Abstract
Nucleosomes containing the histone variant H3.3 tend to be clustered in vivo in the neighborhood of transcriptionally active genes and over regulatory elements. It has not been clear, however, whether H3.3-containing nucleosomes possess unique properties that would affect transcription. We report here that H3.3 nucleosomes isolated from vertebrates, regardless of whether they are partnered with H2A or H2A.Z, are unusually sensitive to salt-dependent disruption, losing H2A/H2B or H2A.Z/H2B dimers. Immunoprecipitation studies of nucleosome core particles (NCPs) show that NCPs that contain both H3.3 and H2A.Z are even less stable than NCPs containing H3.3 and H2A. Intriguingly, NCPs containing H3 and H2A.Z are at least as stable as H3/H2A NCPs. These results establish an hierarchy of stabilities for native nucleosomes carrying different complements of variants, and suggest how H2A.Z could play different roles depending on its partners within the NCP. They also are consistent with the idea that H3.3 plays an active role in maintaining accessible chromatin structures in enhancer regions and transcribed regions. Consistent with this idea, promoters and enhancers at transcriptionally active genes and coding regions at highly expressed genes have nucleosomes that simultaneously carry both H3.3 and H2A.Z, and should therefore be extremely sensitive to disruption.
Keywords: Histone H2A.Z, histone H3.3, nucleosome structure
The ability of nucleosomes to be disrupted or displaced is critical to their biological function. Although this process involves specific enzyme systems that are able to remodel, move, and displace nucleosomes, the intrinsic energy of histone–histone or histone–DNA interactions must also play an important role in determining nucleosome stability. Earlier studies have determined the average stability of nucleosome core particles (NCPs) derived from both yeast and vertebrate sources, by measuring either the resistance to thermal denaturation or the response to salt-induced dissociation. NCPs that are exposed to increasing concentrations of salt will typically lose first the two H2A/H2B dimers, and at higher salt concentrations the H3/H4 tetramer that forms the central organizing complex of the NCP (Burton et al. 1978). These results reflect the average behavior of a genomic sample, and cannot take into account the effects of histone modifications or the contributions of NCPs carrying histone variants. This is of particular interest in the case of variants that can alter the packing of histones within the NCP. The localized distribution of such variants in the genome has been implicated in biological function, specifically in the regulation of gene expression.
Among these variants, considerable attention has been devoted recently to histone H3.3, a variant of histone H3, which differs by four amino acids from H3. In a wide variety of organisms, H3.3 is incorporated into chromatin during interphase, with the help of an interphase-specific chaperone complex (Ahmad and Henikoff 2002; Tagami et al. 2004). In contrast, the major variant H3 is incorporated only during S phase (Ahmad and Henikoff 2002). Several studies have analyzed the genomic distribution of H3.3 (Chow et al. 2005; Mito et al. 2005; Schwartz and Ahmad 2005; Wirbelauer et al. 2005; Jin and Felsenfeld 2006). In Drosophila, genome-wide microarray analysis has shown that on average, H3.3 is distributed preferentially both upstream of and downstream from transcriptionally active genes, with the greatest enrichment immediately downstream from promoters, and with higher levels of incorporation associated with higher levels of bound RNA polymerase II (Mito et al. 2005). In chicken erythroid cells, we have found that incorporation at certain genes, such as the erythroid-specific folate receptor (FR) and vascular endothelial growth factor D (VEGFD) genes, is confined to the upstream regulatory region, while in other genes, H3.3 incorporation spreads over both promoter and coding regions (Jin and Felsenfeld 2006). Locus control regions and other regulatory sites are enriched for H3.3 even when the associated genes are not active (Jin and Felsenfeld 2006), perhaps because they are constitutively transcribed at a low level (Ashe et al. 1997). The distribution of H3.3 suggests that it might play some role in the epigenetic marking of transcriptionally active genes, perhaps by local alterations in chromatin structure.
A second histone variant, H2A.Z, has also been the subject of many recent studies. Incorporation into chromatin of Saccharomyces cerevisiae H2A.Z (Htz1) is mediated by the SWR1 chromatin remodeling complex (Krogan et al. 2003, 2004; Kobor et al. 2004; Mizuguchi et al. 2004), and is replication-independent. Like H3.3, H2A.Z is not distributed uniformly in the genome. The actual distribution and the proposed correlations with function that have been made differ among organisms. In S. cerevisiae, genetic experiments have implicated Htz1 in the prevention of gene silencing caused by spreading of heterochromatin from neighboring telomeres or the HMR mating type locus (Meneghini et al. 2003). At the same time, genome-wide surveys of the distribution of Htz1 show that it is enriched in nucleosomes at promoters; high-resolution analysis reveals that a pair of Htz1 nucleosomes may surround a nucleosome-free region (Guillemette et al. 2005; Li et al. 2005; Raisner et al. 2005; Zhang et al. 2005). It has been suggested that these are promoters of basal or repressed genes and that upon induction there is a preferential loss of the Htz1-containing nucleosomes. In this view, such nucleosomes “poise genes for transcriptional activation.” Other studies, however, see no correlation between Htz1 occupancy and transcription rates (Raisner et al. 2005).
Related, but not identical patterns of H2A.Z have been found in the few metazoan studies available. H2Av, the H2A.Z of Drosophila, is involved in Polycomb-mediated silencing and establishment of centromeric heterochromatin (Swaminathan et al. 2005). In chicken erythroid cells, H2A.Z appears to be concentrated at promoters of developmentally regulated and actively expressed genes (Bruce et al. 2005), while at the human c-myc locus, H2A.Z is always enriched at the promoter, whether or not c-myc expression is induced, but is lost from the coding region after induction (Farris et al. 2005).
The suggestion that nucleosomes carrying H2A.Z in vivo may be more susceptible to disruption has led to questions about the physical stability of NCPs containing histone variants. The stability of NCPs as a function of ionic strength of the solvent has been the subject of numerous investigations over many years. Recently, this has been extended to studies of the properties of NCPs in which H2A.Z replaces H2A, but these studies have not always led to identical conclusions. In some cases, H2A.Z is shown to stabilize nucleosome structure (Park et al. 2004; Thambirajah et al. 2006), while in others it appears that H2A.Z is released from chromatin more readily than is H2A (Suto et al. 2000; Abbott et al. 2001; Zhang et al. 2005).
Because of our interest in the possible roles of these histone variants in chromatin structure, we asked whether NCPs containing H3.3 had physical properties that distinguished them from those containing H3. We find that all NCPs that contain H3.3 are much less stable than H3 NCPs, as measured by susceptibility to salt-dependent dissociation of H2A/H2B or H2A.Z/H2B dimers, suggesting that H3.3 NCPs have the potential to play a regulatory role at promoters and enhancers where disruption of nucleosomes is likely to be important. We extended this study by comparing the stability of NCPs containing both H3.3 and H2A with those containing H3.3 and H2A.Z. We show that H3.3/H2A.Z NCPs are present in vivo, but they are even less stable than NCPs carrying H3.3 and H2A. These results reveal a hierarchy of stabilities that could account for the discrepancies among earlier studies of H2A.Z-containing NCPs. Finally, we show by double chromatin immunoprecipitation (ChIP) that these very unstable NCPs are concentrated in vivo over promoters and enhancers of transcriptionally active genes, as well as over the transcribed regions of some genes that are very active. Our results suggest how H2A.Z could play different regulatory roles in the genome, depending on the identity of its histone partners within the nucleosome. They also suggest a regulatory role for histone H3.3 that is distinct from that of the more abundant H3, and for a special role when it is coupled with H2A.Z.
Results
Composition of native NCP fractions containing H3.3 and H3
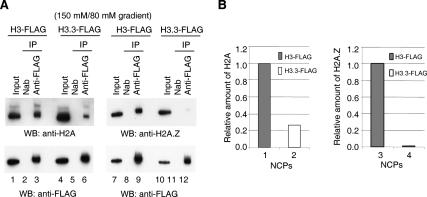
We wished to measure the relative stability of NCPs containing H3.3, as compared with those containing H3. In principle, H3.3 can be paired in NCPs with either H2A or H2A.Z, and therefore it seemed important to determine separately the properties of NCPs containing H3.3/H2A and H3.3/H2A.Z. We used 6C2 cells, an avian erythroleukemia line that had been stably transformed to express either H3.3-Flag or H3-Flag. There has been extensive experience with the use of these tagged histones (Tagami et al. 2004; Jin and Felsenfeld 2006), necessitated by the inaccessibility within the NCP to antibodies against the distinguishing amino acids of H3.3. We prepared NCP monomers from H3-Flag and H3.3-Flag cells by a standard procedure involving micrococcal nuclease (MNase) digestion of nuclei, and suspension in a solvent containing 150 mM NaCl followed by sedimentation on a sucrose gradient containing buffers and 80 mM NaCl (see Materials and Methods). The gradient fractions around the monomer peak were isolated, immunoprecipitated with anti-Flag antibody, and the immunoprecipitates were subjected to Western blotting with either anti-H2A or anti-H2A.Z antibody. As expected, both H2A and H2A.Z could be detected in NCPs containing H3-Flag (Fig. 1A, lanes 1–3,7–9). However, to our surprise, considerably less H2A was found associated with H3.3-Flag (Fig. 1A, lanes 4–6): The H2A:H3.3 ratio was about one-third the H2A:H3 ratio in these immunoprecipitates (Fig. 1B, left panel). Furthermore, almost no H2A.Z was detectable in association with H3.3 (Fig. 1A [lanes 10–12], B [right panel]). This could, in principle, be explained by the low abundance or complete absence in the genome of any NCPs containing both H3.3 and H2A.Z, but as we show below, that is not the case.
Figure 1.
Relative stability of NCPs containing H3 and H3.3. (A) NCPs were prepared from cells expressing either H3-Flag or H3.3-Flag. NCP monomers suspended in a solvent containing 150 mM NaCl were obtained by fractionation on a sucrose gradient (see Materials and Methods) in a solvent containing 80 mM NaCl and buffers. In each case, the sample was immunoprecipitated with antibody to Flag, histones were isolated and fractionated by gel electrophoresis, and Western blots of the samples were probed with antibody to H2A (left) or H2A.Z (right). Input lanes are loaded with an aliquot representing 10% of the starting sample. The label “150 mM/80 mM gradient” indicates the highest NaCl concentration used in NCP preparation and the NaCl concentration in the gradient. (Nab) No antibody control. (B) Comparative recoveries of H2A and H2A.Z from the data in A. The amounts of H2A or H2A.Z in H3-Flag-containing NCPs were set to 1. The relative amount was calculated by comparing the intensity of immunoprecipitated H2A or H2A.Z with that of immunoprecipitated H3-Flag or H3.3-Flag.
H3.3/H2A.Z-containing NCPs are present in nuclei
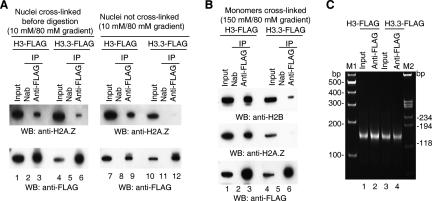
We next addressed the question of whether H3.3/H2A.Z-containing nucleosomes exist in vivo. Nuclei were prepared in 10 mM NaCl to prevent any salt-dependent dissociation of histones (see below) and fixed with formaldehyde before digestion with micrococcal nuclease. The monomer fractions were isolated on a sucrose gradient containing 80 mM NaCl. A second sample of nuclei was treated similarly except that cross-linking was omitted. As shown in Figure 2A, the uncross-linked control displayed the characteristic absence of H2A.Z from the H3.3-Flag IP fraction (lane 12). The cross-linked H3.3 sample, however, contained some H2A.Z (Fig. 2A, lane 6), but relatively less than that complexed to H3-Flag (Fig. 2A, lane 3). The results indicate that complete NCPs containing H3.3 and H2A.Z are present in vivo (although at low levels compared with H3.3/H2A), but that they are relatively unstable, and subject to further disruption during preparation if they are not cross-linked.
Figure 2.
NCPs containing H3.3 are disrupted during purification. (A) Complete NCPs containing H3.3 and H2A.Z are present in vivo. (Left panel) Nuclei isolated from 6C2 cells in 10 mM NaCl were fixed with formaldehyde (see Materials and Methods) and then digested with micrococcal nuclease. NCPs carrying H3.3Flag or H3-Flag were purified on sucrose gradients containing 80 mM NaCl and analyzed for H2A.Z content on Western blots. (Right panel) Control in which formaldehyde treatment was omitted. (B) Experiments like those in Figure 1A, except NCPs were cross-linked with formaldehyde before immunoprecipitation, and Western blots were probed for H2A.Z and H2B. Input lanes are loaded with an aliquot representing 10% of the starting sample. (C) Length distribution of DNA isolated from NCPs. NCPs from cells carrying H3-Flag or H3.3-Flag were immunoprecipitated with anti-Flag, and the nucleosomal DNA was purified and analyzed by gel electrophoresis.
H3.3-Flag-containing nucleosomes are disrupted during purification
This raised the question whether the immunoprecipitation process was somehow preferentially disrupting all NCPs containing H3.3. To address this, NCP monomer fractions prepared as described above were cross-linked with formaldehyde before antibodies were added. The results (Fig. 2B) were similar to those shown in Figure 1A. In addition, histone H2B is also depleted in the H3.3-containing NCPs. This made it clear that disruption of H3.3 NCPs was occurring before the immunoprecipitation, as cross-linking would protect NCPs from disruption during that process.
It seemed possible that disruption was connected with partial degradation of the H3.3 nucleosomes that had resulted from some greater sensitivity to micrococcal nuclease digestion. To address this, nucleosome monomers containing either H3-Flag or H3.3-Flag were immunoprecipitated and deproteinized, and the DNA was analyzed by gel electrophoresis. As shown in Figure 2C, the DNA in both cases consisted almost entirely of ∼145-base-pair (bp) fragments consistent with the presence of undegraded DNA, suggesting that the disruption of H3.3-Flag-containing nucleosomes was not due to their greater sensitivity to micrococcal nuclease digestion.
H3.3-Flag-containing nucleosomes are unusually sensitive to the ionic strength of the solvent
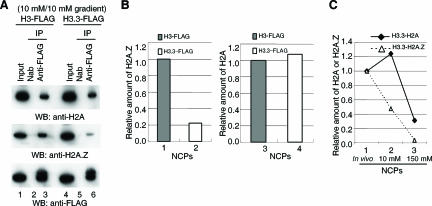
The most obvious candidate as a cause of NCP disruption is unusual sensitivity to the ionic strength of the solvent. NCPs containing the principal histone variants H3 and H2A typically lose first H2A/H2B dimers, then H3/H4 tetramers, as the salt concentration increases. This suggested that the disruption of H3.3-Flag-containing nucleosomes was due to the relatively high concentration of NaCl (150 mM) that we used in the preceding experiments to remove histone H1/H5 (Figs. 1A, 2B). We therefore repeated the digestion and sucrose gradient fractionation steps in solvents containing only 10 mM NaCl (Fig. 3A). At this ionic strength, immunoprecipitation of NCPs revealed the presence of H3.3 NCPs that contained an amount of H2A comparable to that of H3 NCPs (Fig. 3B, right panel). The H2A.Z:H3.3 ratio is now about one-fifth the H2A.Z:H3 ratio (Fig. 3B, left panel), confirming the presence of some NCPs containing both H3.3 and H2A.Z and showing that those NCPs were disrupted in vitro even in solvents containing low NaCl, such as 80 mM (see Fig. 2A, right panel). These results support the conclusion that H3.3 NCPs, regardless of their histone partners, are more unstable than H3 NCPs. Furthermore, as shown in Figure 3C, H3.3/H2A.Z NCPs are even less stable than H3.3/H2A NCPs: As the salt concentration to which H3.3 NCPs are exposed is increased, the fractional loss of H2A.Z is earlier and greater than the loss of H2A.
Figure 3.
Unusual sensitivity of nucleosomes containing H3.3 to the ionic strength of the solvent. (A) Experiments like those in Figure 1, but NCPs were isolated from a sucrose gradient containing 10 mM NaCl without exposure to any higher-ionic-strength solvents. (B) Comparative recoveries of H2A and H2A.Z from the data in A. The relative amounts of histone H2A and H2A.Z in NCPs were calculated as described in Figure 1B. (C) Dynamic changes of relative amount of H2A or H2A.Z in H3.3-containing NCPs during purification. The relative amount of H2A.Z and H2A in H3.3 NCPs in nuclei (shown as in vivo) (Fig. 2A; data not shown) were measured as described and set to 1. These amounts were then compared with those presented in B (10 mM) and in Figure 1B (150 mM).
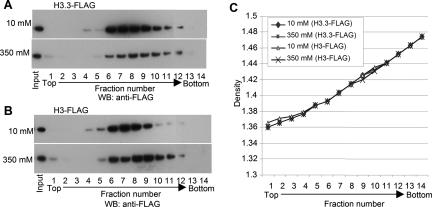
To explore further the question of stability of H3.3-containing nucleosomes, NCP monomer preparations containing either Flag-H3 or Flag-H3.3 were dialyzed into either 10 mM or 350 mM NaCl, cross-linked with formaldehyde, and subjected to equilibrium centrifugation in CsCl. The band at the density corresponding to intact NCPs was isolated, the cross-linking was reversed, and the histones were electrophoresed and analyzed by Western blotting with anti-Flag antibody. The fractions from 4 to 12 (Fig. 4A,B) were considered to contain intact NCPs since in addition to H3, histones H2A and H2B were detectable in those fractions by Western blotting (data not shown). Additionally, anti-Flag bands are distributed with the highest intensity at fractions 7 and 8, densities of ∼1.40 and 1.42, respectively (Fig. 4C), a characteristic of intact NCPs (see Materials and Methods). As shown in Figure 4B, the amounts of H3 found in octamers were similar at the two ionic strengths. In contrast, the amount of H3.3 contained in octamers was considerably reduced at the higher, as compared with the lower, ionic strength (Fig. 4A). These data demonstrate again that H3.3-containing NCPs, regardless of their partners, are more unstable than those containing H3.
Figure 4.
Sensitivity of purified NCPs to the higher ionic strength of the solvent. NCPs carrying H3.3-Flag (A) or H3-Flag (B) were dialyzed into solvents containing 10 mM or 350 mM NaCl, formaldehyde-fixed, and centrifuged to equilibrium in CsCl density gradients. Fractions were collected, histones were fractionated as in Figure 1, and Western blots were probed with anti-Flag antibody. Density increases with increasing fraction number. (C) Density distribution within the gradient.
Loss of histone acetylation does not affect stability of NCPs containing H3.3-Flag
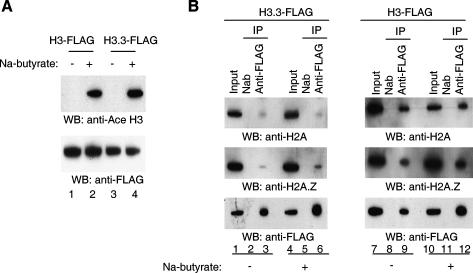
It seemed possible that the relative instability of H3.3 NCPs was connected to high levels of H3.3 histone modifications. Earlier studies have shown, for example, that H3.3 is highly acetylated (McKittrick et al. 2004; Hake et al. 2006). To test this, NCP preparations were repeated, omitting sodium butyrate, which is normally included to inhibit the action of histone deacetylases. In preparations that include butyrate, histone H3 is acetylated (Fig. 5A, lanes 2,4). In the absence of butyrate, histone H3 is unacetylated (Fig. 5A, lanes 1,3). As shown in Figure 5B, loss of acetylation has no effect on the observed relative instability of H3.3 NCPs, as judged by the depletion of both H2A and H2A.Z in the H3.3 immunoprecipitates in the absence of butyrate (Fig. 5B, cf. lanes 3 and 6).
Figure 5.
Loss of histone acetylation does not measurably affect relative stabilities of H3 and H3.3 NCPs. (A) Loss of histone acetylation in the absence of sodium butyrate. NCPs containing either H3-Flag or H3.3-Flag isolated from cells in the absence or presence of sodium butyrate were immunoprecipitated with anti-Flag antibody, and histones were isolated and subjected to Western blot analysis with antibody to acetylated H3. (B) Loss of acetylation does not affect the relative instability of H3.3 NCPs. Immunoprecipitation studies similar to those in Figure 1, in the presence or absence of sodium butyrate.
NCPs containing untagged H3.3 are also unstable
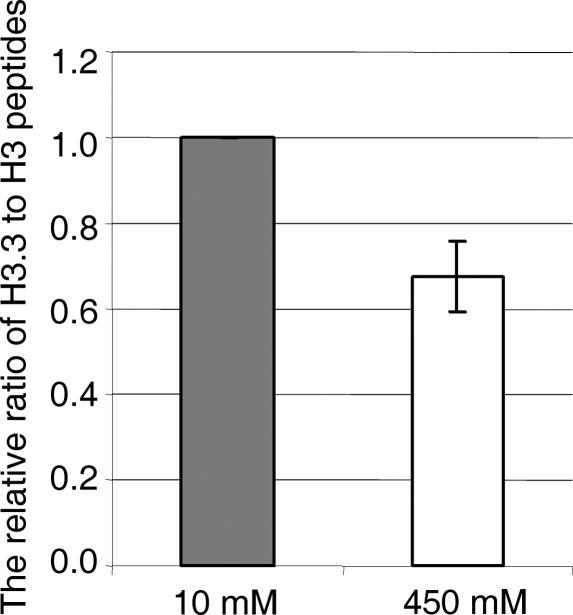
All of the experiments described were carried out with Flag-tagged histones, which have been shown in numerous experimental systems to behave similarly to the corresponding untagged molecules. Furthermore, the Flag tag was present on both H3 and H3.3, indicating that the tag alone was not responsible for the destabilization seen in H3.3 NCPs. Nonetheless, it seemed important to determine whether the Flag element could have contributed to the observed stability differences. For this purpose, the experiment described in Figure 4 was repeated with NCPs from chicken erythrocytes, carrying only wild-type histones. The monomers were prepared in 10 mM NaCl and then dialyzed into 10 mM and 450 mM NaCl, respectively. Both the low- and high-salt samples were formaldehyde cross-linked and banded in CsCl gradients. The octamer fractions were isolated, the cross-linking reversed, and the histones fractionated by electrophoresis. The bands containing H3 variants were excised and subjected to mass spectrometry to determine the ratio of H3.3 to H3 at each ionic strength. As shown in Figure 6, NCPs that had been exposed to the higher salt concentration were relatively depleted in H3.3 compared with NCPs maintained in low salt. Thus, the experiments with Flag-tagged histones accurately reflect the stability of normal genomic NCPs containing untagged H3 or H3.3.
Figure 6.
Sensitivity of the NCPs consisting of untagged histones to the ionic strength of the solvent. NCPs from chicken erythrocytes were dialyzed into 10 mM NaCl and 450 mM NaCl, respectively. The octamer fractions were cross-linked with formaldehyde and isolated by CsCl gradient sedimentation, and the ratios of H3.3 to H3 at each ionic strength were determined by mass spectrometry (see Materials and Methods). The results are the average of two independent experiments. The ratio at 10 mM NaCl was set to 1.
It should be noted that the predominant variant, referred to here as H3, in mammals consists of two subtypes: H3.1 and H3.2, which differ only at amino acid 96 (cysteine vs. serine) (Franklin and Zweidler 1977). In chickens, only H3.2 is present, but the H3-Flag used in this study was derived from human H3.1. Many studies of bulk genomic NCP stability show that the stability of H3.1 and H3.2 NCPs is similar. The results summarized in Figure 6, obtained with histones derived from chicken cells, confirm this.
Location in vivo of nucleosomes containing both H3.3 and H2A.Z
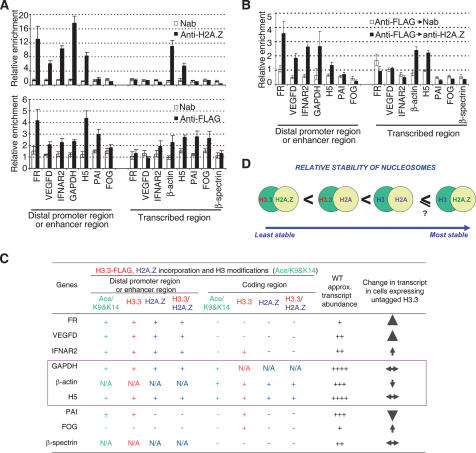
We have shown that H2A.Z and H3.3 can be present within the same nucleosome in vivo. Where are such nucleosomes located? We prepared nucleosome monomers by micrococcal nuclease digestion of 6C2 nuclei exposed only to 10 mM NaCl, followed by sucrose gradient sedimentation at the same salt concentration. Samples were immunoprecipitated with antibody either to Flag or H2A.Z, and the DNA was subjected to quantitative PCR using primers designed for single nucleosome resolution. In an earlier paper (Jin and Felsenfeld 2006), we mapped at lower resolution the distribution of H3.3-containing nucleosomes in 6C2 cells for a variety of genes that are expressed in those cells. As shown in Figure 7A, bottom panel, a similar result was obtained for the distribution of H3.3 monomers. When these data are compared with the distribution of H2A.Z monomers (Fig. 7A, top panel), it is clear that five of six distal promoter and enhancer regions and two of five coding regions that contain H3.3 monomers carry H2A.Z at the same site, at single nucleosome resolution. This strongly suggests that individual nucleosomes at these sites contain both variants. To confirm this, we carried out double ChIP experiments, first with antibody against Flag, followed by a second immunoprecipitation of that fraction with antibody against H2A.Z. As shown in Figure 7B, the distribution of these H3.3/H2A.Z nucleosomes coincides in every case with the results shown in Figure 7A: At every nucleosome site where both H3.3 and H2A.Z are present, there are nucleosomes that have both. The doubly substituted H3.3/H2A.Z nucleosome is found in the distal promoter and enhancer sites of five of seven genes tested: FR, VEGFD, GAPDH, IFNAR2 (IFN-α/β receptor 2), and histone H5, but is found in the transcribed regions of only two of eight genes: β-actin and histone H5.
Figure 7.
(A) ChIP analysis of H3.3-Flag and H2A.Z over distal promoter or enhancer regions and transcribed regions of a variety of genes in 6C2 cells expressing H3.3-Flag. (Open bars) No antibody control; (filled bars) anti-Flag or anti-H2A.Z immunoprecipitation. Error bars reflect three separate measurements. (PAI) Plasminogen activator inhibitor; (FOG) friend of GATA. (B) Double ChIP analysis over same regions. First ChIPs by anti-Flag were followed by second ChIPs by anti-H2A.Z antibodies. (C) Summary of ChIP and double ChIP results; level of Ac/H3K9&K14; relative expression level of those genes surveyed in wild-type 6C2 cells and in the cells overexpressing untagged H3.3. The ChIP data are from A and B, while all others are from our previous study (Jin and Felsenfeld 2006). (N/A) Not applicable. (D) Schematic representation of relative stability of nucleosomes containing different histone variants.
Discussion
Effects of variants
Most measurements of nucleosome stability have been made in vertebrates, where the predominant histone variants are H3 (H3.1/3.2) and H2A. Such measurements on genomic nucleosome populations therefore reflect the properties of those variants. On that basis, it is expected that typical NCPs will be stable at NaCl concentrations up to ∼0.5 M (Park et al. 2004), and that increasing concentrations beyond that will result successively in release of H2A/H2B dimers and H3/H4 tetramers. The data in Figure 1 confirm that monomer fractions containing H3-Flag were still associated with both H2A and H2A.Z after exposure to 150 mM NaCl. When the experiment was repeated with H3.3-Flag, however, the amount of H2A associated with H3.3-Flag under these conditions was considerably less than that found in H3-Flag NCPs (Fig. 1). Histone H2B was similarly depleted (Fig. 2B). Furthermore, almost no H2A.Z was detected in NCPs containing H3.3-Flag. This was not the result of preferential displacement of histones by the anti-Flag antibody, since cross-linking before immunoprecipitation did not alter the result (Fig. 2B). This behavior appears to arise from the preferential sensitivity of H3.3 NCPs, whether containing H2A or H2A.Z, resulting in loss of H2A/2B or H2A.Z/H2B at moderate salt concentrations. Isolation of NCPs in 10 mM rather than 80 mM NaCl resulted in no depletion of H2A from H3.3 NCPs as compared with H3 NCPs, and some H2A.Z was now also detectible in the H3.3 fraction (Fig. 3A). Consistent with this, H3.3/H2A.Z NCPs do exist in vivo (Fig. 2A). The quantitation of the response to salt dissociation (Fig. 3C) clearly indicates that H3.3-containing NCPs are unstable and H3.3-Flag forms even more unstable NCPs with H2A.Z than it does with H2A.
The overall instability of H3.3 NCPs was further confirmed by experiments in which NCPs dialyzed into either 10 mM or 350 mM NaCl were cross-linked and centrifuged to equilibrium in CsCl gradients to isolate intact NCPs. The abundance of H3 NCPs was the same at the two salt concentrations, but there was a clear depletion of H3.3-containing monomers at the higher salt concentration (Fig. 4). This result shows that the instability detected in Figures 1 and 2 is not a property of a small fraction of NCPs that are unusually reactive with anti-Flag antibody. The fact that stable octamers can be isolated at low ionic strength also shows that the stability difference is not simply a reflection of differences in susceptibility to attack by micrococcal nuclease.
The same differential stability observed with purified, then cross-linked NCP monomers could also be seen when nuclei were first formaldehyde cross-linked in 10 mM NaCl and then digested with micrococcal nuclease. However, by this procedure it was possible to detect some H3.3/H2A.Z NCPs, whereas none were observed if formaldehyde cross-linking was omitted (Fig. 2A). Differential stability is therefore not solely a property displayed at the level of mononucleosomes, or a result of the digestion process. Furthermore, although there is evidence that H3.3 in vivo carries high levels of histone acetylation, the observed instability is not dependent on the extent of acetylation (Fig. 5B). Since active marks can also be enriched on H3.1 (Hake et al. 2006; Loyola et al. 2006), it seems unlikely that the stability difference is due solely to the status of post-translational modifications, although these experiments do not rule out the possibility that methylation or other modifications could contribute to stability. In any case, the relative instability of H3.3 NCPs is a property of these particles in vivo.
It did not seem likely that the Flag peptide, present both on H3 and H3.3, could give rise to the observed differential stability of NCPs. We confirmed, by carrying out salt dissociation experiments on NCPs isolated from wild-type chicken erythrocytes (Fig. 6), that the reduced stability of H3.3 NCPs is a property of normal, untagged histones in vivo.
Measurements of nucleosome stability
Our results show that native NCPs containing histone H3.3 are relatively less stable than those containing H3: Histone H2A/H2B or H2A.Z/H2B dimers are more readily dissociated from octamers containing H3.3, and NCPs containing both H3.3 and H2A.Z are even more unstable (Fig. 7D). It should be pointed out that our results concern the dissociation of H2A/H2B and H2A.Z/H2B from the NCP, and do not provide information about the stability of the H3/H4 or H3.3/H4 tetramer. It might be expected that the strength of tetramer binding is affected by local DNA sequences, making a separate contribution to nucleosome stability. However, our earlier results (Jin and Felsenfeld 2006) show that NCPs exposed to 150 mM NaCl retain H3.3 at the upstream regulatory elements shown in Figure 7, so the loss under these conditions of H2A or H2A.Z from H3.3 NCPs is not due to total loss of the octamer.
Recent studies have investigated the structure and stability of NCPs containing H2A.Z. Suto et al. (2000) have solved the structure of an NCP containing cloned histones H2A.Z from mouse, and H2B, H3, and H4 from Xenopus, and shown that there are differences from the structure of NCPs containing H2A. These include H2A.Z–H2A.Z and H2A.Z–H3 contacts in the former that differ from H2A–H2A and H2A–H3 contacts in the latter. Park et al. (2004) have shown that monomers containing histone H3 and H2A.Z are more resistant to salt dissociation than are H3/H2A nucleosomes. They conclude that H2A.Z stabilizes nucleosomes. A similar conclusion has been reached from measurements of NCP stability as the pH is lowered (Thambirajah et al. 2006). These observations are not inconsistent with the results reported here, which show that NCPs containing H2A.Z and H3 are at least as stable as H2A/H3 NCPs.
A more revealing comparison might be made with the results of Zhang et al. (2005), who carried out studies of histone release from S. cerevisiae nuclei washed with solvents of increasingly higher ionic strength, and found that H2A.Z was released at lower salt concentrations than H2A, an apparent contradiction of the results of Park et al. (2004). However, the sole H3 variant in yeast is similar to vertebrate H3.3. Although yeast and vertebrate H2A.Z differ somewhat in amino acid sequence, we suggest that the observed instability of yeast nucleosomes containing H2A.Z is comparable to the behavior of vertebrate NCPs containing H3.3 and H2A.Z.
The majority of yeast NCPs contain H2A, and a similar comparison can be made between the stability of these particles and vertebrate H3.3/H2A NCPs. It has been known for some time that yeast NCPs are less stable than those prepared from vertebrate sources (Lee et al. 1982; Pineiro et al. 1991; White et al. 2001), and the structure of the yeast NCP shows that they are likely to be “subtly destabilized as compared with nucleosomes from higher eukaryotes” (White et al. 2001), consistent with the relatively “open” state of chromatin in yeast. Again, these observations could reflect the fact that all yeast nucleosomes contain a variant of H3.3, and yeast H3.3/H2A nucleosomes are likely to be less stable than their H3/H2A counterparts in other organisms.
Genomic location of H3.3/H2A.Z NCPs
As noted above, H3.3 has been shown to be incorporated preferentially at promoters and enhancers, as well as at sites on some transcribed genes. H2A.Z is also preferentially incorporated at promoters in yeast and at promoters of some developmentally regulated and actively expressed genes (Bruce et al. 2005) in chicken. It seemed important, therefore, to determine the frequency with which H3.3 and H2A.Z were found at the same site. To measure this, we first carried out separate ChIP experiments for H3.3 and H2A.Z at single nucleosome resolution followed by a double ChIP study that purified only those NCP monomers containing both variants. Five of the six genes with distal promoter or enhancer sites that we had identified earlier as containing H3.3 NCPs also contained H2A.Z NCPs (Fig. 7A; summarized in Fig. 7C). At all five of these sites, double ChIP revealed the presence of NCPs incorporating H3.3 and H2A.Z in the same particle. It may be relevant that expression of three of these genes (FR, VEGFD, IFNAR2) is stimulated by ectopic expression of H3.3 (Jin and Felsenfeld 2006). Two other genes, β-actin and histone H5, carry H3.3/H2A.Z NCPs over transcribed regions. We detect high levels of transcript from these two genes (Fig. 7C). Furthermore, all loci that have H3.3/H2A.Z nucleosomes, whether in the transcribed regions or upstream regulatory elements, are marked by high levels of histone H3 Lys 9 and Lys 14 acetylation (Fig. 7C; Jin and Felsenfeld 2006). The presence of an H3.3/H2A.Z nucleosome appears to be correlated with the presence of these marks of highly active chromatin. Thus promoters, enhancers, and coding regions of these transcriptionally active genes are occupied by nucleosomes that are quite unstable, and presumably easily displaced by the machinery of transcriptional activation and transcription.
Roles of histone variants in vivo
The distribution of histone H3.3 in the genome has been investigated and discussed at length. H3.3 is preferentially incorporated near and within transcriptionally active genes, as well as at certain upstream regulatory regions. Unlike H3, incorporation of H3.3 is not confined to the S phase of the cell cycle. It can, therefore, replace histones that are displaced during the transcription process. It has been suggested that H3.3 might serve as an epigenetic mark that would help to maintain transcriptionally active genes in an “open” conformation, and that, in turn, transcription would lead to greater deposition of H3.3 (Henikoff et al. 2004). We have reported that overexpression of H3.3 can raise the expression of a subset of genes that show high levels of incorporated H3.3, but a possible mechanism of action has remained unclear (Jin and Felsenfeld 2006). On the basis of the present results, we conclude that H3.3-containing nucleosomes are intrinsically less stable than those containing H3, and that this may reduce the energy required to move or displace nucleosomes from promoters, enhancers, and gene-coding regions. This is supported by the observation that nucleosome assembly protein I (NAP-I) mediated assembly and disassembly of the H2A.Bbd–H2B dimers from reconstituted NCPs was accomplished more efficiently when the NCPs contained H3.3 (Okuwaki et al. 2005). There has been considerable debate as to whether H3.3 is simply a marker of transcriptionally active regions, deposited on active genes when H3 NCPs are displaced during transcription, or is an active participant in the regulatory process. Our data support the latter view.
Our results also show that nucleosomes containing both H3.3 and H2A.Z are even more unstable than those containing H3.3 and H2A. As noted in the introduction, a wide variety of functions has been attributed to H2A.Z, including involvement in centromeric heterochromatin and Polycomb-mediated silencing, on the one hand, and on the other, the marking of genes so that they can be more easily activated. We suggest that these disparate properties might reflect the identity of the partners with which H2A.Z finds itself engaged within the nucleosome. The combination of H2A.Z with H3.3, localized to active or potentially active genes, might provide promoters with an unusually labile chromatin conformation. Similarly, the presence of a labile H2A.Z/H3.3 nucleosome at the end of a yeast telomere or mating type locus might prevent the extension of a condensed chromatin structure (Meneghini et al. 2003). On the other hand, when H2A.Z is paired with H3, it might, as suggested by Park et al. (2004), form an unusually stable nucleosome. Such nucleosomes would presumably be situated in regions of the genome that are not normally transcribed and are therefore poor in H3.3. Thus different combinations of histone variants could be delivered to different sites in the genome, and with quite different consequences for chromatin structure and gene regulation. Such a role for the histone variants has been proposed recently by Hake and Allis (2006).
These results raise several questions and suggest several directions for future study. First, it will be interesting to see how the structure of nucleosomes containing H3 and H2A differs from ones containing H3.3 complexed with H2A or H2A.Z. A difference in four amino acids distinguishes H3 from H3.3; three are located internally within the histone octamer structure, at sites that, in the solved H3/H2A.Z structure, do not make contact with H2A.Z. Whether this is also true of the H3.3/H2A.Z structure remains to be determined. In any case, it seems that the choice of partners among the histone variants will have an important effect on nucleosome stability and it is likely, therefore, may play an important regulatory role through that mechanism. It will be important to determine not only the location of a histone variant, but the identity of the other variants with which it shares the nucleosome.
Materials and methods
Cell lines
6C2 cells, stably expressing the H3 or H3.3 proteins fused with C-terminal Flag- and HA-epitope tags, were established and cultured as described elsewhere (Jin and Felsenfeld 2006). Cells were maintained in logarithmic-phase growth for all experiments.
Micrococcal nuclease digestions and preparation of mononucleosomes
Nuclei from 6C2 cells were prepared as described (Prioleau et al. 1999). We used 10 mM Tris-HCl (pH 7.4), 10 mM NaCl, 3 mM MgCl2, and 0.4% NP-40 as the lysis buffer. All buffers were supplemented with 10 mM Na-butyrate, 0.5 μg/mL aprotinin, 0.5 μg/mL leupeptin, and 1 μg/mL aprotinin. Nuclei were pelleted and resuspended in the same buffer plus 1 mM CaCl2. In some cases, the suspension of nuclei was treated with 0.2% formaldehyde for 1 min at room temperature to fix nuclei before MNase digestion. The A260 was adjusted to 1.25, and the resuspended nuclei were digested with 12 × 10−2 U/μL MNase (Worthington) for 12 min at 37°C. The reaction was stopped by adding EDTA (pH 8.0) to a final concentration of 10 mM, and the suspension was centrifuged at 2500 rpm for 5 min, retaining supernatant S1. The pellet was resuspended in lysis buffer plus 0.25 mM EDTA, incubated on ice for 15 min, and recentrifuged at 10,000 rpm for 10 min after passing four times through a 20-gauge needle followed by four passes through a 25-gauge needle. The supernatant S2 was combined with S1. In some cases, histones H1/H5 were removed by incubating S1 and S2 with 150 mM NaCl for 20 min at 4°C, followed by centrifugation at 10,000 rpm for 15 min. Mononucleosomes were purified on a 5%–30% sucrose gradient containing indicated 10 mM or 80 mM NaCl, 10 mM Tris-HCl (pH 7.4), and 0.2 mM EDTA.
Immunoprecipitation and Western blot analysis
H3-Flag- and H3.3-Flag-containing nucleosomes were isolated from mononucleosomes prepared from 6C2 cells expressing the H3 or H3.3 proteins fused with C-terminal Flag- and HA-epitope tags by immunoprecipitation on anti-Flag antibody-conjugated agarose. Mononucleosomes (0.7 mL) in immunoprecipitation buffer (10 mM Tris-HCl at pH 7.4, 50 mM or 10 mM NaCl, 0.2 mM EDTA) were incubated with 100 μL of anti-Flag M2 affinity gel overnight at 4°C (Sigma) and then washed five times with immunoprecipitation buffer. Immunoprecipitated H3-Flag- and H3.3-Flag-containing nucleosomes were then eluted by Catch and Release Denaturing elution buffer (Upstate Biotechnology) into a final volume of 140 μL. Twenty microliters of immunoprecipitated proteins were fractionated by SDS-PAGE and electrotransferred to PVDF membranes. Blots were incubated with the indicated primary antibodies and then with HRP-conjugated secondary antibodies. Proteins were visualized by ECL Plus Western blotting detection reagents (Amersham Bioscience). Band intensities were calculated using NIH ImageJ software.
Cesium chloride density gradient sedimentation
Sucrose gradient fractions containing mononucleosomes were dialyzed into 10 mM, 350 mM, or 450 mM NaCl; 10 mM triethanolamine-HCl (pH 7.4); and 0.2 mM EDTA and cross-linked with 0.5% formaldehyde for 15 min at room temperature. Excess cross-linker was removed by dialysis into DB buffer (80 mM NaCl, 10 mM Tris-HCl at pH 7.4, 0.2 mM EDTA). Samples were made up to 1.1 mL and treated with an equal volume of cesium chloride in dialysis buffer having a concentration of 1.06 g/mL, to a final density of ∼1.40 g/mL. An aliquot (2.2 mL) of this solution was centrifuged in a Beckman TLS-55 rotor for 96 h at 20.0°C and 40,000 rpm. Gradients were fractionated from the top into 22 × 100-μL fractions. An aliquot (30 μL) of each fraction was used to determine the refractive index, which was converted into a density based on an experimentally determined calibration curve using densities measured at 20°C on an Anton Paar DMA 5000 densimeter. The remaining 70 μL were dialyzed into DB buffer and reverse cross-linked in LSD loading buffer by heating for 10 min at 100°C. All fractions were then separated on SDS-PAGE and electrotransferred to PVDF membranes. Blots were incubated with the indicated primary antibodies and then with HRP-conjugated secondary antibodies. Proteins were visualized by Super Signal West Femto Maximum Sensitivity Substrate (Pierce).
Mass spectrometry
The intact monomers were isolated (see Results), and the protein bands corresponding to histone H3 were cut out from SDS-PAGE gels and subjected to protein sequence analysis by LC-MS/MS at the Taplin Biological Mass Spectrometry Facility at Harvard Medical School. The excised gel pieces were digested in-gel with trypsin followed by chymotrypsin (Shevchenko et al. 1996). The dried samples were reconstituted in 5–10 μL of HPLC solvent A (2.5% acetonitrile, 0.1% formic acid). A nano-scale reverse-phase HPLC capillary column was created by packing 5-μm C18 spherical silica beads into a fused silica capillary (100 μm inner diameter × ∼12 cm length) with a flame-drawn tip (Peng and Gygi 2001). After equilibrating the column, each sample was pressure-loaded offline onto the column. The column was then reattached to the HPLC system. A gradient was formed, and peptides were eluted with increasing concentrations of solvent B (97.5% acetonitrile, 0.1% formic acid). As each peptide was eluted, it was subjected to electrospray ionization and then entered into an LTQ-FT mass spectrometer (ThermoFinnigan). Eluting peptides were detected, isolated, and fragmented to produce a tandem mass spectrum of specific fragment ions for each peptide. Peptide sequences (and hence protein identity) were determined by matching protein or translated nucleotide databases with the acquired fragmentation pattern by the software program Sequest (ThermoFinnigan). Only the peptides FQSSAVMALQEASEAY (m/z = 874.40) and FQSAAIGALQEASEAY (m/z = 828.40), specific to H3 and H3.3, respectively, were selectively analyzed. To determine the ratio of H3 and H3.3 between different samples, the peak areas corresponding to the abovementioned peptides were compared.
ChIP and double ChIP
ChIP was carried out as described elsewhere (Jin and Felsenfeld 2006) except that only mononucleosomes prepared by micrococcal nuclease digest were used in this study. For double ChIP, purified mononucleosomes were cross-linked with 0.5% formaldehyde for 15 min at room temperature, and the reaction was stopped by adding glycine to a final concentration of 0.125 M. Excess cross-linker was removed by dialysis into DB buffer. We followed the procedure of anti-Flag antibody immuno-affinity purification (Nakatani and Ogryzko 2003) with minor modifications for first immunoprecipitation with anti-Flag M2 affinity gel. Four milliliters of cross-linked mononucleosomes in DB buffer (30 μg/mL) were incubated with 200 μL of anti-Flag M2 affinity gel for 3 h at 4°C and then loaded onto a column (Bio-Rad Poly Prep). After a strict wash of the beads with washing buffer (150 mM NaCl, 20 mM Tris-HCl at pH 7.4, 0.2 mM EDTA, 0.1% Tween), 100 μL of Flag-elution buffer (50 μg/mL Flag peptide in washing buffer) were loaded on the beads and incubated for 20 min at room temperature. The elution step was repeated to get maximum recovery of the first immunoprecipitates—namely, Flag-H3.3-containing mononucleosomes. The eluates were then subjected to the second immunoprecipitation with anti-H2A.Z. After purification, immunoprecipitated DNA was analyzed by quantitative real-time PCR. TaqMan probes and primers and the regions surveyed are described elsewhere (Jin and Felsenfeld 2006). A detailed description of the methods used for calculation can be found elsewhere (Litt et al. 2001).
Antibodies
The antibodies used were as follows: anti-Flag M2 Monoclonal antibody (F3165), anti-histone H2A.Z (07-594), anti-histone H3 (05-499), anti-acetyl-histone H3 (06-599), anti-histone H2A (7-146), and anti-histone H2B (07-371). All antibodies were purchased from Upstate Biotechnology except the anti-Flag antibody, which was from Sigma.
Acknowledgments
We thank Rodolfo Ghirlando for advice and help in CsCl gradient centrifugation, Julie Wallace and other members of the Felsenfeld laboratory for criticism of the manuscript, and the Taplin Biological Mass Spectrometry Facility at Harvard Medical School for mass spectrometry analysis. This researchwas supported by the Intramural Research Program of the NationalInstitutes of Health (National Institute of Diabetes and Digestiveand Kidney Diseases).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1547707
References
- Abbott D.W., Ivanova V.S., Wang X., Bonner W.M., Ausio J., Ivanova V.S., Wang X., Bonner W.M., Ausio J., Wang X., Bonner W.M., Ausio J., Bonner W.M., Ausio J., Ausio J. Characterization of the stability and folding of H2A.Z chromatin particles: Implications for transcriptional activation. J. Biol. Chem. 2001;276:41945–41949. doi: 10.1074/jbc.M108217200. [DOI] [PubMed] [Google Scholar]
- Ahmad K., Henikoff S., Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- Ashe H.L., Monks J., Wijgerde M., Fraser P., Proudfoot N.J., Monks J., Wijgerde M., Fraser P., Proudfoot N.J., Wijgerde M., Fraser P., Proudfoot N.J., Fraser P., Proudfoot N.J., Proudfoot N.J. Intergenic transcription and transinduction of the human β-globin locus. Genes & Dev. 1997;11:2494–2509. doi: 10.1101/gad.11.19.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce K., Myers F.A., Mantouvalou E., Lefevre P., Greaves I., Bonifer C., Tremethick D.J., Thorne A.W., Crane-Robinson C., Myers F.A., Mantouvalou E., Lefevre P., Greaves I., Bonifer C., Tremethick D.J., Thorne A.W., Crane-Robinson C., Mantouvalou E., Lefevre P., Greaves I., Bonifer C., Tremethick D.J., Thorne A.W., Crane-Robinson C., Lefevre P., Greaves I., Bonifer C., Tremethick D.J., Thorne A.W., Crane-Robinson C., Greaves I., Bonifer C., Tremethick D.J., Thorne A.W., Crane-Robinson C., Bonifer C., Tremethick D.J., Thorne A.W., Crane-Robinson C., Tremethick D.J., Thorne A.W., Crane-Robinson C., Thorne A.W., Crane-Robinson C., Crane-Robinson C. The replacement histone H2A.Z in a hyperacetylated form is a feature of active genes in the chicken. Nucleic Acids Res. 2005;33:5633–5639. doi: 10.1093/nar/gki874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton D.R., Butler M.J., Hyde J.E., Phillips D., Skidmore C.J., Walker I.O., Butler M.J., Hyde J.E., Phillips D., Skidmore C.J., Walker I.O., Hyde J.E., Phillips D., Skidmore C.J., Walker I.O., Phillips D., Skidmore C.J., Walker I.O., Skidmore C.J., Walker I.O., Walker I.O. The interaction of core histones with DNA: Equilibrium binding studies. Nucleic Acids Res. 1978;5:3643–3663. doi: 10.1093/nar/5.10.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C.M., Georgiou A., Szutorisz H., Maia e Silva A., Pombo A., Barahona I., Dargelos E., Canzonetta C., Dillon N., Georgiou A., Szutorisz H., Maia e Silva A., Pombo A., Barahona I., Dargelos E., Canzonetta C., Dillon N., Szutorisz H., Maia e Silva A., Pombo A., Barahona I., Dargelos E., Canzonetta C., Dillon N., Maia e Silva A., Pombo A., Barahona I., Dargelos E., Canzonetta C., Dillon N., Pombo A., Barahona I., Dargelos E., Canzonetta C., Dillon N., Barahona I., Dargelos E., Canzonetta C., Dillon N., Dargelos E., Canzonetta C., Dillon N., Canzonetta C., Dillon N., Dillon N. Variant histone H3.3 marks promoters of transcriptionally active genes during mammalian cell division. EMBO Rep. 2005;6:354–360. doi: 10.1038/sj.embor.7400366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris S.D., Rubio E.D., Moon J.J., Gombert W.M., Nelson B.H., Krumm A., Rubio E.D., Moon J.J., Gombert W.M., Nelson B.H., Krumm A., Moon J.J., Gombert W.M., Nelson B.H., Krumm A., Gombert W.M., Nelson B.H., Krumm A., Nelson B.H., Krumm A., Krumm A. Transcription-induced chromatin remodeling at the c-myc gene involves the local exchange of histone H2A.Z. J. Biol. Chem. 2005;280:25298–25303. doi: 10.1074/jbc.M501784200. [DOI] [PubMed] [Google Scholar]
- Franklin S.G., Zweidler A., Zweidler A. Non-allelic variants of histones 2a, 2b and 3 in mammals. Nature. 1977;266:273–275. doi: 10.1038/266273a0. [DOI] [PubMed] [Google Scholar]
- Guillemette B., Bataille A.R., Gevry N., Adam M., Blanchette M., Robert F., Gaudreau L., Bataille A.R., Gevry N., Adam M., Blanchette M., Robert F., Gaudreau L., Gevry N., Adam M., Blanchette M., Robert F., Gaudreau L., Adam M., Blanchette M., Robert F., Gaudreau L., Blanchette M., Robert F., Gaudreau L., Robert F., Gaudreau L., Gaudreau L. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 2005 doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake S.B., Allis C.D., Allis C.D. Histone H3 variants and their potential role in indexing mammalian genomes: The ‘H3 barcode hypothesis.’. Proc. Natl. Acad. Sci. 2006;103:6428–6435. doi: 10.1073/pnas.0600803103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake S.B., Garcia B.A., Duncan E.M., Kauer M., Dellaire G., Shabanowitz J., Bazett-Jones D.P., Allis C.D., Hunt D.F., Garcia B.A., Duncan E.M., Kauer M., Dellaire G., Shabanowitz J., Bazett-Jones D.P., Allis C.D., Hunt D.F., Duncan E.M., Kauer M., Dellaire G., Shabanowitz J., Bazett-Jones D.P., Allis C.D., Hunt D.F., Kauer M., Dellaire G., Shabanowitz J., Bazett-Jones D.P., Allis C.D., Hunt D.F., Dellaire G., Shabanowitz J., Bazett-Jones D.P., Allis C.D., Hunt D.F., Shabanowitz J., Bazett-Jones D.P., Allis C.D., Hunt D.F., Bazett-Jones D.P., Allis C.D., Hunt D.F., Allis C.D., Hunt D.F., Hunt D.F. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J. Biol. Chem. 2006;281:559–568. doi: 10.1074/jbc.M509266200. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Furuyama T., Ahmad K., Furuyama T., Ahmad K., Ahmad K. Histone variants, nucleosome assembly and epigenetic inheritance. Trends Genet. 2004;20:320–326. doi: 10.1016/j.tig.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Jin C., Felsenfeld G., Felsenfeld G. Distribution of histone H3.3 in hematopoietic cell lineages. Proc. Natl. Acad. Sci. 2006;103:574–579. doi: 10.1073/pnas.0509974103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobor M.S., Venkatasubrahmanyam S., Meneghini M.D., Gin J.W., Jennings J.L., Link A.J., Madhani H.D., Rine J., Venkatasubrahmanyam S., Meneghini M.D., Gin J.W., Jennings J.L., Link A.J., Madhani H.D., Rine J., Meneghini M.D., Gin J.W., Jennings J.L., Link A.J., Madhani H.D., Rine J., Gin J.W., Jennings J.L., Link A.J., Madhani H.D., Rine J., Jennings J.L., Link A.J., Madhani H.D., Rine J., Link A.J., Madhani H.D., Rine J., Madhani H.D., Rine J., Rine J. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004 doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N.J., Keogh M.C., Datta N., Sawa C., Ryan O.W., Ding H., Haw R.A., Pootoolal J., Tong A., Canadien V., Keogh M.C., Datta N., Sawa C., Ryan O.W., Ding H., Haw R.A., Pootoolal J., Tong A., Canadien V., Datta N., Sawa C., Ryan O.W., Ding H., Haw R.A., Pootoolal J., Tong A., Canadien V., Sawa C., Ryan O.W., Ding H., Haw R.A., Pootoolal J., Tong A., Canadien V., Ryan O.W., Ding H., Haw R.A., Pootoolal J., Tong A., Canadien V., Ding H., Haw R.A., Pootoolal J., Tong A., Canadien V., Haw R.A., Pootoolal J., Tong A., Canadien V., Pootoolal J., Tong A., Canadien V., Tong A., Canadien V., Canadien V., et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- Krogan N.J., Baetz K., Keogh M.C., Datta N., Sawa C., Kwok T.C., Thompson N.J., Davey M.G., Pootoolal J., Hughes T.R., Baetz K., Keogh M.C., Datta N., Sawa C., Kwok T.C., Thompson N.J., Davey M.G., Pootoolal J., Hughes T.R., Keogh M.C., Datta N., Sawa C., Kwok T.C., Thompson N.J., Davey M.G., Pootoolal J., Hughes T.R., Datta N., Sawa C., Kwok T.C., Thompson N.J., Davey M.G., Pootoolal J., Hughes T.R., Sawa C., Kwok T.C., Thompson N.J., Davey M.G., Pootoolal J., Hughes T.R., Kwok T.C., Thompson N.J., Davey M.G., Pootoolal J., Hughes T.R., Thompson N.J., Davey M.G., Pootoolal J., Hughes T.R., Davey M.G., Pootoolal J., Hughes T.R., Pootoolal J., Hughes T.R., Hughes T.R., et al. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc. Natl. Acad. Sci. 2004;101:13513–13518. doi: 10.1073/pnas.0405753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.P., Baxter H.J., Guillemette J.G., Lawford H.G., Lewis P.N., Baxter H.J., Guillemette J.G., Lawford H.G., Lewis P.N., Guillemette J.G., Lawford H.G., Lewis P.N., Lawford H.G., Lewis P.N., Lewis P.N. Structural studies on yeast nucleosomes. Can. J. Biochem. 1982;60:379–388. doi: 10.1139/o82-045. [DOI] [PubMed] [Google Scholar]
- Li B., Pattenden S.G., Lee D., Gutierrez J., Chen J., Seidel C., Gerton J., Workman J.L., Pattenden S.G., Lee D., Gutierrez J., Chen J., Seidel C., Gerton J., Workman J.L., Lee D., Gutierrez J., Chen J., Seidel C., Gerton J., Workman J.L., Gutierrez J., Chen J., Seidel C., Gerton J., Workman J.L., Chen J., Seidel C., Gerton J., Workman J.L., Seidel C., Gerton J., Workman J.L., Gerton J., Workman J.L., Workman J.L. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl. Acad. Sci. 2005;102:18385–18390. doi: 10.1073/pnas.0507975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt M.D., Simpson M., Recillas-Targa F., Prioleau M.N., Felsenfeld G., Simpson M., Recillas-Targa F., Prioleau M.N., Felsenfeld G., Recillas-Targa F., Prioleau M.N., Felsenfeld G., Prioleau M.N., Felsenfeld G., Felsenfeld G. Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J. 2001;20:2224–2235. doi: 10.1093/emboj/20.9.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyola A., Bonaldi T., Roche D., Imhof A., Almouzni G., Bonaldi T., Roche D., Imhof A., Almouzni G., Roche D., Imhof A., Almouzni G., Imhof A., Almouzni G., Almouzni G. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol. Cell. 2006;24:309–316. doi: 10.1016/j.molcel.2006.08.019. [DOI] [PubMed] [Google Scholar]
- McKittrick E., Gafken P.R., Ahmad K., Henikoff S., Gafken P.R., Ahmad K., Henikoff S., Ahmad K., Henikoff S., Henikoff S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl. Acad. Sci. 2004;101:1525–1530. doi: 10.1073/pnas.0308092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneghini M.D., Wu M., Madhani H.D., Wu M., Madhani H.D., Madhani H.D. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- Mito Y., Henikoff J.G., Henikoff S., Henikoff J.G., Henikoff S., Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G., Shen X., Landry J., Wu W.H., Sen S., Wu C., Shen X., Landry J., Wu W.H., Sen S., Wu C., Landry J., Wu W.H., Sen S., Wu C., Wu W.H., Sen S., Wu C., Sen S., Wu C., Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- Nakatani Y., Ogryzko V., Ogryzko V. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 2003;370:430–444. doi: 10.1016/S0076-6879(03)70037-8. [DOI] [PubMed] [Google Scholar]
- Okuwaki M., Kato K., Shimahara H., Tate S., Nagata K., Kato K., Shimahara H., Tate S., Nagata K., Shimahara H., Tate S., Nagata K., Tate S., Nagata K., Nagata K. Assembly and disassembly of nucleosome core particles containing histone variants by human nucleosome assembly protein I. Mol. Cell. Biol. 2005;25:10639–10651. doi: 10.1128/MCB.25.23.10639-10651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.J., Dyer P.N., Tremethick D.J., Luger K., Dyer P.N., Tremethick D.J., Luger K., Tremethick D.J., Luger K., Luger K. A new fluorescence resonance energy transfer approach demonstrates that the histone variant H2AZ stabilizes the histone octamer within the nucleosome. J. Biol. Chem. 2004;279:24274–24282. doi: 10.1074/jbc.M313152200. [DOI] [PubMed] [Google Scholar]
- Peng J., Gygi S.P., Gygi S.P. Proteomics: The move to mixtures. J. Mass Spectrom. 2001;36:1083–1091. doi: 10.1002/jms.229. [DOI] [PubMed] [Google Scholar]
- Pineiro M., Puerta C., Palacian E., Puerta C., Palacian E., Palacian E. Yeast nucleosomal particles: Structural and transcriptional properties. Biochemistry. 1991;30:5805–5810. doi: 10.1021/bi00237a025. [DOI] [PubMed] [Google Scholar]
- Prioleau M.N., Nony P., Simpson M., Felsenfeld G., Nony P., Simpson M., Felsenfeld G., Simpson M., Felsenfeld G., Felsenfeld G. An insulator element and condensed chromatin region separate the chicken β-globin locus from an independently regulated erythroid-specific folate receptor gene. EMBO J. 1999;18:4035–4048. doi: 10.1093/emboj/18.14.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner R.M., Hartley P.D., Meneghini M.D., Bao M.Z., Liu C.L., Schreiber S.L., Rando O.J., Madhani H.D., Hartley P.D., Meneghini M.D., Bao M.Z., Liu C.L., Schreiber S.L., Rando O.J., Madhani H.D., Meneghini M.D., Bao M.Z., Liu C.L., Schreiber S.L., Rando O.J., Madhani H.D., Bao M.Z., Liu C.L., Schreiber S.L., Rando O.J., Madhani H.D., Liu C.L., Schreiber S.L., Rando O.J., Madhani H.D., Schreiber S.L., Rando O.J., Madhani H.D., Rando O.J., Madhani H.D., Madhani H.D. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz B.E., Ahmad K., Ahmad K. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes & Dev. 2005;19:804–814. doi: 10.1101/gad.1259805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M., Wilm M., Vorm O., Mann M., Vorm O., Mann M., Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Suto R.K., Clarkson M.J., Tremethick D.J., Luger K., Clarkson M.J., Tremethick D.J., Luger K., Tremethick D.J., Luger K., Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat. Struct. Biol. 2000;7:1121–1124. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- Swaminathan J., Baxter E.M., Corces V.G., Baxter E.M., Corces V.G., Corces V.G. The role of histone H2Av variant replacement and histone H4 acetylation in the establishment of Drosophila heterochromatin. Genes & Dev. 2005;19:65–76. doi: 10.1101/gad.1259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H., Ray-Gallet D., Almouzni G., Nakatani Y., Ray-Gallet D., Almouzni G., Nakatani Y., Almouzni G., Nakatani Y., Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- Thambirajah A.A., Dryhurst D., Ishibashi T., Li A., Maffey A.H., Ausio J., Dryhurst D., Ishibashi T., Li A., Maffey A.H., Ausio J., Ishibashi T., Li A., Maffey A.H., Ausio J., Li A., Maffey A.H., Ausio J., Maffey A.H., Ausio J., Ausio J. H2A.Z stabilizes chromatin in a way that is dependent on core histone acetylation. J. Biol. Chem. 2006;281:20036–20044. doi: 10.1074/jbc.M601975200. [DOI] [PubMed] [Google Scholar]
- White C.L., Suto R.K., Luger K., Suto R.K., Luger K., Luger K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 2001;20:5207–5218. doi: 10.1093/emboj/20.18.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirbelauer C., Bell O., Schubeler D., Bell O., Schubeler D., Schubeler D. Variant histone H3.3 is deposited at sites of nucleosomal displacement throughout transcribed genes while active histone modifications show a promoter-proximal bias. Genes & Dev. 2005;19:1761–1766. doi: 10.1101/gad.347705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Roberts D.N., Cairns B.R., Roberts D.N., Cairns B.R., Cairns B.R. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]