates TIM at one or a small number of “stability-critical” phosphorylation sites that enable TIM to accumulate in the cytoplasm. The stabilized TIM binds to and stabilizes PER in the cytoplasm. PER is further stabilized by PP2A, which also promotes PER nuclear translocation. TIM nuclear expression is promoted by SGG phosphorylation, which does not have a major effect on TIM stability, likely because the “stability-critical” phosphorylation site is protected by PP1. TIM/PER are continually stabilized by PP1 during their nuclear translocation and accumulation, and they then inhibit their own transcription by repressing CLK and CYC. However, our data do not exclude additional indirect PP1 regulation of TIM/PER as reported for PP5 in the mammalian clock (Partch et al. 2006), nor do they rule out the involvement of additional clock target(s) of PP1.
Figure 7.
A model for post-translational regulation of the Drosophila circadian clock by multiple phosphorylation events. Dephosphorylation of TIM by PP1 prevents TIM from proteasome-mediated degradation, which enables TIM to accumulate and to stabilize PER. PP1-stabilized TIM/PER are then subject to further phosphorylation/dephosphorylation by SGG and PP2A, both of which regulate nuclear expression of TIM/PER. Question marks indicate unknown mechanisms or processes that need to be experimentally validated. For instance, it is unclear whether TIM and PER translocate to the nucleus separately or as heterodimers, whether DBT phosphorylates TIM or SGG phosphorylates PER in vivo as implied by our in vitro and cell culture experiments, and which kinase is the counterpart of PP1 that phosphorylates TIM at the “stability-critical” site.
Regulation of PP1 and its circadian function
Although PP1 is no longer viewed as a simple housekeeping gene (Honkanen and Golden 2002), a steady state of PP1c levels seems critical for an organism, as PP1c is encoded by multiple genes in most eukaryotic species (Lin et al. 1999). In flies, overexpression of NIPP1 using stronger and more widespread drivers such as tim-Gal4, elav-Gal4, and actin-Gal4 causes lethality (data not shown). In addition, we found that the expression of the Drosophila PP1c isoforms in S2R+ cells is regulated such that the total PP1c transcript level remains stable despite the loss or reduction of one PP1c mRNA (Fig. 1C). While it is beyond the scope of this study, it would be interesting to explore the mechanism underlying this phenomenon and to determine whether this regulation of PP1c expression exists in other fly cells.
The functional diversity of PP1 is exerted via its association with a large variety of regulatory subunits (Honkanen and Golden 2002). PP1 regulatory subunits not only confer in vivo substrate specificity by directing PP1c to various subcellular loci for its substrates, but also allow the activity of PP1 to be modulated in response to intracellular signals and extracellular stimuli (Egloff et al. 1997). It is possible that some adaptor proteins/PP1 regulatory subunits facilitate the interaction between PP1 and TIM documented here through co-IP experiments (Fig. 5B). In addition, although none of the PP1c isoforms is rhythmically expressed in the fly head (Supplementary Fig. S7), the regulatory subunit(s) targeting PP1 to “clock substrates” may oscillate. The paradigm for the cyclic phosphatase activity concept is PP2A, whose regulatory subunits TWS and WDB are expressed with a robust circadian rhythm and affect Drosophila behavioral rhythms (Sathyanarayanan et al. 2004).
The mechanisms by which TIM stabilizes PER are not known, but it is possible that they involve phosphorylation. Perhaps most importantly, PER is stable and nuclear in tim01 flies if the kinase DBT is also knocked down (Cyran et al. 2005), suggesting that, in the absence of TIM, PER is subject to excessive destabilizing phosphorylation. Thus, TIM may stabilize PER either by decreasing phosphorylation by DBT, or by increasing dephosphorylation by a phosphatase such as PP2A or PP1. Since DBT accumulation is not under circadian control and it is found in complexes with PER at all times in vivo (Kloss et al. 2001), it is likely that the phosphatase activity is dynamic and limiting, regulating the rhythmic abundance of PER. Our data suggest that PP1 is the primary phosphatase involved in the stabilizing effect of TIM on PER, as TIM is not more resistant than PER to PP2A inhibition (Supplementary Fig. S6) and does not appear to affect dephosphorylation of PER by PP2A (Fig. 6B). Given that PER does not contain an RVxF-binding motif as found in TIM, it is tempting to speculate that TIM is a target as well as a regulatory subunit of PP1, which may target PP1c to PER and up-regulate local PP1 activity to antagonize the destabilizing action of clock kinases on PER. We suggest that identification of the circadian-relevant PP1 regulatory subunit(s) will provide profound insight into the post-translational regulation of the clock.
Conservation of PP1 function in the clock
Our study demonstrates that PP1 plays an essential role in the regulation of the Drosophila clock. PP1 is one of the most conserved eukaryotic proteins, and it often performs similar essential functions in different species. Indeed, studies in the dinoflagellate and the fungus Neurospora have also implied a clock function for PP1. Consistent with the long period phenotype caused by inhibiting PP1 in flies, short pulses of phosphatase inhibitors in dinoflagellates cause phase delays (Comolli et al. 1996), and PP1 appears to be the dominant phosphatase mediating this circadian function (Comolli et al. 2003). In Neurospora, PP1 regulates the stability of the clock component FREQUENCY (FRQ) (Yang et al. 2004). And recently, PP1 was reported to regulate degradation of the mammalian clock protein PER2 (Gallego et al. 2006). Together, multiple studies indicate an evolutionarily conserved role for PP1 in the circadian clock.
Materials and methods
RNAi and qPCR
Isoform-specific PP1c dsRNA was generated by in vitro transcription of DNA fragments containing sequences from the untranslated region (UTR) of each PP1c and a T7 promoter at both the 5′ and 3′ ends (see the Supplemental Material for primer sequences). dsRNA against GFP was previously described (Sathyanarayanan et al. 2004). One microgram of dsRNA was added to the culture medium 5 h after transfection and was incubated with cells for 4 d before harvest for Western blot analysis. For qPCR, total RNA was isolated from S2R+ cells using RNeasy Mini kit (Qiagen), or from fly heads using Trizol (Invitrogen). After DNase treatment, RT reactions were performed using the cDNA Archive kit (Applied Biosystems) with random primers. The cDNA was then used for SYBR green-based real-time PCR (ABI Prism) with PP1c isoform-specific primers (see the Supplemental Material). actin was used to normalize the mRNA levels of each PP1c or of tim in the fly head experiments.
Western blot analysis
Cells or fly heads were collected as previously described (Sathyanarayanan et al. 2004) and then lysed in 4× LDS Sample Buffer (Invitrogen). Lysates were separated on 3%–8% tris-acetate gels (Invitrogen) and subject to Western blot. Primary antibodies were used at the following dilutions: anti-TIM (UPR8), 1:1000; anti-V5 (Invitrogen), 1:1000; anti-HA (Covance), 1:500; anti-PER (332), 1:25,000; anti-HSP70 (Sigma), 1:20,000.
Fly lines, locomotor activity measurements, and statistics
Transgenic flies carrying UAS-sgg (stock #5361) and GMR-GAL4.w[−] (stock #9146) were obtained from the Bloomington Stock Center. Other fly lines were kindly provided by several researchers (see Acknowledgments). Flies were entrained to a LD cycle at 25°C. Locomotor activity of individual flies was monitored under DD for 10–14 d and analyzed as previously described (Sathyanarayanan et al. 2004). The length of the circadian period, τ, was calculated using χ2 periodogram analysis, and the relative power of rhythmicity was indicated as Fast Fourier Transformation (FFT) value. Statistical significance was determined by two-tailed Student’s t-test with unequal variance at P < 0.05.
Immunohistochemistry
For larvae brain immunostaining, populations of developing per01 flies were kept in DD. Third instar larvae brains were briefly dissected in dim red light and incubated in Schneider’s culture medium containing vehicle DMSO or TTM (4 μM), alone or along with MG132 (100 μM) in darkness for 4 h at 18°C with gentle shaking. Brain tissue was then fixed with 4% paraformaldehyde and further dissected with lights on. Adult fly brains were dissected and processed as described (Sathyanarayanan et al. 2004). Samples were incubated with primary antibodies diluted as anti-PDF (HH74), 1:1000; and anti-TIM (UPR8), 1:1000. Ten to 20 fly brain hemispheres were examined per condition. Immunofluorescent images were obtained with a Leica confocal microscope and LNs were identified by PDF staining.
Co-IP
Transfected S2R+ cells were lysed in lysis buffer (10 mM HEPES at pH 7.5, 100 mM KCl, 1 mM EDTA, 10% Glycerol, 0.1% Triton X-100, 5 mM DTT, protease inhibitors). Cell lysates were incubated with rabbit anti-V5 antibody (Bethyl Laboratories) and Protein G Dynabeads (Invitrogen) in immunoprecipitation buffer (10 mM HEPES at pH 7.5, 300 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.3% Triton X-100, 0.01% SDS, protease inhibitor) overnight at 4°C.
In vitro dephosphorylation assay
Immunoprecipitated TIM was in vitro phosphorylated with γ-P32 ATP by CK1δ or GSK-3β (see the Supplemental Material) and then incubated with the indicated units of PP1 (New England Biolabs) for 30 min at 30°C in 20 μL of PP1 reaction buffer (50 mM HEPES at pH 7.0, 100 μM EDTA, 5 mM DTT, 0.25% Tween 20, 1 mM MnCl2, protease inhibitors, 50 μM 5-IT). Reactions were terminated by adding 4× LDS Sample Buffer (Invitrogen) and run on 3%–8% tris-acetate gels (Invitrogen). Equal loading of TIM was confirmed using Silver Stain Plus kit (Bio-Rad), and the P32 signal was detected by autoradiography.
Acknowledgments
We are very grateful to L. Alphey and D. Bennett (University of Oxford) for providing the UAS-HA-nipp1 and several UAS-HA-pp1c transgenic fly strains, M.W. Young for the timUL strain, J. Blau for the tim-UAS-Gal4 strain, S.M. Reppert for the pAct-per-HA construct, J. Price for the anti-PER antibody, and F. Lin for the pIZ-tim-GFP construct. We thank Y. Quan for invaluable help and suggestions, K. Koh and X. Zheng for helpful discussions, W.J. Joiner and A. Crocker for critical comments on the manuscript, and other members of the laboratory for useful discussions. The work was supported by NIH grant, NS48471. A.S. is an Investigator of the HHMI.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1541607
References
- Akten B., Jauch E., Genova G.K., Kim E.Y., Edery I., Raabe T., Jackson F.R., Jauch E., Genova G.K., Kim E.Y., Edery I., Raabe T., Jackson F.R., Genova G.K., Kim E.Y., Edery I., Raabe T., Jackson F.R., Kim E.Y., Edery I., Raabe T., Jackson F.R., Edery I., Raabe T., Jackson F.R., Raabe T., Jackson F.R., Jackson F.R. A role for CK2 in the Drosophila circadian oscillator. Nat. Neurosci. 2003;6:251–257. doi: 10.1038/nn1007. [DOI] [PubMed] [Google Scholar]
- Allada R., White N.E., So W.V., Hall J.C., Rosbash M., White N.E., So W.V., Hall J.C., Rosbash M., So W.V., Hall J.C., Rosbash M., Hall J.C., Rosbash M., Rosbash M. A mutant Drosophila homolog of mammalian clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- Ashmore L.J., Sathyanarayanan S., Silvestre D.W., Emerson M.M., Schotland P., Sehgal A., Sathyanarayanan S., Silvestre D.W., Emerson M.M., Schotland P., Sehgal A., Silvestre D.W., Emerson M.M., Schotland P., Sehgal A., Emerson M.M., Schotland P., Sehgal A., Schotland P., Sehgal A., Sehgal A. Novel insights into the regulation of the timeless protein. J. Neurosci. 2003;23:7810–7819. doi: 10.1523/JNEUROSCI.23-21-07810.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett D., Szoor B., Gross S., Vereshchagina N., Alphey L., Szoor B., Gross S., Vereshchagina N., Alphey L., Gross S., Vereshchagina N., Alphey L., Vereshchagina N., Alphey L., Alphey L. Ectopic expression of inhibitors of protein phosphatase type 1 (PP1) can be used to analyze roles of PP1 in Drosophila development. Genetics. 2003;164:235–245. doi: 10.1093/genetics/164.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat R., Xue Y., Berg S., Hellberg S., Ormo M., Nilsson Y., Radesater A.C., Jerning E., Markgren P.O., Borgegard T., Xue Y., Berg S., Hellberg S., Ormo M., Nilsson Y., Radesater A.C., Jerning E., Markgren P.O., Borgegard T., Berg S., Hellberg S., Ormo M., Nilsson Y., Radesater A.C., Jerning E., Markgren P.O., Borgegard T., Hellberg S., Ormo M., Nilsson Y., Radesater A.C., Jerning E., Markgren P.O., Borgegard T., Ormo M., Nilsson Y., Radesater A.C., Jerning E., Markgren P.O., Borgegard T., Nilsson Y., Radesater A.C., Jerning E., Markgren P.O., Borgegard T., Radesater A.C., Jerning E., Markgren P.O., Borgegard T., Jerning E., Markgren P.O., Borgegard T., Markgren P.O., Borgegard T., Borgegard T., et al. Structural insights and biological effects of glycogen synthase kinase 3-specific inhibitor AR-A014418. J. Biol. Chem. 2003;278:45937–45945. doi: 10.1074/jbc.M306268200. [DOI] [PubMed] [Google Scholar]
- Brand A.H., Perrimon N., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Ceulemans H., Bollen M., Bollen M. Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol. Rev. 2004;84:1–39. doi: 10.1152/physrev.00013.2003. [DOI] [PubMed] [Google Scholar]
- Chatfield K., Eastman A., Eastman A. Inhibitors of protein phosphatases 1 and 2A differentially prevent intrinsic and extrinsic apoptosis pathways. Biochem. Biophys. Res. Commun. 2004;323:1313–1320. doi: 10.1016/j.bbrc.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Chen C.S., Weng S.C., Tseng P.H., Lin H.P., Chen C.S., Weng S.C., Tseng P.H., Lin H.P., Chen C.S., Tseng P.H., Lin H.P., Chen C.S., Lin H.P., Chen C.S., Chen C.S. Histone acetylation-independent effect of histone deacetylase inhibitors on Akt through the reshuffling of protein phosphatase 1 complexes. J. Biol. Chem. 2005;280:38879–38887. doi: 10.1074/jbc.M505733200. [DOI] [PubMed] [Google Scholar]
- Comolli J., Taylor W., Rehman J., Hastings J.W., Taylor W., Rehman J., Hastings J.W., Rehman J., Hastings J.W., Hastings J.W. Inhibitors of serine/threonine phosphoprotein phosphatases alter circadian properties in Gonyaulax polyedra. Plant Physiol. 1996;111:285–291. doi: 10.1104/pp.111.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comolli J.C., Fagan T., Hastings J.W., Fagan T., Hastings J.W., Hastings J.W. A type-1 phosphoprotein phosphatase from a dinoflagellate as a possible component of the circadian mechanism. J. Biol. Rhythms. 2003;18:367–376. doi: 10.1177/0748730403254103. [DOI] [PubMed] [Google Scholar]
- Cyran S.A., Yiannoulos G., Buchsbaum A.M., Saez L., Young M.W., Blau J., Yiannoulos G., Buchsbaum A.M., Saez L., Young M.W., Blau J., Buchsbaum A.M., Saez L., Young M.W., Blau J., Saez L., Young M.W., Blau J., Young M.W., Blau J., Blau J. The double-time protein kinase regulates the subcellular localization of the Drosophila clock protein period. J. Neurosci. 2005;25:5430–5437. doi: 10.1523/JNEUROSCI.0263-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePaoli-Roach A.A. Synergistic phosphorylation and activation of ATP-Mg-dependent phosphoprotein phosphatase by F A/GSK-3 and casein kinase II (PC0.7) J. Biol. Chem. 1984;259:12144–12152. [PubMed] [Google Scholar]
- Egloff M.P., Johnson D.F., Moorhead G., Cohen P.T., Cohen P., Barford D., Johnson D.F., Moorhead G., Cohen P.T., Cohen P., Barford D., Moorhead G., Cohen P.T., Cohen P., Barford D., Cohen P.T., Cohen P., Barford D., Cohen P., Barford D., Barford D. Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J. 1997;16:1876–1887. doi: 10.1093/emboj/16.8.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre B., Turowski P., Hemmings B.A., Turowski P., Hemmings B.A., Hemmings B.A. Differential inhibition and posttranslational modification of protein phosphatase 1 and 2A in MCF7 cells treated with calyculin-A, okadaic acid, and tautomycin. J. Biol. Chem. 1997;272:13856–13863. doi: 10.1074/jbc.272.21.13856. [DOI] [PubMed] [Google Scholar]
- Gallego M., Kang H., Virshup D.M., Kang H., Virshup D.M., Virshup D.M. Protein phosphatase 1 regulates the stability of the circadian protein PER2. Biochem. J. 2006;399:169–175. doi: 10.1042/BJ20060678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasser F.A., Konig S., Konig S. Phosphorylation of SV40 large T antigen at threonine residues results in conversion to a lower apparent molecular weight form. Arch. Virol. 1992;126:313–320. doi: 10.1007/BF01309704. [DOI] [PubMed] [Google Scholar]
- Grima B., Lamouroux A., Chelot E., Papin C., Limbourg-Bouchon B., Rouyer F., Lamouroux A., Chelot E., Papin C., Limbourg-Bouchon B., Rouyer F., Chelot E., Papin C., Limbourg-Bouchon B., Rouyer F., Papin C., Limbourg-Bouchon B., Rouyer F., Limbourg-Bouchon B., Rouyer F., Rouyer F. The F-box protein slimb controls the levels of clock proteins period and timeless. Nature. 2002;420:178–182. doi: 10.1038/nature01122. [DOI] [PubMed] [Google Scholar]
- Harms E., Kivimae S., Young M.W., Saez L., Kivimae S., Young M.W., Saez L., Young M.W., Saez L., Saez L. Posttranscriptional and posttranslational regulation of clock genes. J. Biol. Rhythms. 2004;19:361–373. doi: 10.1177/0748730404268111. [DOI] [PubMed] [Google Scholar]
- Honkanen R.E., Golden T., Golden T. Regulators of serine/threonine protein phosphatases at the dawn of a clinical era? Curr. Med. Chem. 2002;9:2055–2075. doi: 10.2174/0929867023368836. [DOI] [PubMed] [Google Scholar]
- Kaneko M., Helfrich-Forster C., Hall J.C., Helfrich-Forster C., Hall J.C., Hall J.C. Spatial and temporal expression of the period and timeless genes in the developing nervous system of Drosophila: Newly identified pacemaker candidates and novel features of clock gene product cycling. J. Neurosci. 1997;17:6745–6760. doi: 10.1523/JNEUROSCI.17-17-06745.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss B., Rothenfluh A., Young M.W., Saez L., Rothenfluh A., Young M.W., Saez L., Young M.W., Saez L., Saez L. Phosphorylation of period is influenced by cycling physical associations of double-time, period, and timeless in the Drosophila clock. Neuron. 2001;30:699–706. doi: 10.1016/s0896-6273(01)00320-8. [DOI] [PubMed] [Google Scholar]
- Ko H.W., Jiang J., Edery I., Jiang J., Edery I., Edery I. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature. 2002;420:673–678. doi: 10.1038/nature01272. [DOI] [PubMed] [Google Scholar]
- Koh K., Zheng X., Sehgal A., Zheng X., Sehgal A., Sehgal A. JETLAG resets the Drosophila circadian clock by promoting light-induced degradation of TIMELESS. Science. 2006;312:1809–1812. doi: 10.1126/science.1124951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka R.J., Pittendrigh C., Orr D., Pittendrigh C., Orr D., Orr D. Reciprocal behaviour associated with altered homeostasis and photosensitivity of Drosophila clock mutants. J. Neurogenet. 1989;1:1–10. doi: 10.3109/01677068909107096. [DOI] [PubMed] [Google Scholar]
- Lesage B., Beullens M., Nuytten M., Van Eynde A., Keppens S., Himpens B., Bollen M., Beullens M., Nuytten M., Van Eynde A., Keppens S., Himpens B., Bollen M., Nuytten M., Van Eynde A., Keppens S., Himpens B., Bollen M., Van Eynde A., Keppens S., Himpens B., Bollen M., Keppens S., Himpens B., Bollen M., Himpens B., Bollen M., Bollen M. Interactor-mediated nuclear translocation and retention of protein phosphatase-1. J. Biol. Chem. 2004;279:55978–55984. doi: 10.1074/jbc.M411911200. [DOI] [PubMed] [Google Scholar]
- Lin Q., Buckler IV E.S., Muse S.V., Walker J.C., Buckler IV E.S., Muse S.V., Walker J.C., Muse S.V., Walker J.C., Walker J.C. Molecular evolution of type 1 serine/threonine protein phosphatases. Mol. Phylogenet. Evol. 1999;12:57–66. doi: 10.1006/mpev.1998.0560. [DOI] [PubMed] [Google Scholar]
- Lin J.M., Kilman V.L., Keegan K., Paddock B., Emery-Le M., Rosbash M., Allada R., Kilman V.L., Keegan K., Paddock B., Emery-Le M., Rosbash M., Allada R., Keegan K., Paddock B., Emery-Le M., Rosbash M., Allada R., Paddock B., Emery-Le M., Rosbash M., Allada R., Emery-Le M., Rosbash M., Allada R., Rosbash M., Allada R., Allada R. A role for casein kinase 2α in the Drosophila circadian clock. Nature. 2002;420:816–820. doi: 10.1038/nature01235. [DOI] [PubMed] [Google Scholar]
- Lin J.M., Schroeder A., Allada R., Schroeder A., Allada R., Allada R. In vivo circadian function of casein kinase 2 phosphorylation sites in Drosophila PERIOD. J. Neurosci. 2005;25:11175–11183. doi: 10.1523/JNEUROSCI.2159-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinek S., Inonog S., Manoukian A.S., Young M.W., Inonog S., Manoukian A.S., Young M.W., Manoukian A.S., Young M.W., Young M.W. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell. 2001;105:769–779. doi: 10.1016/s0092-8674(01)00383-x. [DOI] [PubMed] [Google Scholar]
- McCluskey A., Sim A.T., Sakoff J.A., Sim A.T., Sakoff J.A., Sakoff J.A. Serine–threonine protein phosphatase inhibitors: Development of potential therapeutic strategies. J. Med. Chem. 2002;45:1151–1175. doi: 10.1021/jm010066k. [DOI] [PubMed] [Google Scholar]
- Meyer P., Saez L., Young M.W., Saez L., Young M.W., Young M.W. PER–TIM interactions in living Drosophila cells: An interval timer for the circadian clock. Science. 2006;311:226–229. doi: 10.1126/science.1118126. [DOI] [PubMed] [Google Scholar]
- Naidoo N., Song W., Hunter-Ensor M., Sehgal A., Song W., Hunter-Ensor M., Sehgal A., Hunter-Ensor M., Sehgal A., Sehgal A. A role for the proteasome in the light response of the timeless clock protein. Science. 1999;285:1737–1741. doi: 10.1126/science.285.5434.1737. [DOI] [PubMed] [Google Scholar]
- Oliver C.J., Shenolikar S., Shenolikar S. Physiologic importance of protein phosphatase inhibitors. Front. Biosci. 1998;3:D961–D972. doi: 10.2741/a336. [DOI] [PubMed] [Google Scholar]
- Parker L., Gross S., Beullens M., Bollen M., Bennett D., Alphey L., Gross S., Beullens M., Bollen M., Bennett D., Alphey L., Beullens M., Bollen M., Bennett D., Alphey L., Bollen M., Bennett D., Alphey L., Bennett D., Alphey L., Alphey L. Functional interaction between nuclear inhibitor of protein phosphatase type 1 (NIPP1) and protein phosphatase type 1 (PP1) in Drosophila: Consequences of over-expression of NIPP1 in flies and suppression by co-expression of PP1. Biochem. J. 2002;368:789–797. doi: 10.1042/BJ20020582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partch C.L., Shields K.F., Thompson C.L., Selby C.P., Sancar A., Shields K.F., Thompson C.L., Selby C.P., Sancar A., Thompson C.L., Selby C.P., Sancar A., Selby C.P., Sancar A., Sancar A. Posttranslational regulation of the mammalian circadian clock by cryptochrome and protein phosphatase 5. Proc. Natl. Acad. Sci. 2006;103:10467–10472. doi: 10.1073/pnas.0604138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J.L., Dembinska M.E., Young M.W., Rosbash M., Dembinska M.E., Young M.W., Rosbash M., Young M.W., Rosbash M., Rosbash M. Suppression of PERIOD protein abundance and circadian cycling by the Drosophila clock mutation timeless. EMBO J. 1995;14:4044–4049. doi: 10.1002/j.1460-2075.1995.tb00075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J.L., Blau J., Rothenfluh A., Abodeely M., Kloss B., Young M.W., Blau J., Rothenfluh A., Abodeely M., Kloss B., Young M.W., Rothenfluh A., Abodeely M., Kloss B., Young M.W., Abodeely M., Kloss B., Young M.W., Kloss B., Young M.W., Young M.W. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- Rahgavan S., Williams I., Aslam H., Thomas D., Ször B., Morgan G., Gross S., Turner J., Fernandes J., Vijayraghavan K., Williams I., Aslam H., Thomas D., Ször B., Morgan G., Gross S., Turner J., Fernandes J., Vijayraghavan K., Aslam H., Thomas D., Ször B., Morgan G., Gross S., Turner J., Fernandes J., Vijayraghavan K., Thomas D., Ször B., Morgan G., Gross S., Turner J., Fernandes J., Vijayraghavan K., Ször B., Morgan G., Gross S., Turner J., Fernandes J., Vijayraghavan K., Morgan G., Gross S., Turner J., Fernandes J., Vijayraghavan K., Gross S., Turner J., Fernandes J., Vijayraghavan K., Turner J., Fernandes J., Vijayraghavan K., Fernandes J., Vijayraghavan K., Vijayraghavan K., et al. Protein phosphatase 1β is required for the maintenance of muscle attachments. Curr. Biol. 2000;10:269–272. doi: 10.1016/s0960-9822(00)00364-x. [DOI] [PubMed] [Google Scholar]
- Rothenfluh A., Young M.W., Saez L., Young M.W., Saez L., Saez L. A TIMELESS-independent function for PERIOD proteins in the Drosophila clock. Neuron. 2000;26:505–514. doi: 10.1016/s0896-6273(00)81182-4. [DOI] [PubMed] [Google Scholar]
- Rutila J.E., Suri V., Le M., So V., Rosbash M., Hall J.C., Suri V., Le M., So V., Rosbash M., Hall J.C., Le M., So V., Rosbash M., Hall J.C., So V., Rosbash M., Hall J.C., Rosbash M., Hall J.C., Hall J.C. CYCLE is a second bHLH–PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell. 1998;93:805–814. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- Sakashita G., Shima H., Komatsu M., Urano T., Kikuchi A., Kikuchi K., Shima H., Komatsu M., Urano T., Kikuchi A., Kikuchi K., Komatsu M., Urano T., Kikuchi A., Kikuchi K., Urano T., Kikuchi A., Kikuchi K., Kikuchi A., Kikuchi K., Kikuchi K. Regulation of type 1 protein phosphatase/inhibitor-2 complex by glycogen synthase kinase-3β in intact cells. J. Biochem. 2003;133:165–171. doi: 10.1093/jb/mvg020. [DOI] [PubMed] [Google Scholar]
- Sathyanarayanan S., Zheng X., Xiao R., Sehgal A., Zheng X., Xiao R., Sehgal A., Xiao R., Sehgal A., Sehgal A. Posttranslational regulation of Drosophila PERIOD protein by protein phosphatase 2A. Cell. 2004;116:603–615. doi: 10.1016/s0092-8674(04)00128-x. [DOI] [PubMed] [Google Scholar]
- Scully A.L., Kay S.A., Kay S.A. Time flies for Drosophila. Cell. 2000;100:297–300. doi: 10.1016/s0092-8674(00)80665-0. [DOI] [PubMed] [Google Scholar]
- Shafer O.T., Rosbash M., Truman J.W., Rosbash M., Truman J.W., Truman J.W. Sequential nuclear accumulation of the clock proteins period and timeless in the pacemaker neurons of Drosophila melanogaster. J. Neurosci. 2002;22:5946–5954. doi: 10.1523/JNEUROSCI.22-14-05946.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppeck J.E., II, Gauss C.M., Chamberlin A.R., Gauss C.M., Chamberlin A.R., Chamberlin A.R. Inhibition of the Ser-Thr phosphatases PP1 and PP2A by naturally occurring toxins. Bioorg. Med. Chem. 1997;5:1739–1750. doi: 10.1016/s0968-0896(97)00146-6. [DOI] [PubMed] [Google Scholar]
- Suri V., Lanjuin A., Rosbash M., Lanjuin A., Rosbash M., Rosbash M. TIMELESS-dependent positive and negative autoregulation in the Drosophila circadian clock. EMBO J. 1999;18:675–686. doi: 10.1093/emboj/18.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatmari E., Habas A., Yang P., Zheng J.J., Hagg T., Hetman M., Habas A., Yang P., Zheng J.J., Hagg T., Hetman M., Yang P., Zheng J.J., Hagg T., Hetman M., Zheng J.J., Hagg T., Hetman M., Hagg T., Hetman M., Hetman M. A positive feedback loop between glycogen synthase kinase 3β and protein phosphatase 1 after stimulation of NR2B NMDA receptors in forebrain neurons. J. Biol. Chem. 2005;280:37526–37535. doi: 10.1074/jbc.M502699200. [DOI] [PubMed] [Google Scholar]
- Takano A., Isojima Y., Nagai K., Isojima Y., Nagai K., Nagai K. Identification of mPer1 phosphorylation sites responsible for the nuclear entry. J. Biol. Chem. 2004;279:32578–32585. doi: 10.1074/jbc.M403433200. [DOI] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L., Sleeman J.E., Lamond A.I., Sleeman J.E., Lamond A.I., Lamond A.I. Dynamic targeting of protein phosphatase 1 within the nuclei of living mammalian cells. J. Cell Sci. 2001;114:4219–4228. doi: 10.1242/jcs.114.23.4219. [DOI] [PubMed] [Google Scholar]
- Ubukata M., Cheng X., Isono K., Cheng X., Isono K., Isono K. The structure of tautomycin, a regulator of eukaryotic cell growth. J. Chem. Soc. Chem. Commun. 1990;3:244–246. [Google Scholar]
- Vanselow K., Vanselow J.T., Westermark P.O., Reischl S., Maier B., Korte T., Herrmann A., Herzel H., Schlosser A., Kramer A., Vanselow J.T., Westermark P.O., Reischl S., Maier B., Korte T., Herrmann A., Herzel H., Schlosser A., Kramer A., Westermark P.O., Reischl S., Maier B., Korte T., Herrmann A., Herzel H., Schlosser A., Kramer A., Reischl S., Maier B., Korte T., Herrmann A., Herzel H., Schlosser A., Kramer A., Maier B., Korte T., Herrmann A., Herzel H., Schlosser A., Kramer A., Korte T., Herrmann A., Herzel H., Schlosser A., Kramer A., Herrmann A., Herzel H., Schlosser A., Kramer A., Herzel H., Schlosser A., Kramer A., Schlosser A., Kramer A., Kramer A. Differential effects of PER2 phosphorylation: Molecular basis for the human familial advanced sleep phase syndrome (FASPS) Genes & Dev. 2006;20:2660–2672. doi: 10.1101/gad.397006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulsteke V., Beullens M., Waelkens E., Stalmans W., Bollen M., Beullens M., Waelkens E., Stalmans W., Bollen M., Waelkens E., Stalmans W., Bollen M., Stalmans W., Bollen M., Bollen M. Properties and phosphorylation sites of baculovirus-expressed nuclear inhibitor of protein phosphatase-1 (NIPP-1) J. Biol. Chem. 1997;272:32972–32978. doi: 10.1074/jbc.272.52.32972. [DOI] [PubMed] [Google Scholar]
- Wakula P., Beullens M., Ceulemans H., Stalmans W., Bollen M., Beullens M., Ceulemans H., Stalmans W., Bollen M., Ceulemans H., Stalmans W., Bollen M., Stalmans W., Bollen M., Bollen M. Degeneracy and function of the ubiquitous RVXF motif that mediates binding to protein phosphatase-1. J. Biol. Chem. 2003;278:18817–18823. doi: 10.1074/jbc.M300175200. [DOI] [PubMed] [Google Scholar]
- Xu Y., Toh K.L., Jones C.R., Shin J.Y., Fu Y.H., Ptacek L.J., Toh K.L., Jones C.R., Shin J.Y., Fu Y.H., Ptacek L.J., Jones C.R., Shin J.Y., Fu Y.H., Ptacek L.J., Shin J.Y., Fu Y.H., Ptacek L.J., Fu Y.H., Ptacek L.J., Ptacek L.J. Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell. 2007;128:59–70. doi: 10.1016/j.cell.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Sehgal A., Sehgal A. Role of molecular oscillations in generating behavioral rhythms in Drosophila. Neuron. 2001;29:453–467. doi: 10.1016/s0896-6273(01)00218-5. [DOI] [PubMed] [Google Scholar]
- Yang Y., He Q., Cheng P., Wrage P., Yarden O., Liu Y., He Q., Cheng P., Wrage P., Yarden O., Liu Y., Cheng P., Wrage P., Yarden O., Liu Y., Wrage P., Yarden O., Liu Y., Yarden O., Liu Y., Liu Y. Distinct roles for PP1 and PP2A in the Neurospora circadian clock. Genes & Dev. 2004;18:255–260. doi: 10.1101/gad.1152604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.W. The molecular control of circadian behavioral rhythms and their entrainment in Drosophila. Annu. Rev. Biochem. 1998;67:135–152. doi: 10.1146/annurev.biochem.67.1.135. [DOI] [PubMed] [Google Scholar]
- Yuan Q., Lin F., Zheng X., Sehgal A., Lin F., Zheng X., Sehgal A., Zheng X., Sehgal A., Sehgal A. Serotonin modulates circadian entrainment in Drosophila. Neuron. 2005;47:115–127. doi: 10.1016/j.neuron.2005.05.027. [DOI] [PubMed] [Google Scholar]
- Zeng H., Qian Z., Myers M.P., Rosbash M., Qian Z., Myers M.P., Rosbash M., Myers M.P., Rosbash M., Rosbash M. A light entrainment mechanism for the Drosophila circadian clock. Nature. 1996;380:129–135. doi: 10.1038/380129a0. [DOI] [PubMed] [Google Scholar]