Abstract
To facilitate the measurement of intramolecular distances in solvated RNA systems, a combination of spin-labeling, electron paramagnetic resonance (EPR), and molecular dynamics (MD) simulation is presented. The fairly rigid spin label 2,2,5,5-tetramethyl-pyrrolin-1-yloxyl-3-acetylene (TPA) was base and site specifically introduced into RNA through a Sonogashira palladium catalyzed cross-coupling on column. For this purpose 5-iodo-uridine, 5-iodo-cytidine and 2-iodo-adenosine phosphoramidites were synthesized and incorporated into RNA-sequences. Application of the recently developed ACE® chemistry presented the main advantage to limit the reduction of the nitroxide to an amine during the oligonucleotide automated synthesis and thus to increase substantially the reliability of the synthesis and the yield of labeled oligonucleotides. 4-Pulse Electron Double Resonance (PELDOR) was then successfully used to measure the intramolecular spin–spin distances in six doubly labeled RNA-duplexes. Comparison of these results with our previous work on DNA showed that A- and B-Form can be differentiated. Using an all-atom force field with explicit solvent, MD simulations gave results in good agreement with the measured distances and indicated that the RNA A-Form was conserved despite a local destabilization effect of the nitroxide label. The applicability of the method to more complex biological systems is discussed.
INTRODUCTION
RNA and its structural diversity has gained major attention in the last 10 years, in particular through the finding of RNAi as a natural antiviral mechanism of cells (1,2) and of riboswitches, a class of RNAs in bacteria, specialized in translational regulation (3). Furthermore, the folding of RNA and its conformational changes, induced by interactions with proteins, metal ions or small molecules, are essential for its biological function. This, in conjunction with a growing number of X-ray structures, makes RNA an ever increasingly interesting target for drug interactions and design.
In order to rationally approach RNA as a 3D target, simple, fast and accurate methods to gain structural and dynamical information are necessary. Electron Paramagnetic Resonance (EPR) has already proved its efficiency in characterizing the structural environment of paramagnetic centers (4–7), as well as the global arrangement of domains in proteins and protein complexes (8–13). Yet, EPR-based studies on the local structure of, e.g. metal ion binding sites (14,15) or of tertiary structure elements in RNA (16) and RNA/protein complexes (17,18) are rare. One reason for the lack of EPR studies related to tertiary RNA structures is that they require site directed and efficient labeling of RNA domains with nitroxides and subsequent measurements of the distance between these nitroxides. It is only recently that strategies were developed to spin label the phosphate backbone (17,19), the sugar moiety (16,20) or the uridine base (21–23) of RNA. Furthermore, pulsed EPR sequences like pulsed electron double resonance (PELDOR) (24–26) or double quantum coherence EPR (DQC-EPR) (27) had to be introduced, which are capable to reliably and precisely measure spin–spin distance of up to 8 nm (28) and overcome thereby the distance limit of ∼2 nm for continuous wave EPR techniques (29). First applications of PELDOR (30,31) and DQC (32) to duplex RNAs have been reported.
Despite these advances, each of the RNA spin labeling strategies mentioned above has its disadvantages and limitations. The major disadvantage of spin labeling RNA bases is the restriction to uridine. Either a 5-iodouridine (21) or a 4-thiouridine (22,23) is incorporated into the RNA during the automated phosphoramidite synthesis and then coupled with the acetylenic nitroxide derivative 2,2,5,5-tetramethyl-pyrrolin-1-yloxyl-3-acetylene (TPA) or a methanethio-sulfonate nitroxide (MTSSL), respectively. Advantageous of the TPA labeling is the chemically stable and geometrically fairly rigid acetylenic linker, whereas the disulfide bridge formed by MTSSL is chemically unstable and leads to the loss of the N3 imino proton, inducing structural distortions. The spin labeling of sugar moieties is also restricted with respect to label sites, at the moment to the 2′ site of pyrimidines (20). The largest flexibility with respect to the choice of label site is given by spin labeling a specific phosphate group using phosphorothioates in combination with a iodomethylnitroxide (19). However, in this case the 2′ site of the nucleotide 5′ to the label side has to be protected or replaced with a 2′ deoxyribose to avoid strand cleavage and the mixture of RP and SP diastereomers makes a translation of the measured distance into RNA structure more difficult. With respect to PELDOR, it should be mentioned that a parameter free and reliable extraction of a distance from the time trace requires the observation of a dipolar modulation. This can be achieved if most of the sample is labeled (>80%) and the distance distribution is small. Thus, highly efficient labeling strategies with rigid labels, a broad flexibility with respect to label sites and small structural perturbations are needed.
Here, we report an extension of RNA base specific labeling to cytosine and the purine adenine, in addition to a considerable increase of the yield of TPA labeled RNAs using ACetoxyEthyl orthoester (ACE®) chemistry. PELDOR measurements for each duplex-RNA yielded a dipolar modulation, from which the distance between the spin labels distances could be extracted. Furthermore, molecular dynamics (MD) simulations gave results in good agreement with the measured distances and indicated that the TPA label induces only a small and local structural distortion.
RESULTS AND DISCUSSION
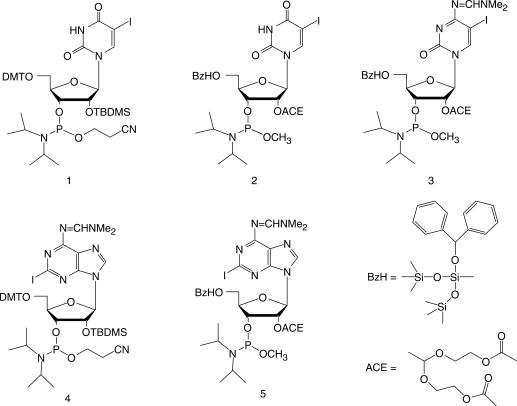
As we already had learned from our DNA work (33), the introduction of acetylenic spin labels postsynthetically on 5-iodo-2′-deoxyuridine during the oligonucleotide solid-phase synthesis presents several advantages in comparison to the derivatization in solution: the required amount of spin label is smaller, the yields are better and a simple washing step removes all excess reagents. Furthermore, the method can be extended to various labels, e.g. pyrene for fluorescence studies (34) or Fluorescence Resonance Energy Transfer (FRET) measurements (35). In order to successfully apply this method to RNA, we synthesized a series of iodinated RNA-building blocks (Figure 1).
Figure 1.
Phosphoramidites prepared for incorporation of iodinated bases into protected RNA oligomers.
In addition to the pyrimidine bases U and C iodinated at the 5 position, we also modified a purine base A at the 2 position, which brings more flexibility in the choice of the labeled nucleotide and therefore of the spin-label position in the strand. This flexibility is particularly important for future EPR studies or NMR based ‘Paramagnetic Relaxation Enhancement’ measurements on biological systems. We took also into consideration the orientation of the nitroxide spin label in the RNA, which can be directed for duplexes either into the major (for U, C and potentially for A) or into the minor groove [for A and G (36)]. Therefore, RNA–protein interactions, for example, could be studied without any interference of the label with the structure of the complex.
Initially, we decided to use the current standard phosphoramidite chemistry with the acid-labile 4,4′-dimethoxytrityl group (DMT) and the fluoride-labile tert-butyldimethylsilyl (TBDMS) group for protection of the 5′-OH and 2′-OH, respectively (37,38). 5-Iodouridine-phosphoramidite 1 is commercially available but can also be easily obtained in three steps from 5-iodouridine using standard methods. The key step for the synthesis of 5-iodocytidine was the iodination of the partially protected cytidine with iodic acid and iodine (39,40) (for the scheme and numbering of the compounds see supporting information Scheme 1). Standard deprotection/protection steps led to the phosphoramidite. For the minor groove modification, the nucleoside 2-iodoadenosine was synthesized in four steps from guanosine according to procedures described in the literature (41–43). In particular, the iodination was performed with iodine, copper iodide and methylene iodide via a radical mechanism. The phosphoramidite was obtained without difficulties after protection of the exocyclic amino group with formamidine (21).
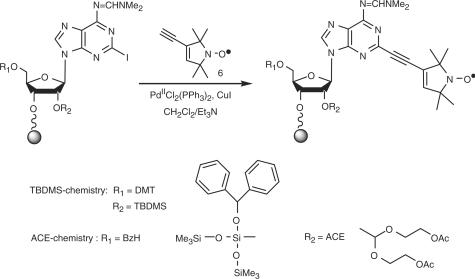
The phosphoramidites 1 and 4 were coupled successfully during synthesis of RNA 12 mers with the same efficiency as the phosphoramidites of the natural nucleobases. Key reaction for the derivatization on solid-phase was the Sonogashira palladium(II)-catalyzed cross-coupling reaction of the above iodo compounds with TPA 6 (Figure 2).
Figure 2.
Sonogashira cross-coupling during the solid-phase synthesis (example of 2-iodoadenosine).
Direct transfer of the procedure reported for DNA failed for RNA, in part due to its lower reactivity: three successive cross-couplings were necessary to achieve nearly quantitative yields in the case of 5-iodo-uridine in RNA (44), instead of two for 5-iodo-desoxyuridine in DNA. Hereby we envisaged also a serious side reaction for some of the RNA-building blocks, in particular for A. Detailed analysis showed it to be the reduction of the nitroxide to the corresponding amine. For example, a molecular mass of 16 g mol−1 less than the calculated one (minus oxygen) was often observed in MALDI-MS, as well as a minor peak corresponding to a mass of −30 g mol−1 (cleavage of NO). Purification by anion-exchange HPLC did not allow a complete separation of these side-products. When checking the RNA-synthesis cycle routinely performed for the oligonucleotides synthesis using TBDMS as a 2′-protecting group, we observed the same problem with the oxidation step iodine in pyridine/water as already suggested by Gannett et al. (45). The spin label could be first oxidized to the nitrone by the halogen and then react with water as proton donor to yield the hydroxylamine. Under the acidic conditions for the cleavage of the DMT group with dichloroacetic acid (DCA), this hydroxylamine could be protonated, the leaving water could give itself the necessary hydride for the formation of the amine (46).
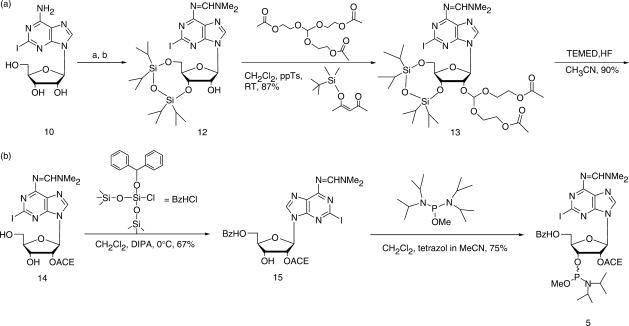
As we recently have had excellent results with the newly developed ACE chemistry (47–49), we changed our protocol to this procedure. This chemistry does not use iodine for the oxidation but water-free t-butyl hydroperoxide instead. In addition, the 5′-deprotection in each step is accomplished by fluoride, which is a much milder reagent than the acid DCA (46). The acid is responsible for further degradation of the oxidized nitroxide label. Thus the combination of the mild oxidation agent t-butyl hydroperoxide with the neutral fluoride is advantageous for the survival of the nitroxide spin label during RNA synthesis. The ACE protected 5-iodouridine-phosphoramidite 2 was purchased from Dharmacon, Chicago, IL, USA, whereas 2-iodoadenosine-phosphoramidite 5 and 5-iodocytidine-phosphoramidite 3 were synthesized in five steps from the protected nucleoside as previously described (50). The synthesis of 5 is shown in Scheme 1.
Scheme 1.
Synthetic pathway to 2-iodoadenosine phosphoramidite (ACE® chemistry); (a) HC(OMe)2NMe2, DMF, 50°C, 69% (compound 11); (b) TIPS-Cl2, pyridine, 0°C, 72%.
The 5′ and 3′ hydroxyl groups were simultaneously protected with the Markiewicz silyl group (51) in a 72% yield for 2-iodoadenosine and 71% for 5-iodocytidine. Selective introduction of the ACE-group at the 2′-OH was then performed with tris(2-acetoxyethyl)orthoformate under acidic catalysis (pyridinium-para-toluenesulfonate, ppTs). Once the reaction started, 4-tert-butyldimethylsiloxy-3-penten-2-one was added to increase the speed of the reaction by shifting the equilibrium towards the product. The completion of the reaction is achieved within 36–48 h after addition of the ketone, which led to very good yields of about 90% for both nucleobases. After quantitative deprotection of the Markiewicz group with a freshly prepared tetramethylethylenediamine (TEMED), HF solution in acetonitrile, the 5′-hydroxyl group was protected at 0°C with benzhydryloxy-bis(trimethylsilyloxy)chlorosilane under basic conditions (67% for 2-iodoadenosine and 79% for 5-iodocytidine). A pure phosphoramidite was formed by reaction of the 3′-hydroxyl group with methyl-N,N,N′,N-tetraisopropylphosphor-diamidite, activated with tetrazole.
Phosphoramidites 3 and 5 were incorporated into RNA sequences (Table 1) on a 0.2 μmol scale, and labeled with the nitroxide TPA by the same method and the same amount of reagents as in case of the TBDMS-chemistry (1 μmol synthesis). In this case, only two successive Sonogashira cross-couplings with the spin label were necessary to obtain quantitative yields. Analysis of the identity and purity of the synthesized oligonucleotides were performed with MALDI-Tof mass spectrometry, analytical HPLC and enzymatic digestion (see supporting information).
Table 1.
Spin-labeled RNA and their corresponding masses; the chemistry used for the preparation is indicated by a cross, a: RNA6 is non-self-complementary
| RNA | Sequence | Method | Calc. mass (g mol−1) | Meas. mass (g mol−1) | |
|---|---|---|---|---|---|
| TBDMS | ACE | ||||
| 1 | 3′ CGA CUA UAG UCG | × | 3958.6 | 3963.4 | |
| 5′ GCU GAU AUC AGC | |||||
| 2 | 3′ CUG ACU AGU CAG | × | 3958.6 | 3962.1 | |
| 5′ GAC UGA UCA GUC | |||||
| 3 | 3′ GCU GAC UAU AGU CAG C 5′ | × | 5244.3 | 5248.4 | |
| 5′ CGA CUG AUA UCA GUC G 3′ | |||||
| 4 | 3′ CGA CAU AUG UCG 5′ | × | × | 3958.6 | 3960.0 |
| 5′ GCU GUA UAC AGC 3′ | |||||
| 5 | 3′ GCA CAU ACG UAU GUG C 5′ | × | 5244.3 | 5244.5 | |
| 5′ CGU GUA UGC AUA CAC G 3′ | |||||
| 6a | 3′ CGA GUG AUA CAU CGC 5′ | × | 4938.1 | 4938.5 | |
| 5′ GCU CAC UAU GUA GCG 3′ | 4915.1 | 4920.6 | |||
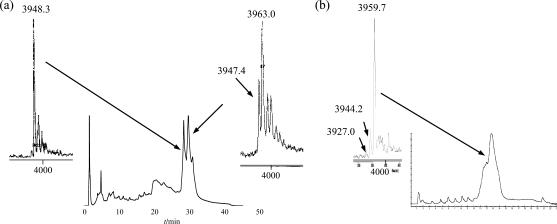
To stress the advantage of using the ACE chemistry, we compared the results obtained by both methods for RNA 4, modified with a spin-labeled adenosine. Using the TBDMS chemistry, a significant amount of reduced oligonucleotide was observed, as shown on the HPLC-chromatogram in Figure 3, and a second purification was necessary to obtain a pure enough sample for the EPR measurement, but which therefore lowered the yield. The ACE chemistry enabled us to achieve much better yields, as the proportion of reduced RNA was significantly decreased already after the first HPLC run yielding comparable MS-spectra (Figure 3). According to experiments in solution, this side reaction is due to the treatment of the oligonucleotides with disodium-2-carbamoyl-2-cyanoethylene-1,1-dithiolate-trihydrate to deprotect the phosphate groups at the end of the oligonucleotide synthesis. For all the labeled RNAs, reproducible results could be obtained with the ACE chemistry. Yields were 7–10 OD for a 12-mer (35–50%) to be compared with 4–10 OD with the TBDMS chemistry on a five times larger scale (4–10%).
Figure 3.
(a) HPLC-chromatogram of RNA 4 synthesized with the TBDMS chemistry and the corresponding MS spectra of the separated fractions as indicated with an arrow. The reduced oligonucleotide strand shows a mass of 3947–3948 g mol−1 and the spin labeled strand of 3963 g mol−1. (b) HPLC-chromatogram and MS spectrum of RNA 4, synthesized with the ACE® chemistry without second HPLC purification. The calculated mass of the spin labeled RNA 4 is 3958.6 g mol−1.
To determine if the spin label disturbs the RNA structure, UV-melting and Circular Dichroism (CD) studies were performed. The UV-melting curves showed a destabilization of the duplexes between 1.5 and 5.1°C, slightly higher than for spin labeled DNA-duplexes (33), which probably results from the geometry of the A-helix, with a very deep but narrow major groove and a very wide but shallow minor groove. CD spectroscopy confirmed that the A-helix is conserved with a similar ellipticity for both modified and unmodified RNAs. Both data together indicate that TPA does not significantly perturb the A-form RNA structure (for detailed data see supporting information).
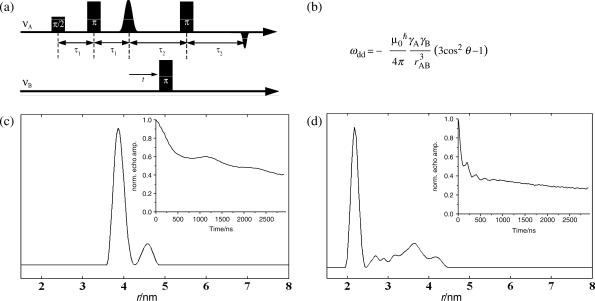
We then subjected RNA-duplexes 1–6 to 4-pulse-ELDOR measurements (Figure 4a). The PELDOR pulse sequence recovers the magnetic dipole coupling ωdd between two electron spins A and B from which the spin–spin distance rAB can be calculated according to the equation given in Figure 4b. The principle of the pulse sequence is the following: The detection sequence is applied at a microwave frequency νA which is in resonance with the A spin and creates an refocused echo. The amplitude of this echo is monitored as function of the time position t of an inversion pulse applied at a microwave frequency νB, which is in resonance with the B spin. This stimulated flip of the B spin induces a sudden change in the Larmor frequency of the spin A by ±ωdd, so that the A spins precess with this altered frequency in the transversal plane, leading to a non-perfect refocusing of the A spins. By variation of the time position t of the inversion pulse, the dephasing angle can be changed which induces a periodic modulation of the A-spin echo intensity by the dipolar coupling ωdd. The observation of this dipolar modulation is crucial for a reliable and parameter free distance calculation.
Figure 4.
(a) 4-Pulse ELDOR sequence. (b) Relation between the dipolar coupling ωdd and the spin–spin distance rAB. μ0 is the vacuum permeability, γ are the magnetogyric ratios of the spins A and B, ħ is the Planck's constant divided by 2Π and θAB is the angle between rAB and the external magnetic field. (c) Plot of the Tikhonov regularization and of the corresponding PELDOR time trace (inset) for RNA 3 and (d) for RNA 4.
Figure 4 shows as an example the measured PELDOR time traces and the corresponding distances obtained by Tikhonov regularization (52–54) for RNA 3 and RNA 4. Both RNAs show clearly visible oscillations in the time traces and one dominant peak in the Tikhonov regularizations at 38.7 and 21.9 Å for RNA 3 and 4, respectively. The peaks of weak intensity at larger distances are most likely due to a slight orientation selection, which is compensated by the Tikhonov regularization by adding more distances. An additional explanation may be coaxially end-to-end stacking of the helices (see supporting information) (30).
Also all other RNAs exhibit visible oscillations (supporting information) and the extracted mean distances are listed in Table 2. The observation of the dipolar modulation for each RNA proves that the labeling efficiency is high and that the label is sufficiently rigid to yield small distance distributions. Compared to earlier PELDOR studies on RNA (30,31), the dipolar oscillations are deeper for the spin-label TPA used here, which might be attributed to its increased rigidity. Accordingly, the modulation depth is comparable with TPA-labeled duplex DNAs (33). A recent PELDOR study on duplex DNAs labeled at the phosphate backbone also shows oscillations of comparable depth (55).
Table 2.
RNA sequences and the corresponding N–O/N–O distances from PELDOR and Molecular Dynamics (MD) simulations
| RNA | Sequence | R(PELDOR) [Å] | r(MD) [Å] |
|---|---|---|---|
| 1 | 3′CGA CU20A19 UAG UCG | 19.3 ± 1.2 [DNA 23.3 ± 0.6] | 18.0 (2.4) [DNA 21.4 (1.6)] |
| 5′GCU GAU6 AU8C AGC | |||
| 2 | 3′CUG ACU AGU CAG | 33.7 ± 3.9 [DNA 34.7 ± 1.4] | 30.5 (2.4) [DNA 33.0 (2.7)] |
| 5′GAC UGA UCA GUC | |||
| 3 | 3′GCUGACUAUAGUCAGC | 38.7 ± 1.3 [DNA 44.8 ± 5.0] | 36.2 (3.1) [DNA 43.3 (2.5)] |
| 5′CGACUGAUAUCAGUCG | |||
| 4 | 3′CGACAUAUGUCG | 21.9 ± 0.8 | 24.7 (0.8) |
| 5′GCUGUAUACAGC | |||
| 5 | 3′GCACAUACGUAUGUGC | 33.6 ± 2.6 | 34.3 (1.8) |
| 5′CGUGUAUGCAUACACG | |||
| 6 | 3′GCAGUGAUACAUCGC | 26.9 ± 1.3 | 24.6 (2.4) |
| 5′GCUCACUAUGUAGCG |
The PELDOR distances are given along with the experimental error determined from the full width of the peak at half height. The MD distances are given including the width of the distance distribution in brackets. For RNA 1–3, the mean distances for the sequence analog B-form DNA (33) is given in square brackets. The superscript numbers in the sequence of RNA 1 are used in the discussion of the MD part to identify the bases.
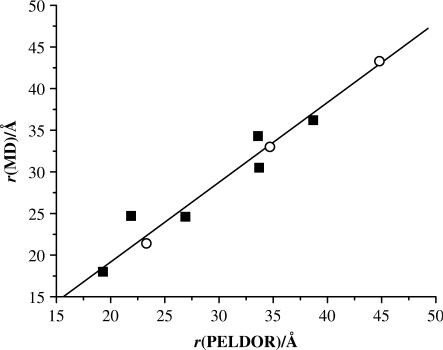
To be able to translate the measured N–O/N–O distances into RNA structures, we performed 50 ns all-atom MD simulations of all doubly labeled RNAs in explicit water solvent. The simulations yielded single-peaked distance distributions (measured between the oxygen atoms), whose mean and width are reported in Table 2. Mean distances from the PELDOR measurements compare well with the ones from the MD simulations started from generic A-form RNA duplexes. A linear fit through all data points yields r(MD) = 0.93 · r(PELDOR) + 1.0 Å with a standard deviation of 1.9 Å and a correlation coefficient of 0.976 (Figure 5). This correlation indicates that the RNA and DNA duplexes retain their conformations in frozen aqueous buffer solution if 20% ethylene glycol is added. Furthermore, the PELDOR experiments allow us to distinguish between A- and B-form helices, as long as the peak width is smaller than the distance difference between both conformations. Thus, RNA/DNA 1 and 3 can clearly be assigned to A- and B-conformations, whereas the distance difference is below this limit for RNA/DNA 2.
Figure 5.
Correlation of the PELDOR and MD distances for RNAs 1–6 (squares) and DNAs 1–3 (open circles).
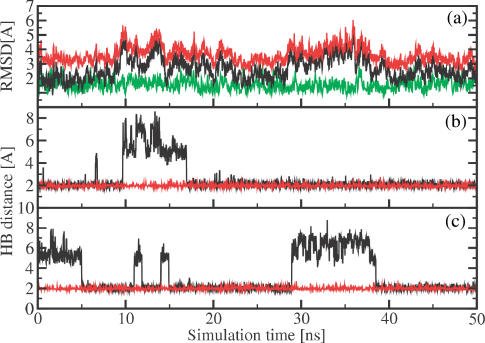
Apart from the distances, MD simulations may provide detailed information on the structure and conformational dynamics of these RNA systems (56–62). Here, we are interested in to what extent the label disturbs the RNA structure and dynamics. As a representative example, Figure 6a shows the time evolution of the root mean squared deviations (RMSD) of the MD trajectory obtained for RNA 1. The black line displays the RMSD from the standard A-form, while the red line reflects the RMSD from the standard B-form of RNA 1. During the 50 ns simulation, the RMSD from the A-form (2.7 Å in average) is always smaller than the RMSD from the B-form (3.7 Å in average). Note that there are no transitions between the A and B-form. The time evolutions of the RMSDs pertaining to the two RNA forms are correlated, indicating that the structure simultaneously moves away from both the A and the B-form that is, it approaches a non-standard structure of RNA. Besides the overall comparison of the structures with standard A-form or B-form by RMSD values, the classification of a dinucleotide step as A-form or B-form based on the positioning of phosphorus atoms with respect to the middle frame was performed using the program X3DNA (63). All dinucleotide steps maintain A-form quite well (∼80%), except for step 6 (i.e. between base pairs U6:A19 and A7:U18), which is only to 20% in A-form (Figure 6).
Figure 6.
Molecular dynamics simulation results obtained for RNA 1. (a) Time evolution of the RMSD from the standard A-form (black line) and B-form (red line) for labeled RNA 1. For comparison, the RMSD from the A-form obtained for unlabeled RNA 1 is also shown (green line). (b) and (c): Hydrogen bonding as monitored by (b) the distance between the H3 atom of uracil 6 and the N1 atom of adenine 19 and (c) the distance between the N1 atom of adenine 7 and the H3 atom of uracil 18. While the hydrogen bonds of labeled RNA 1 (black lines) open occasionally, the hydrogen bonds of unlabeled RNA 1 (red lines) remain stable.
To study to what extent the spin labels affect the overall structure of RNA 1, the green line in Figure 6a shows the RMSD from the A-form obtained for unlabeled RNA 1. As may be expected, the latter is smaller (1.6 Å in average) than in the case of the spin-labeled RNA 1 (2.7 Å in average). The RMSD from the B-form of unlabeled RNA 1 (data not shown) looks quite similar with an average of 3.9 Å. Moreover, the time evolution of the RMSD of unlabeled RNA 1 misses the characteristic modulations seen in the RMSD of labeled RNA 1, e.g. at times t = 10 and 30 ns. A closer analysis reveals that the latter are caused by occasional openings of hydrogen bonds between canonical Watson–Crick pairs, in particular in the vicinity of the labeled bases uracil 8 and uracil 20 of RNA 1. To illustrate this effect for the two base pairs between the two labeled bases, Figure 6b shows the distance between the H3 atom of uracil 6 and the N1 atom of adenine 19 and Figure 6c shows the distance between the N1 atom of adenine 7 and the H3 atom of uracil 18 (for the numbering of the bases see Table 2). When a hydrogen bond is formed the distance remains around 2.0 Å, while the distance may increase up to 7–8 Å, when the hydrogen bond is broken. It is interesting to note that the spin labels hardly perturb the two labeled bases uracil 8 and uracil 20, but mainly affect the structure of dinucleotide step 6 including the base pairs U6:A19 and A7:U18. For comparison, Figure 6 also shows (red lines) the corresponding results obtained for unlabeled RNA 1, which shows no indication of hydrogen bond opening. The opening of the Watson–Crick hydrogen bonds may be a reason for the observed lower melting temperatures found for the spin-labeled RNAs.
In conclusion, the spin-label TPA was introduced into RNA by Sonogashira cross-coupling on column during oligonucleotide solid phase synthesis utilizing the bases A, U and C.
Application of the recently developed ACE chemistry presented the main advantage to limit the reduction of the nitroxide TPA into the corresponding amine during the oligonucleotide synthesis and thereby to increase substantially the reliability of the synthesis and the yield of labeled oligonucleotides. Thus, we are able now to introduce the spin label either into the major or minor groove of duplex RNA and to adenine, uridine and cytosine. The combination of site specific labeling with the advantage of the rigid label renders this method advantageous for distance measurements.
PELDOR experiments enabled us to measure the intramolecular distances in the six doubly labeled RNA-duplexes with significant modulations depth of up to 40% and allowed us to distinguish A-form RNA from B-form DNA duplexes. MD simulations on the same oligonucleotides gave results in good agreement with the measured distances and showed that the destabilization effect of the label is only local. Thus the combination of this spin-label strategy with PELDOR and MD opens a way to study structures in complex RNA folds and how they change upon binding of metals, small organic ligands or proteins.
EXPERIMENTAL PART
Chemical synthesis
The reactions were monitored by thin-layer chromatography (TLC) analysis on silica gel aluminum plates (silica gel 60 F254, 0.2 mm, Merck, Columbus, OH, USA). Column chromatography was performed on silica gel (40–63 μm, 230–400 mesh, Merck). Technical solvents were used after distillation for chromatography; absolute solvents, dried over molecular sieve, were purchased from FLUKA. 1H, 13C and 31P NMR spectra were recorded with a Bruker AMX250/DPX 250 at 250 MHz or AMX400 at 400 MHz as indicated. Electron Spray Ionization (ESI) masses were collected on a VG Platform II (Fisons Instruments, San Carlos, CA, USA). Elemental analyses were performed on CHN-O-Rapid from Foss-Heraeus, Hanau, Germany.
5-Iodo-uridine-phosphoramidites 1 and 2
Commercially available from Glen Research, Sterling, VA, USA and Dharmacon, respectively.
2-Iodo-adenosine-phosphoramidites 4 and 5
All reactions were carried out under a protective argon atmosphere.
2′,3′,5′-Tri-O-acetyl-guanosine 7
A mixture of 35 g (0.12 mol) guanosine, 70 ml (0.74 mol, 6.2 eq) acetic anhydride in 140 ml of dried dimethylformamide/pyridine 5/2 was heated at 75°C for 4 h. The resulting solution was cooled to 4°C, at which the product crystallized overnight. After filtration, washing with isopropanol and drying overnight, 46 g (91%) 2′,3′,5′-tri-O-acetyl-guanosine were obtained.
Rf (CH2Cl2/MeOH: 9/1): 0.64; 1H NMR (250 MHz, DMSO-d6): δ[ppm] 10.80 (brs, 1H, NH), 7.95 (s, 1H, H8), 6.57 (s, 2H, NH2), 6.01 (d, J = 6 Hz, 1H, H1′), 5.81 (dt, J = 6.0 Hz, 1H, H2′), 5.52 (dd, J = 5.8 Hz, 1H, H3′), 4.42–4.24 (m, 3H, H4′ and H5′), 2.12 (s, 3H, OAc), 2.06 (s, 3H, OAc), 2.05 (s, 3H, OAc); 13C NMR (62.9 MHz, DMSO-d6): δ[ppm] 170.1–169.4–169.2 (3C=O), 156.7 (C6), 153.7 (C2), 151.1 (C4), 136.7 (C8), 116.8 (C5), 84.4 (C1′), 79.5 (C4′), 72.0 (C2′), 70.3 (C3′), 63.1 (C5′), 20.5–20.3–20.1 (3CH3); ESI-MS (+): calc. 409.1, found 410.0 [MH]+.
2′,3′,5′-Tri-O-acetyl-1′-deoxy-1′-(2-amino-6-chloropurine)-β-D-ribofuranose 8
To a solution of pre-dried 2′,3′,5′-tri-O-acetyl-guanosine (30 g, 73 mmol) in 150 ml abs acetonitrile were added at room temperature, in this order, 24.3 g (0.14 mol, 2 eq) tetraethylammoniumchloride, pre-dried overnight at 80°C over P2O5, 9.3 ml N,N-dimethylaniline (73 mmol, 1 eq) and 41 ml freshly distilled phosphoryl chloride (0.43 mol, 6 eq). After stirring for 10 min under reflux (the oil bath was preheated to 100°C), volatile materials were evaporated immediately in vacuo. The resulting oily yellow foam was dissolved in 300 ml chloroform and 200 ml cold water and stirred for 15 min under ice-cooling. The layers were separated and the aqueous phase was extracted with 3 × 100 ml chloroform. The combined organic phases were washed with 6 × 50 ml of cold water and 6 × 100 ml of a 5% NaHCO3 aqueous solution. After drying over Na2SO4, 150 ml isopropanol was added to the organic phase, which was then slowly evaporated to 100 ml. The product crystallized at 4°C, was filtered, washed with isopropanol and dried overnight in vacuo. The crude product could directly be used for the next step without any further purification. Yield: 19 g (61%). Rf (CH2Cl2/MeOH: 95/5): 0.52; 1H NMR (250 MHz, CDCl3): δ[ppm] 7.84 (s, 1H, H8), 5.95 (d, J = 4.8 Hz, 1H, H1′), 5.89 (dt, J = 5.1 Hz, 1H, H2′), 5.67 (dd, J = 4.9 Hz, 1H, H3′), 4.41–4.25 (m, 3H, H4′ and H5′), 2.07 (s, 3H, OAc), 2.03 (s, 3H, OAc), 2.01 (s, 3H, OAc); 13C NMR (62.9 MHz, CDCl3): δ[ppm] 170.5–169.7–169.4 (3C=O), 159.2 (C6), 153.1 (C2), 151.8 (C4), 140.7 (C8), 126.7 (C5), 86.6 (C1′), 79.9 (C4′), 72.7 (C2′), 70.5 (C3′), 64.0 (C5′), 20.7–20.5–20.4 (3CH3); ESI-MS (+): calc. 427.1, found 428.0 [MH]+.
2′,3′,5′-Tri-O-acetyl-1′-deoxy-1′-(6-chloro-2-iodopurine)-β-D-ribofuranose 9
Iodine (11.3 g, 44 mmol), diiodomethane (36 ml, 10 eq), copper iodide (9.3 g, 49 mmol) and isopentyl nitrite (17.8 ml, 0.13 mol) were added to a solution of 19.0 g of 2′,3′,5′-tri-O-acetyl-1′-deoxy-1′-(2-amino-6-chloropurine)-β-D-ribofuranose (44 mmol) in 200 ml abs THF. The suspension was stirred at reflux for 45 min. After cooling at RT, the mixture was filtered off and the solution evaporated. Followed a purification through a flash column chromatography (CH2Cl2/MeOH: 99/1). Yield: 19.4 g (82%). Rf (CH2Cl2/MeOH: 98/2): 0.28; 1H NMR (250 MHz, CDCl3): δ[ppm] 8.16 (s, 1H, H8), 6.25 (m, 1H, H1′), 5.73 (m, 1H, H2′), 5.53 (m, 1H, H3′), 4.39 (m, 1H, H4′), 4.34 (m, 1H, H5′), 2.11 (s, 3H, OAc), 2.08 (s, 3H, OAc), 2.04 (s, 3H, OAc); 13C NMR (62.9 MHz, CDCl3): δ[ppm] 170.4–169.9–169.6 (3C=O), 152.0 (C6), 150.9 (C4), 143.2 (C2), 132.2 (C8), 117.0 (C5), 84.7 (C1′), 80.8 (C4′), 73.3 (C2′), 70.6 (C3′), 62.9 (C5′), 20.8–20.6–20.4 (3CH3); ESI-MS (+): calc. 538.0, found 538.9 [MH]+.
2-Iodo-adenosine 10
To 750 ml of abs ethanol saturated with ammonia at 0°C was added 2′,3′,5′-tri-O-acetyl-1′-deoxy-1′-(6-chloro-2-iodopurine)-β-D-ribofuranose (7.16 g, 13.3 mmol). The solution was stirred 1 h at 0°C and 24 h at RT. The solvent was then removed under reduced pressure and the residue treated with a NaOCH3 solution (25 mM) for 1 h at RT to remove the last acetyl group. After neutralization with DOWEX 50W-8, filtration of the resin and evaporation of the solvent, the crude product was purified by crystallization in water. Yield: 4.45 g (85%). Rf (CH2Cl2/MeOH: 9/1): 0.24; 1H NMR (400 MHz, DMSO-d6): δ[ppm] 8.29 (s, 1H, H8), 7.73 (brs, 2H, NH2), 5.80 (d, 1H, H1′, J = 6.2 Hz), 5.46 (d, J = 6.2 Hz, 1H, 2′-OH), 5.20 (d, J = 3.7 Hz, 1H, 3′-OH), 5.04 (t, J = 6.3 Hz, 1H, 5′-OH), 4.52 (dd, J = 6.2 Hz and 11.2 Hz, 1H, H2′), 4.11 (m, 1H, H3′), 3.93 (m, 1H, H4′), 3.67–3.51 (m, 2H, H5′); 13C NMR (100.6 MHz, DMSO-d6): δ[ppm] 155.9 (C6), 149.7 (C4), 136.4 (C8), 121.3 (C2), 120.8 (C5), 87.2 (C1′), 85.8 (C4′), 73.6 (C2′), 70.7 (C3′), 61.4 (C5′); ESI-MS (−): calc. 393.1, found 391.9 [M-H]−.
N,N-Dimethyl-N′-formamidine-2-iodo-adenosine 11
A mixture of 2-iodo-adenosine (4.44 g, 11.3 mmol), 7.5 ml N,N-dimethylformamide-dimethylacetal (56.5 mmol, 5 eq) in 90 ml abs dimethylformamide (DMF) was heated at 50°C for 1 h. After evaporation of the solvent, the obtained oil was purified through a flash chromatography (CH2Cl2/MeOH: 95/5). Yield 3.5 g (69%). Rf (CH2Cl2/MeOH: 9/1): 0.36; 1H NMR (250 MHz, DMSO-d6): δ[ppm] 8.83 (s, 1H, CH), 8.42 (s, 1H, H8), 5.87–5.50–5.23 (3 s, 3H, 3OH), 5.03 (d, 1H, H1′, J = 5.25 Hz), 4.57 (m, 1H, H2′), 4.47–4.09 (m, 1H, H3′), 3.96 (m, 1H, H4′), 3.72–3.35 (m, 2H, H5′), 3.25 (s, 3H, CH3), 3.24 (s, 3H, CH3); 13C NMR (62.9 MHz, DMSO-d6): δ[ppm] 159.3 (CH), 158.4 (C6), 151.9 (C4), 141.0 (C2), 125.6 (C8), 120.3 (C5), 87.2 (C1′), 86.8 (C4′), 73.5 (C2′), 70.4 (C3′), 61.4 (C5′), 34.6 (2CH3); ESI-MS (+): calc. 448.0, found 448.8 [MH]+; Anal. calc. for C13H17IN6O4: C 34.84%, H 3.82%, N 18.75%; found: C 34.60%, H 4.10%, N 18.50%.
3′,5′-Tetraisopropyldisiloxane-N,N-dimethyl-N′-formamidine-2-iodo-adenosine 12
N,N′-dimethyl-N-formamidine-2-iodo-adenosine (600 mg, 1.33 mmol) was dissolved in 5 ml abs pyridine and cooled at 0°C. 1,3-Dichloro-1,1,3,3-tetraisopropyldisiloxane (0.46 ml, 1.1 eq) was added dropwise for 1 h. After further stirring for 30 min at 0°C, the reaction was quenched with 0.2 ml water, the solution concentrated in vacuo and coevaporated with toluene. The residue was dissolved in 30 ml CH2Cl2 and washed with 5% aqueous NaHCO3. After separation of the two layers, the aqueous phase was extracted once with 15 ml CH2Cl2. The combined organic phases were washed with 20 ml saturated aqueous NaCl. The separated aqueous phase was extracted with 15 ml CH2Cl2. The organic phases were combined, dried with Na2SO4, filtered and evaporated. A flash chromatography (EtOAc/n-hex: 8/2 to 10/0) afforded 660 mg product (72%). Rf (CH2Cl2/MeOH: 98/2): 0.33; 1H NMR (250 MHz, DMSO-d6): δ[ppm] 8.80 (s, 1H, CH), 8.25 (s, 1H, H8), 5.86 (1 d, 1H, OH), 5.62 (d, 1H, H1′), 4.66–4.56 (m, 2H, H2′ and H3′), 4.01–3.97 (m, 3H, H4′and H5′), 3.23 (s, 3H, CH3), 3.14 (s, 3H, CH3), 1.15–0.90 (m, 28H, Si-iPr); 13C NMR (62.9 MHz, DMSO-d6): δ[ppm] 159.4 (CH), 158.2 (C6), 152.7 (C4), 141.1 (C2), 125.8 (C8), 120.3 (C5), 89.4 (C1′), 80.8 (C4′), 73.3 (C2′), 70.8 (C3′), 61.1 (C5′), 40.9 and 34.8 (2CH3), 17.3–16.9 (CH(CH3)2), 13.9–12.4 (CH(CH3)2); ESI-MS (+): calc. 690.19, found 691.2 [MH]+; Anal. calc. for C25H43IN6O5Si2: C 43.47%, H 6.27%, N 12.17%; found: C 43.44%, H 6.33%, N 12.03%.
2′-O-bis-(Acetoxyethyloxy)methylester-3′,5′-tetraisopropyldisiloxane-N,N-dimethyl-N′-formamidine-2-iodo-adenosine 13
A solution of 12 (970 mg, 1.4 mmol), tris(2-acetoxyethyl)orthoformate (1.04 g, 2.3 eq), 3 ml abs dichloromethane and pyridinium p-toluenesulfonate (70 mg, 0.2 eq) was stirred at RT. As soon as the reaction has progressed well (∼6 h after TLC analysis), 4-tert-butyldimethylsiloxy-3-penten-2-one (0.6 ml, 1.8 eq) was added, the solution stirred for 40 h at RT and finally neutralized with 105 μl TEMED (0.5 eq). Followed a direct flash chromatography (EtOAc/n-hex/MeOH/TEMED: 50/50/1/0.5) to yield a yellow foam (1.11 g, 87%). Rf (CH2Cl2/MeOH: 95/5): 0.42; 1H NMR (250 MHz, CDCl3): δ[ppm] 8.80 (s, 1H, CH), 8.00 (s, 1H, H8), 6.04 (m, 1H, H1′), 5.65 (s, 1H, CH(ACE)), 4.61–4.55 (m, 1H, H2′), 4.43–4.41 (m, 1H, H3′), 4.30–4.08 (m, 6H, CH2(ACE), H4′and H5′), 3.99–3.80 (m, 5H, H5′ and CH2(ACE)), 3.18 (s, 3H, CH3), 3.15 (s, 3H, CH3), 1.99 and 1.93 (2 s, 6H, OAc), 1.92–1.05 (m, 28H, Si-iPr); 13C NMR (62.9 MHz, CDCl3): δ[ppm] 170.9 (C=O), 159.7 (CH), 158.5 (C6), 151.1 (C4), 139.4 (C2), 126.7 (C8), 120.0 (C5), 112.3 (CH(ACE)), 88.6 (C1′), 81.4 (C4′), 76.5 (C2′), 69.1 (C3′), 63.6, 63.3, 63.2, 61.0 (CH2(ACE)), 59.9 (C5′), 41.6 and 36.5 (2CH3 formamidine), 20.9 (CH3(ACE)), 17.6–16.9 (CH(CH3)2), 13.3–12.7 (CH(CH3)2); MALDI-MS (+): calc. 908.3, found 909.3 [MH]+ and 931.2 [MNa]+.
2′-O-bis-(Acetoxyethyloxy)methylester-N,N-dimethyl-N′-formamidine-2-iodo-adenosine 14
TEMED/HF was freshly and separately prepared: 0.89 ml TEMED (5.9 mmol) was dissolved in 2 ml acetonitrile at 0°C and 0.15 ml hydrofluoric acid (48% in water, 4.1 mmol) was added slowly in 2 min. This mixture was stirred for 5 min at 0°C and added dropwise in 5 min at RT to a solution of 1.08 g 2′-O-ACE-3′,5′-O-TIPS-N,N-dimethyl-N′-formamidine-2-iodo-adenosine 13 in 2 ml acetonitrile. After 2 h stirring, the solution was concentrated in vacuo, the oily residue was dissolved in 5 ml CH2Cl2, 1 ml n-hexane and 0.1 ml TEMED and purified through a flash chromatography (95/5/0.5: EtOAc/MeOH/TEMED). Yield: 711 mg (90%). Rf (EtOAc/MeOH: 95/5): 0.13; 1H NMR (250 MHz, CDCl3): δ[ppm] 8.81 (s, 1H, CH), 7.78 (s, 1H, H8), 5.84 (d, J = 7.8 Hz, 1H, H1′), 5.17 (s, 1H, CH(ACE)), 4.96 (m, 1H, H2′), 4.45 (m, 1H, H3′), 4.29 (m, 1H, H4′), 4.06–3.92 (m, 5H, CH2(ACE) and H5′), 3.72–3.45 (m, 5H, H5′ and CH2(ACE)), 3.21 (s, 3H, CH3), 3.20 (s, 3H, CH3 formamidine), 1.99 and 1.93 (2 s, 6H, OAc); 13C NMR (62.9 MHz, CDCl3): δ[ppm] 170.8 and 170.8 (C=O), 160.2 (CH), 158.7 (C6), 150.9 (C4), 141.6 (C2), 127.6 (C8), 119.1 (C5), 112.7 (CH(ACE)), 89.1 (C1′), 87.7 (C4′), 76.6 (C2′), 71.9 (C3′), 63.4 (CH2(ACE)), 63.3 (C5′), 62.8 and 62.6 (CH2(ACE)), 41.7 and 35.6 (CH3 formamidine), 20.9 (CH3(ACE)); ESI-MS (−): calc. 666.1, found: 700.9 [MCl]−.
2′-O-bis-(Acetoxyethyloxy)methylester-5′-O-bis(trimethylsilyloxy)benzhydryloxysilyl-N,N-dimethyl-N′-formamidine-2-iodo-adenosine 15
Compound 14 (660 mg, 0.99 mmol) and diisopropylamine (169 μl, 1 eq) were dissolved in 5 ml abs dichloromethane and cooled to 0°C. Simultaneously, 210 μl diisopropylamine were added dropwise in 1 min to a solution of benzhydryloxy-bis(trimethylsilyloxy)chlorsilane (BzHCl, 630 mg, 1.5 eq) in 2 ml abs CH2Cl2. After 5 min stirring at RT, this last solution was added dropwise and in portions to the previous one: first 0.5 eq BzHCl in 15 min, then twice 0.2 eq in 10 min and finally portions of 0.1 eq until the completion of the reaction (TLC analysis). The mixture was washed with 10 ml 8% aqueous NaHCO3 and 10 ml brine. The layers were separated and the organic phase dried with Na2SO4 and evaporated. The crude product was purified through a flash chromatography (EtOAc/n-hex/acetone/Et3N: 35/45/20/0.5), which afforded 700 mg (67%) of 15. Rf (CH2Cl2/MeOH: 95/5): 0.36; 1H NMR (250 MHz, CDCl3): δ[ppm] 8.92 (s, 1H, CH), 8.04 (s, 1H, H8), 7.31–7.12 (m, 10H, phenyl), 6.09 (d, 1H, H1′, J = 4.5 Hz), 5.88 (s, 1H, CH-Ph), 5.33 (s, 1H, CH(ACE)), 4.58 (m, 1H, H2′), 4.21–4.00 (m, 6H, H3′, H4′ and CH2(ACE)), 3.88–3.82 (m, 1H, H5′), 3.76–3.64 (m, 5H, H5′ and CH2(ACE)), 3.21 (s, 3H, CH3 formamidine), 3.15 (s, 3H, CH3 formamidine), 1.97 (s, 6H, OAc), −0.02 to 0.01 (2 s, 18H, SiMe3); 13C NMR (62.9 MHz, CDCl3): δ[ppm] 169.3 (C=O), 158.4 (CH), 157.7 (C6), 150.2 (C4), 142.5 (CAr), 138.3 (C2), 126.7 (C8), 126.7, 124.9, 124.9, 124.7 (Ph-H), 118.5 (C5), 111.3 (CH(ACE)), 85.5 (C1′), 83.0 (C4′), 76.7 (CH-Ph), 75.4 (C2′), 68.8 (C3′), 61.6 (CH2(ACE)), 61.6 (C5′), 61.4 and 60.8 (CH2(ACE)), 40.01 and 33.8 (CH3 formamidine), 19.4 and 19.3 (CH3(ACE)), 0.00 (SiMe3); ESI-MS (+): calc. 1054.3, found: 1055.3 [MH]+; Anal. calc. for C41H59IN6O13Si3: C 46.67%, H 5.64%, N 7.97%; found: C 46.63%, H 5.88%, N 7.32%.
2′-O-bis-(Acetoxyethyloxy)methylester-3′-O-(N,N-diisopropylamino)methoxyphosphinyl-5′-O-bis-(trimethylsilyloxy)benzhydryloxysilyl-N,N-dimethyl-N′-formamidine-2-iodo-adenosine 5
About 216 μl Methyl-N,N,N′,N′-tetraisopropylphosphordiamidite (0.75 mol) and 666 μl of a 0.45 M tetrazole solution in acetonitrile (0.5 eq) were solved in 2 ml abs dichloromethane. After 5 min at RT, this solution was added dropwise to a solution of 2′-O-ACE-5′-O-BzH-N,N-dimethyl-N′-formamidine-2-iodo-adenosine (632 mg, 0.60 mmol) in 2 ml abs dichloromethane, cooled at 0°C. After 5 min at 0°C and 11 h at RT, the reaction mixture was quenched with 175 μl ethanol (5 eq) and evaporated. A purification with a flash chromatography (n-hex/acetone/Et3N: 70/30/0.5) yielded 546 mg of the phosphoramidite (75%). Rf (n-hex/acetone: 70/30): 0.20 and 0.11; 1H NMR (250 MHz, CDCl3): δ[ppm] 9.00 (s, 2H, CH), 8.11 and 8.08 (2 s, 2H, H8), 7.25–7.37 (2 m, 20, Phenyl), 6.20 (s, 2H, CH–Ph), 5.95–5.96 (m, 2H, 2H1′), 5.40 and 5.48 (2 s, 2H, CH(ACE)), 4.72 (m, 2H, H2′), 4.50 (m, 2H, 2H3′), 4.12–4.18 (m, 10H, 2H4′ and 2CH2(ACE)), 3.61–3.81 (m, 14H, 2H5′, 2H5″, 2CH2(ACE) and 4CH(iPr)), 3.34–3.43 (m, 6H, OCH3), 3.26 (s, 6H, CH3 formamidine), 3.18 (s, 6H, CH3 formamidine), 2.00–2.06 (4 s, 12H, CH3(ACE)), 1.16–1.22 (m, 24H, iPr), 0.04–0.11 (s, 36H, SiMe3); 31P NMR (162 MHz, CDCl3): δ[ppm] 150.82 and 150.71 (1/1); ESI-MS (+): calc. 1215.4, found 1215.8.
5′-O-(4,4′-Dimethoxytriphenylmethyl)-N,N-dimethyl-N′-formamidine-2-iodo adenosine 16
About 1.50 g N,N-dimethyl-N′-formamidine-2-iodo-adenosine (3.34 mmol) were dissolved in 35 ml abs DMF. A solution of 4,4′-dimethoxytriphenylmethylchloride (1.39 g, 1.2 eq) in 7 ml abs pyridine was added dropwise in three portions. After 3 h at RT, the reaction was quenched with 5 ml methanol, the solution evaporated in vacuo and coevaporated with toluene. The obtained foam was taken up in dichloromethane and washed with a saturated NaHCO3 aqueous solution. The two layers were separated and the organic phase extracted with CH2Cl2. The combined organic phases were dried with Na2SO4, filtered and evaporated. A flash chromatography yielded 1.69 g of the purified product (67%). Rf (CH2Cl2/MeOH: 95/5): 0.39; 1H NMR (250 MHz, CDCl3): δ[ppm] 8.87 (s, 1H, CH), 7.87 (s, 1H, H8), 7.26–7.07 (m, 9H, DMT), 6.70–6.66 (m, 4H, DMT), 5.87 (d, J = 6.0 Hz, 1H, H1′), 4.71 (dd, J = 5.8 Hz, 1H, H2′), 4.34–4.31 (m, 2H, H3′ and H4′), 3.70 and 3.69 (2 s, 6H, OCH3), 3.39–3.22 (m, 2H, H5′), 3.19 (s, 3H, CH3), 3.18 (s, 3H, CH3); 13C NMR (62.9 MHz, CDCl3): δ[ppm] 159.0 (C6), 158.5 (CH), 151.2 (C4), 144.5 (C2), 140.0 (C8), 135.6 (DMT), 130.0–125.6 (DMT), 119.6 (C5), 113.1 (DMT), 91.8 (C1′), 85.3 (C4′), 74.7 (C2′), 72.4 (C3′), 63.8 (C5′), 55.2 (OMe), 35.5 and 41.1 (2CH3); ESI-MS (+): calc. 750.2, found 751.2 [MH]+; Anal. calc. for C34H35IN6O6: C 54.41%, H 4.70%, N 11.20%; found: C 54.00%, H 4.84%, N 10.92%.
5′-O-(4,4′-Dimethoxytriphenylmethyl)-2′-O-(tert-butyldimethylsilyl)-N,N-dimethyl-N′-formamidine-2-iodo-adenosine 17
To a solution of 1.82 g of 5′-O-(4,4′-dimethoxytriphenylmethyl)-N,N-dimethyl-N′-formamidine-2-iodo-adenosine in 40 ml of a mixture tetrahydrofurane (THF)/pyridine: 1/1 were added under argon, silver nitrate (536 mg, 1.3 eq) and t-butyltrimethylsilylchloride (1 M in THF, 3.4 ml, 1.4 eq). The suspension was stirred 7 h at RT in the dark. After filtration over Celite®, washing with CH2Cl2, the clear solution was evaporated. The residue was dissolved in CH2Cl2, the organic phase washed with a saturated NaHCO3 aqueous solution. The aqueous phase was then extracted twice with dichloromethane. The combined organic phases were finally dried with Na2SO4, filtered, concentrated in vacuo and coevaporated with toluene. A flash chromatography (EtOAc/n-hex: 7/3 then 100/0) enabled to separate the two isomers. The 2′-O-isomer was obtained in a 61% yield, the unwanted 3′-O-isomer in a 10% yield. Rf (CH2Cl2/MeOH: 95/5): 0.39; 1H NMR (250 MHz, CDCl3): δ[ppm] 8.90 (s, 1H, CH), 8.01 (s, 1H, H8), 7.47–7.16 (m, 9H, DMT), 6.86–6.80 (m, 4H, DMT), 6.03 (d, J = 5.0 Hz, 1H, H1′), 4.85 (dd, J = 4.9 Hz, 1H, H2′), 4.32–4.22 (m, 2H, H3′ and H4′), 3.79 (s, 6H, OCH3), 3.55–3.41 (m, 2H, H5′), 3.27 (s, 3H, CH3), 3.23 (s, 3H, CH3), 0.88 (s, 9H, tBu), 0.05 and −0.05 (2 s, 6H, SiCH3); 13C NMR (62.9 MHz, CDCl3): δ[ppm] 159.1 (C6), 158.4 (CH), 151.8 (C4), 144.5 (C2), 136.7 (C8), 135.6–135.5 (DMT), 130.0–126.3 (DMT), 119.9 (C5), 113.1–113.2 (DMT), 88.2 (C1′), 84.1 (C4′), 75.9 (C2′), 71.5 (C3′), 63.4 (C5′), 55.1 (OMe), 35.4 and 41.4 (2CH3), 25.6 and 25.5 (SitBu), −5.3 and −4.9 (SiMe); ESI-MS (+): calc. 864.8, found 865.4 [MH]+; Anal. calc. for C40H49IN6O6Si: C 55.55%, H 5.71%, N 9.72%; found: C 55.46%, H 5.92%, N 9.49%.
3′-O-(2-Cyanethoxydiisopropylphosphine)-5′-O-(4,4′-dimethoxytriphenylmethyl)-2′-O-(tert butyldimethylsilyl)-N,N-dimethyl-N′-formamidine-2-iodo-adenosine 4
A solution of 705 mg 17 (0.83 mmol) and 420 μl diisopropylethylamine (3 eq) in 20 ml abs dichloromethane was cooled to 0°C. About 280 μl 2-cyanoethyldiisopropylchlorophosphoramidite (1.5 eq) were then added under argon. The mixture was stirred for 5 min at 0°C then 2 h at RT and finally diluted with 20 ml CH2Cl2. The organic phase was washed twice with a saturated NaHCO3 solution, dried with Na2SO4, filtered and concentrated in vacuo. The crude product was purified through a flash chromatography (EtOAc/n-hex: 8/2). Yield: 745 mg (86%). Rf (EtOAc/n-hex: 80/20): 0.43 and 0.38; 1H NMR (250 MHz, CDCl3): δ[ppm] 9.03 (s, 2H, CH), 8.16 and 8.14 (2 s, 2H, H8), 7.61–7.37 (m, 18H, DMT), 6.98–6.93 (m, 8H, DMT), 6.14 (m, 2H, H1′), 5.09 (m, 2H, H2′), 4.49–4.46 (m, 4H, H3′ and H4′), 3.93 (s, 6H, OCH3), 3.92 (s, 6H, OCH3), 3.76–3.72 (m, 8H, OCH2 and H5′), 3.40 (s, 3H, CH3), 3.36 (s, 3H, CH3), 2.82, 2.48 (m, 4H, CH2CN), 1.31–1.21 (m, 24H, iPr), 1.18 and 1.02 (2 s, 18H, SitBu), 0.16–0.04 (m, 12H, SiMe2); 31P NMR (162 MHz, CDCl3): δ[ppm] 151.13 and 149.78; ESI-MS(+): calc. 1064.4, found: 1065.1 [MH]+.
5-Iodo-cytidine-phosphoramidite 3
2′,3′,5′-Tri-O-acetyl-cytidine, hydrochloride 18
Acetylchloride (17.5 ml, 0.24 mol) was added to a solution of 10 g cytidine (40 mmol) in 65 ml acetic acid. After stirring overnight at RT, the solution was concentrated and the obtained white powder recrystallized in ethanol. Yield: 13 g (78%). Rf (CH2Cl2/MeOH: 9/1): 0.64; 1H NMR (250 MHz, DMSO-d6): δ[ppm] 10.19 (brs, 1H, NH), 9.00 (brs, 1H, NH), 8.04 (d, J = 7.8 Hz, 1H, H6), 6.30 (d, J = 5.8 Hz 1H, H5), 5.92 (d, J = 4.3Hz, 1H, H1′), 5.50 (dd, J = 4.5 and 5.8 Hz, 1H, H2′), 5.34 (m, 1H, H3′), 4.29 (m, 3H, H4′-H5′), 2.07 (s, 9H, OAc); 13C NMR (62.9 MHz, DMSO-d6): δ[ppm] 170.0 and 169.3 (3C=O), 159.3 (C4), 146.9 (C2), 145.0 (C6), 94.8 (C5), 89.1 (C1′), 79.3 (C4′), 72.4 (C2′), 69.3 (C3′), 62.7 (C5′), 20.5–20.3–20.2 (3CH3); ESI-MS (+): calc. 405.1, found 370.1 [MCl]−.
2′,3′,5′-Tri-O-acetyl-5-iodo-cytidine (19)
Iodine (4.11 g, 14.4 mmol) was added to a suspension of 2′,3′,5′-tri-O-acetyl-cytidine, hydrochloride (10.0 g, 24 mmol) in 90 ml CH3COOH/CCl4: 1/1. After heating to 40°C, iodic acid (5.2 g, 0.9 eq) was added and the mixture stirred at 40°C for 48 h. The suspension was concentrated in vacuo and the residue taken up in 200 ml dichloromethane. The organic phase was washed with 100 ml 5% aqueous NaHCO3, dried with Na2SO4 and evaporated. A flash chromatography (CH2Cl2/MeOH: 98/2 to 90/10) yielded 10.09 g of 19 (69%). Rf (CH2Cl2/MeOH: 95/5): 0.45; 1H NMR (250 MHz, DMSO-d6): δ[ppm] 8.09 (s, 1H, H6), 8.05 (s, 1H, NH), 6.83 (s, 1H, NH), 5.82 (d, J = 4.3 Hz, 1H, H1′), 5.45 (dd, J = 4.5 and 6.3 Hz, 1H, H2′), 5.38 (m, 1H, H3′), 4.34 (m, 3H, H4′), 4.22 (m, 2H, H5′), 2.07 (s, 9H, OAc); 13C NMR (62.9 MHz, DMSO-d6): δ[ppm] 170.0 and 169.3 (3C=O), 164.9 (C4), 163.9 (C2), 148.6 (C6), 89.8 (C1′), 78.9 (C4′), 72.4 (C2′), 69.7 (C3′), 63.0 (C5′), 57.7 (C5), 20.6 and 20.3 (3CH3); ESI-MS (+): calc. 495.0, found 495.9 [MH]+.
5-Iodo-cytidine 20
About 7.61 g 2′,3′,5′-tri-O-acetyl-5-iodo-cytidine (15.4 mmol) were dissolved in 20 ml of a sodium methanolate solution (0.1 M in methanol) and stirred for 1 h at RT. The mixture was neutralized with DOWEX 50W-X8 (H+), filtered and evaporated in vacuo. Pure 5-iodo-cytidine could be obtained without any further purification in a 86% yield (4.84 g). Rf (CH2Cl2/MeOH: 8/2): 0.36; 1H NMR (250 MHz, DMSO-d6): δ[ppm] 8.43 (s, 1H, H6), 7.81 (s, 2H, NH2), 5.74 (m, 1H, H1′), 3.96 (m, 2H, H2′and H3′), 3.85 (m, 1H, H4′), 3.69 (m, 2H, H5′); 13C NMR (62.9 MHz, DMSO-d6): δ[ppm] 163.7 (C4), 154.1 (C2), 147.6 (C6), 89.4 (C1′), 84.0 (C4′), 74.3 (C2′), 68.7 (C3′), 59.8 (C5′), 56.7 (C5); ESI-MS (−): calc. 369.0, found 368.1 [M-H]−.
N,N-dimethyl-N′-formamidine-5-iodo-cytidine 21
5-Iodo-cytidine 20 (4.07 g, 11.5 mmol) was dissolved under argon in 30 ml abs dimethylformamide. N,N-dimethylformamide-dimethylacetal (8.1 ml, 5 eq) was added and the solution heated at 50°C. After completion of the reaction (∼2 h), the mixture was concentrated and purified through a flash chromatography (CH2Cl2/MeOH: 9/1). Yield: 4.44 g (95%).Rf (CH2Cl2/MeOH: 9/1): 0.33; 1H NMR (250 MHz, DMSO-d6): δ[ppm] 8.61 (s, 1H, CH), 8.58 (s, 1H, H6), 5.73 (d, 1H, H1′), 5.45–5.26–5.01 (3 brs, 3H, 3OH), 3.98 (m, 2H, H2′and H3′), 3.89 (m, 1H, H4′), 3.67 (m, 2H, H5′), 3.21 (s, 3H, CH3), 3.14 (s, 3H, CH3); 13C NMR (62.9 MHz, DMSO-d6): δ[ppm] 163.9 (C4), 158.3 (CH), 154.4 (C2), 147.7 (C6), 89.8 (C1′), 84.0 (C4′), 74.4 (C2′), 68.7 (C3′), 68.7 (C5′), 59.6 (C5), 34.9 (CH3); ESI-MS (+): calc. 424.0, found 425.0 [MH]+.
3′,5′-O-Tetraisopropyldisiloxane-N,N-dimethyl-N′-formamidine-5-iodo-cytidine 22
N,N-dimethyl-N′-formamidine-5-iodo-cytidine (1.0 g, 2.4 mmol) was dissolved in 10 ml abs pyridine and cooled at 0°C. 1,3-Dichloro-1,1,3,3-tetraisopropyldisiloxane (810 μl, 1.1 eq) was added dropwise in 10 min. After further stirring for 30 min at RT, the reaction was quenched with 0.2 ml water, the solution concentrated in vacuo and coevaporated with toluene. The residue was dissolved in 30 ml CH2Cl2 and washed with 30 ml 5% aqueous NaHCO3. After separation of the two layers, the aqueous phase was extracted once with 15 ml CH2Cl2. The combined organic phases were washed with 15 ml saturated aqueous NaCl. The separated aqueous phase was extracted with 10 ml CH2Cl2. The organic phases were combined, dried with Na2SO4, filtered and evaporated. A flash chromatography (CH2Cl2/MeOH: 97/3) followed to yield 1.11 g product (71%). Rf (CH2Cl2/MeOH: 95/5): 0.30; 1H NMR (250 MHz, CDCl3): δ[ppm] 8.76 (s, 1H, CH), 8.14 (s, 1H, H6), 5.74 (m, 1H, H1′), 4.42–4.36 (m, 1H, H2′), 4.29–4.23 (m, 2H, H3′ and H4′), 4.20–3.98 (m, 2H, H5′), 3.24 (s, 3H, CH3 formamidine), 3.21 (s, 3H, CH3 formamidine), 1.12–1.02 (m, 28H, iPr); 13C NMR (62.9 MHz, CDCl3): δ[ppm] 169.0 (C4), 160.0 (CH), 156.3 (C2), 146.8 (C6), 92.2 (C1′), 82.1 (C4′), 75.2 (C2′), 69.14 (C3′), 69.09 (C5′), 60.4 (C5), 41.4 and 35.5 (CH3 formamidine), 17.7–16.9 (CH(CH3)2), 13.4–12.6 (CH(CH3)2); ESI-MS (+): calc. 666.2, found 667.1 [MH]+; Anal. calc. for C24H43IN4O6Si2: C 43.24%, H 6.50%, N 8.40%; found: C 43.41%, H 6.70%, N 8.26%.
2′-O-bis-(Acetoxyethoxy)methylester-3′,5′-O-tetraisopropyldisiloxane-N,N-dimethyl-N′-formamidine-5-iodo-cytidine 23
A solution of 22 (967 mg, 1.45 mmol), tris(2-acetoxyethyl)orthoformate (1.08 g, 2.3 eq), 4 ml abs dichloromethane and pyridinium p-toluenesulfonate (73 mg, 0.2 eq) was stirred at RT. As soon as the reaction has progressed well (overnight after TLC analysis), 4-tert-butyldimethylsiloxy-3-penten-2-one (0.6 ml, 1.8 eq) was added, the solution stirred for 48 h at RT and finally neutralized with 105 μl TEMED (0.5 eq). Followed a direct flash chromatography (EtOAc/n-hex/MeOH/TEMED: 50/49/1/0.5 to yield 23 (1.12 g, 87%). Rf (CH2Cl2/MeOH: 95/5): 0.30; 1H NMR (250 MHz, CDCl3): δ[ppm] 8.67 (s, 1H, CH), 8.16 (s, 1H, H6), 5.81 (m, 1H, H1′), 5.74 (s, 1H, CH(ACE)), 4.20–4.12 (m, 8H, CH2(ACE)), 4.04–3.77 (m, 5H, H2′/H3′/H4′/H5′), 3.16 (s, 3H, CH3 formamidine), 3.15 (s, 3H, CH3 formamidine), 1.99 (s, 6H, CH3(ACE)), 1.04–0.91 (m, 28H, iPr); 13C NMR (62.9 MHz, CDCl3): δ[ppm] 171.0 and 171.0 (C=O), 169.1 (C4), 158.9 (CH), 155.2 (C2), 146.1 (C6), 112.1 (CH ACE), 90.2 (C1′), 81.8 (C4′), 76.6 (C2′), 69.1 (C3′), 67.7 (C5′), 63.8–63.4–62.2–60.9 (CH2 ACE), 59.2 (C5), 41.4 and 35.5 (CH3 formamidine), 21.0 and 20.9 (CH3(ACE)), 17.8–16.9 (CH(CH3)2), 13.5–12.7 (CH(CH3)2); ESI-MS (+): calc. 884.3, found 885.2 [MH]+; Anal. calc. for C33H57IN4O12Si2: C 44.79%, H 6.49%, N 6.33%; found: C 44.96%, H 6.54%, N 6.28%.
2′-O-bis-(Acetoxyethyloxy)methylester-N,N-dimethyl-N′-formamidine-5-iodo-cytidine 24
TEMED, HF was freshly and separately prepared: 0.85 ml tetramethylethylendiamine (5.7 mmol) was dissolved in 2 ml acetonitrile at 0°C and 0.14 ml hydrofluoric acid (48% in water, 4.1 mmol, 3.5 eq) was added slowly in 2 min. This mixture was stirred for 5 min at 0°C and added dropwise in 5 min at RT to a solution of 1.0 g 2′-O-ACE-3′,5′-O-TIPS-N,N-dimethyl-N′-formamidine-5-iodo-cytidine in 4 ml acetonitrile. After 2 h stirring, the solution was concentrated in vacuo, the oily residue was dissolved in 10 ml CH2Cl2, 2 ml hexane and 0.2 ml TEMED and purified through a flash chromatography (93/7/0.5: EtOAc/MeOH/TEMED). Yield: 726 mg (quantitative); Rf (CH2Cl2/MeOH: 95/5): 0.16; 1H NMR (250 MHz, CDCl3): δ[ppm] 8.65 (s, 1H, CH), 8.10 (s, 1H, H6), 5.56 (d, 1H, H1′), 5.46 (s, 1H, CH(ACE)), 4.74 (m, 1H, H2′), 4.31 (m, 1H, H3′), 4.17–4.11 (m, 4H, CH2(ACE)), 4.10 (m, 1H, H4′), 3.97–3.91 (m, 1H, H5′), 3.78–3.70 (m, 5H, H5′ and CH2(ACE)), 3.16 (2 s, 6H, CH3 formamidine), 2.00 (s, 6H, CH3(ACE)); 13C NMR (62.9 MHz, CDCl3): δ[ppm] 170.9 (C=O), 169.1 (C4), 159.8 (CH), 156.0 (C2), 149.4 (C6), 113.0 (CH ACE), 93.3 (C1′), 85.7 (C4′), 76.2 (C2′), 69.5 (C3′), 63.3–63.1–63.0–62.8 (CH2(ACE)), 62.7 (C5′), 41.7 and 35.6 (CH3 formamidine), 21.0 and 20.9 (CH3(ACE)); ESI-MS (+): calc. 642.10, found 643.0 [MH]+; Anal. calc. for C21H31IN4O11: C 39.26%, H 4.86%, N 8.72%; found: C 39.48%, H 5.09%, N 8.52%.
2′-O-bis-(Acetoxyethyloxy)methylester-5′-O-bis-(trimethylsilyloxy)benzhydryloxysilyl-N,N-dimethyl-N′-formamidine-5-iodo-cytidine 25
24 (635 mg, 0.97 mmol) and diisopropylamine (136 μl, 1 eq) were dissolved in 5 ml abs dichloromethane and cooled to 0°C. Simultaneously, 204 μl diisopropylamine were added dropwise in 1 min to a solution of benzhydryloxy-bis(trimethylsilyloxy)chlorsilane (BzHCl, 617 mg, 1.5 eq) in 2 ml abs CH2Cl2. After 5 min stirring at RT, this last solution was added dropwise and in portions to the previous one: first 0.5 eq BzHCl in 15 min, then twice 0.2 eq in 10 min and finally portions of 0.1 eq until the completion of the reaction (TLC analysis). The mixture was washed with 20 ml 8% aqueous NaHCO3 and 20 ml brine. The layers were separated and the organic phase dried with Na2SO4 and evaporated. The crude product was purified through a flash chromatography (EtOAc/n-hex/acetone/Et3N: 50/30/20/0.5 then 60/20/20/0.5), which yielded 797 mg (79%) of 21. Rf (CH2Cl2/MeOH: 95/5): 0.28; 1H NMR (250 MHz, CDCl3): δ[ppm] 8.65 (s, 1H, CH); 8.10 (s, 1H, H6); 7.05–7.33 (m, 10H, Phenyl); 5.92 (s, 1H, CH–Ph); 5.77 (m, 1H, H1′); 5.58 (s, 1H, CH(ACE)); 4.18–4.10 (m, 3H, H2′, H3′, H4′ and H5′); 4.06–4.03 (m, 1H, H5′); 3.70–3.86 (m, 8H, CH2(ACE)); 3.13 (s, 3H, CH3 formamidine); 3.07 (s, 3H, CH3 formamidine); 1.95 (2 s, 6H, CH3(ACE)); 0.03 (s, 18H, SiMe3); 13C NMR (62.9 MHz, CDCl3): δ[ppm] 169.2 (C=O), 167.3 (C4), 157.3 (CH), 153.7 (C2), 145.0 (C6), 142.5 and 142.4 (CAr), 126.5, 126.5, 125.5, 125.4, 124.9 and 124.7 (CHAr), 111.6 (CH (ACE)), 88.0 (C1′), 82.2 (C4′), 76.9 (CHAr), 75.2 (C2′), 67.6 (C3′), 66.1 (C5′), 61.5, 61.5, 61.4 and 61.4 (CH2(ACE)), 59.7 (C5), 39.7 and 33.8 (CH3 formamidine), 19.2 (CH3(ACE)), 0.0 (SiMe3); ESI-MS (+): calc. 1030.2, found 1031.2 [MH]+ and 1053.3 [MNa]+; Anal. calc. for C41H61IN6O14Si3: C 46.59%, H 5.77%, N 5.43%; found C 46.34%, H 5.94%, N 5.43%.
2′-O-bis-(Acetoxyethyloxy)methylester-3′-O-(N,N-diisopropylamino)methoxyphosphinyl-5′-O-bis-(trimethylsilyloxy)benzhydryloxysilyl-N,N-dimethyl-N′-formamidine-5-iodo-cytidine 3
About 0.8 ml of a 0.45 M tetrazole solution in acetonitrile was added to a solution of methyl-N,N,N′,N′-tetraisopropylphosphordiamidite (260 μl, 1.25 eq) in 2 ml abs dichloromethane and stirred 5 min at RT. This activated phosphorylating agent was added dropwise to a solution of 2′-O-ACE-5′-O-BzH-N,N-dimethyl-N′-formamidine-5-iodo-cytidine (741 mg, 0.72 mmol) in 2 ml abs dichloromethane, previously cooled to 0°C. After 5 min at 0°C and 10 h at RT, the reaction was quenched with 330 μl ethanol (5 eq) and the solution evaporated. A flash chromatography (n-hexane/acetone/Et3N: 55/45/0.5) led to a pure phosphoramidite. Yield 608 mg (76%). Rf (n-hexane/acetone: 55/45): 0.31; 1H NMR (250 MHz, CDCl3): δ[ppm] 8.74 (s, 2H, CH), 8.20 and 8.19 (2 s, 2H, H6), 7.25–7.40 (2 m, 20, phenyl), 6.07 (s, 2H, CH-Ph), 6.00–6.02 (m, 2H, 2H1′), 5.75 and 5.69 (2 s, 2H, CH(ACE)), 4.31–4.32 (m, 2H, H2′), 4.16–4.27 (m, 12H, 2H3′, 2H4′ and 2CH2(ACE)), 3.80–3.94 (m, 12H, 2H5′ and 2CH2(ACE)), 3.57–3.61 (m, 2H, iPr), 3.31–3.38 (m, 6H, OCH3), 3.26 (s, 6H, CH3 formamidine), 3.18 (s, 6H, CH3 formamidine), 2.06–2.07 (4 s, 12H, CH3(ACE)), 1.15–1.20 (m, 24H, iPr), 0.04–0.12 (s, 36H, SiMe3); 31P NMR (162 MHz, CDCl3): δ[ppm] 150.81 and 150.60; ESI-MS (+): calc. 1191.3, found 1192.4 [MH]+.
Oligonucleotide synthesis
The oligonucleotides were synthesized on a 1 μmol scale on a EXPEDITE synthesizer from Perseptive Biosystems, Foster City, CA, USA, with phosphoramidites purchased from Biospring, Frankfurt am Main, Germany (TBDMS chemistry) or on a 0.2 μmol scale on a rebuilt ABI 392 synthesizer (Applied Biosystems, Foster City, CA, USA) with phosphoramidites purchased from Dharmacon (ACE chemistry). Every RNA synthesis was stopped after incorporation of the iodinated phosphoramidite without deprotecting the 5′-hydroxyl group (DMTon). The column was removed from the synthesizer and maintained under argon atmosphere. In the mean time 9.5 mg copper(I) iodide were dissolved in dried and deoxygenated CH2Cl2/Et3N (1.75/0.75 ml). About 150 μl of this solution were added under argon to a mixture of PdII(PPh3)2Cl2 (2.1 mg) and TPA (2 mg). The orange solution was given into the column and moved in it back and forth using two syringes. After a reaction time of 2.5 h the column was washed with 10 ml abs CH2Cl2, dried for 10 min under vacuum and flushed with argon. The Sonogashira cross-coupling was performed twice or three times depending on the labeled base and on the chosen chemistry for the oligonucleotide synthesis. Note that the amounts of reagents were not reduced for the 0.2 μmol synthesis. Then the column was reinstalled on the synthesizer to end the synthesis of the oligonucleotide.
When using the TBDMS chemistry, the oligonucleotides were cleaved from the controlled pore glass (CPG) and the amino groups deprotected with a mixture of ammonia (32%)/MeOH (3/1) over 24 h. The TBDMS groups were cleaved with triethylamine, HF over 24 h. After precipitation in abs. ethanol (−20°C over night), the RNA strands were purified via anion-exchange chromatography (Dionex NucleoPacTM PA 100 column, 250 × 9 mm, flow 5 ml min−1) on a JASCO-HPLC.
When using the ACE chemistry, the methyl group on the phosphate was first cleaved on-column with a 0.4 M solution of disodium-2-carbamoyl-2-cyanoethylene-1,1-dithiolate-trihydrate (S2Na2) in DMF/H2O: 98/2 in 30 min. Then, the oligonucleotides were cleaved from the solid support and deprotected with methylamine (40% in water): 10 min at 55°C for the unmodified RNA-strands and 12 h at RT for the spin labeled ones. Followed a purification through anion-exchange HPLC.
The oligonucleotides were desalted with PD-10 Sephadex columns from Amersham Biosciences, Piscataway, NJ, USA and finally characterized with a MALDI-Tof VOYAGER DE-PRO mass spectrometer from Applied Biosystems. In case of the ACE chemistry the final deprotection of the 2′-ACE groups was performed under sterile conditions with a TEMED-acetic acid buffer pH 3.8, 30 min at 60°C for all the RNA strands.
Analysis
Calf intestine alkaline phosphatase (Sigma-Aldrich, St. Louis, MO, USA) and Penicillium citrinum nuclease P1 (Roche, Nutley, NJ, USA) were used for the enzymatic digestion of ODNs (see (33) for the procedure and supplementary data for HPLC diagram). UV-melting curves (Tm) of the duplexes, dissolved in phosphate buffer (10 mM Na2HPO4, 10 mM NaH2PO4, 140 mM NaCl, 2.5 μM duplex, pH 7), were recorded on a Cary UV-vis spectrophotometer equipped with a Peltier thermostat from Varian. The UV absorption was measured at a wavelength of 260 nm, while the temperature was increased with a heating rate of 0.5°C min−1. CD spectra were measured at a temperature of 20 C between 350 and 200 nm on a JASCO J-710 spectropolarimeter with a Peltier thermostat. The duplexes were dissolved in the same buffer and at the same concentration as for the Tm measurements.
PELDOR
All EPR samples had a volume of 100 μl and contained 0.1 mM duplex in phosphate buffer (140 mM NaCl, 10 mM Na2HPO4, 10 mM NaH2PO4, 20% ethylene glycol, pH 7). The solutions were transferred into sterile standard quartz EPR tubes and shock frozen in liquid nitrogen. The 4-Pulse ELDOR experiments were performed on an ELEXSYS E580 pulsed X-band EPR spectrometer using a flex line probehead housing a dielectric ring resonator all from Bruker, Fällanden, Switzerland. The temperature was adjusted with a temperature control system (ITC) in combination with a helium cryostat both from Oxford. For the PELDOR measurements a second microwave source was coupled into the microwave bridge using a commercially available set-up from Bruker. The pumping pulse was applied at the resonance frequency ν0 = νB of the resonator and the detection pulses at a frequency νA, 80 MHz higher than νB. The resonator used exhibits in overcoupled conditions a resonance frequency ν0 of 9.7 GHz, a quality factor Q of about 100, a conversion factor κ of 4 μT/W and a bandwidth of 97 MHz. Accordingly, even with a frequency offset of 80 MHz between νA = ν0 and νB both pulses are still within the bandwidth of the resonator. The pulse lengths used for the detection pulses were 32 ns and for the pumping pulse 12 ns. The amplitude of the detection pulses was chosen to optimize the refocused echo (global power 1 kW, attenuation in the +x-channel 8.0 (π/2-pulse), attenuation in the −x-channel 8.1 (π-pulses)). The amplitude of the inversion pulse with frequency νB was adjusted to a π-pulse using the pulse sequence π(νB-second source)-T-π/2(νB-main source)-τ-π(νB-main source) as far as possible, which is usually at about 8 dB of the second source. B0 was set to a field value so that the detection pulses excited the low field site of the nitroxide field sweep spectrum, whereas the inversion pulse selected the central mI = 0 transition of gzz together with the mI = 0, ± 1 transitions of gxx and gyy. The spectra were recorded at 40 K with an experiment repetition time of 14 ms, a video amplifier bandwidth of 25 MHz, a video amplifier gain of 63 dB and a 2-step phase-cycle. Usually 800 scans and 10 shots per point were accumulated. The inversion pulse was stepped in increments of 20 ns leading to a total of 166 points.
Tikhonov regularizations of the acquired time traces were performed with the program ‘DEER 2006’ from G. Jeschke available at http://www.mpip-mainz.mpg.de/~jeschke/distance.html). An exponential background fitted to the last 2/3 of the time trace was subtracted prior to the regularization. The regularization parameter was for all regularizations between 1 and 10.
Molecular dynamics simulations
All MD simulations were performed with the GROMACS (64) program package. The AMBER98 force field (65) was employed, which was implemented by us in GROMACS. To specify the potential-energy function for the TPA spin-label, density functional theory calculations at the B3LYP/6-31 + G(d) level were performed, using Gaussian program (66). All non-standard force-field parameters of TPA (in particular, partial charges, bond lengths and bond angles) were then derived employing the AMBER strategy of force-field development (67). Force-field parameters will be provided upon request, see (68) for a similar study. In each simulation, the RNA was solvated in a rectangular box of TIP3P water, keeping a minimum distance of 10 Å between the solute and each face of the box. To neutralize the system, sodium counterions were added and water molecules were removed if they overlapped with the sodium ions. The largest simulation (RNA 5) contained 32 823 atoms in a box of the dimension 72 × 66 × 69 Å3. The equation of motion was integrated by using a leapfrog algorithm with a time step of 2 fs. Covalent bond lengths involving hydrogen atoms were constrained by the SHAKE algorithm with a relative geometric tolerance of 0.0001. A cut off of 10 Å was used for the non-bonding van der Waals interactions, and the non-bonded interaction pair-list was updated every 10 fs. Periodic boundary conditions were applied, and the particle mesh Ewald method was used to treat electrostatic interactions. The solute and solvent were separately weakly coupled to external temperature baths at 300 K with a temperature-coupling constant of 0.5 ps (0.01 during the first 100 ps). The total system was also weakly coupled to an external pressure bath at 1 atm using a coupling constant of 5 ps. All systems were minimized and equilibrated with the same protocol, using the program MDRUN in double precision. Assuming that the RNAs are initially in ideal A-form, the whole system was first minimized for 1000 steps. A 100 ps MD run of the water molecules and counter ions with fixed solute was then performed, followed by a 100 ps MD run without position constrains of the solute. The simulation was then continued for 50 ns, where the coordinates were saved every picosecond for analysis. One nanosecond of simulation time required about 12 h using four CPUs on a dual-core Opteron Linux cluster. The relatively long simulation time is necessary to account for the opening and closing of base-pair hydrogen bonds, which occurs on a 10 ns time scale (see Figure 6).
SUPPLEMENTARY DATA
Details about the oligonucleotide analyses (enzymatic digestion, Tm-values, CD) and PELDOR spectra for RNA 1-6. This material is available free of charge via the Internet at NAR online.
Funding to pay the Open Access publication charges for this article was provided by Deutsche Forschungsgemeinschaft (SFB 579)
Conflicts of statement. None declared.
ACKNOWLEDGEMENTS
This work was supported by the SFB 579 RNA-ligand-interactions. Y.M. acknowledges support from a Singapore Ministry of Education (MoE), University Research Committee (URC, RG65/06) grant, and G. S. gratefully acknowledge support by the Frankfurt Center for Scientific Computing and the Fonds der Chemischen Industrie.
REFERENCES
- 1.Denli A.M., Hannon G.J. RNAi: an ever-growing puzzle. Trends Biochem. Sci. 2003;28:196–201. doi: 10.1016/S0968-0004(03)00058-6. [DOI] [PubMed] [Google Scholar]
- 2.Voinnet O. RNA silencing as a plant immune system against viruses. Trends Genet. 2001;17:449–459. doi: 10.1016/s0168-9525(01)02367-8. [DOI] [PubMed] [Google Scholar]
- 3.Winkler W.C., Breaker R.R. Genetic control by metabolite-binding riboswitches. ChemBioChem. 2003;4:1024–1032. doi: 10.1002/cbic.200300685. [DOI] [PubMed] [Google Scholar]
- 4.Ubbink M., Worrall J.A.R., Canters G.W., Groenen E.J.J., Huber M. Paramagnetic resonance of biological metal centers. Annu. Rev. Biophys. Biomol. Struc. 2002;31:393–422. doi: 10.1146/annurev.biophys.31.091701.171000. [DOI] [PubMed] [Google Scholar]
- 5.Prisner T.F., Rohrer M., MacMillan F. Pulsed EPR spectroscopy: biological applications. Annu. Rev. Phys. Chem. 2001;52:279–313. doi: 10.1146/annurev.physchem.52.1.279. [DOI] [PubMed] [Google Scholar]
- 6.Jeschke G. EPR techniques for studying radical enzymes. Biochim. Biophys. Acta Bioenerg. 2005;1707:91–102. doi: 10.1016/j.bbabio.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 7.Lubitz W. EPR in photosynthesis. In: Murphy DM, editor. Electron Paramagnetic Resonance. Vol. 19. London: Royal Society of Chemistry; 2004. pp. 174–242. [Google Scholar]
- 8.Klare J.P., Gordeliy V.I., Labahn J., Büldt G., Steinhoff H.-J., Engelhard M. The archaeal sensory rhodopsin II/transducer complex: a model for transmembrane signal transfer. FEBS Lett. 2004;564:219–224. doi: 10.1016/S0014-5793(04)00193-0. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y.-S., Sompornpisut P., Perozo E. Structure of the KcsA channel intracellular gate in the open state. Nature Struct. Biol. 2001;8:883–887. doi: 10.1038/nsb1001-883. [DOI] [PubMed] [Google Scholar]
- 10.Park S.Y., Borbat P.P., Gonzalez-Bonet G., Bhatnagar J., Pollard A.M., Freed J.H., Bilwes A.M., Crane B.R. Reconstruction of the chemotaxis receptor-kinase assembly. Nat. Struc. Mol. Biol. 2006;13:400–407. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- 11.Borbat P.P., Mchaourab H.S., Freed J.H. Protein structure determination using long-distance constraints from double-quantum coherence ESR: study of T4 lysozyme. J. Am. Chem. Soc. 2002;124:5304–5314. doi: 10.1021/ja020040y. [DOI] [PubMed] [Google Scholar]
- 12.Hustedt E.J., Beth A.H. Structural information from CW-EPR spectra of dipolar coupled nitroxide spin labels. In: Berliner LJ, Eaton SS, Eaton GR, editors. Biological Magnetic Resonance. Vol. 19. New York: Kluwer Academic/Plenum Publisher; 2000. p. 155. [Google Scholar]
- 13.Hubbel W.L., Cafiso D.S., Altenbach C. Identifying conformational changes with site-directed spin labeling. Nature Struct. Biol. 2000;7:735. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 14.Kisseleva N., Khvorova A., Westhof E., Schiemann O. Binding of manganese(II) to a tertiary stabilized hammerhead ribozyme as studied by electron paramagnetic resonance spectroscopy. RNA. 2005;11:1–6. doi: 10.1261/rna.7127105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vogt M., DeRose V.J. Coordination environment of a site-bound metal ion in the hammerhead ribozyme determined by 15N and 2H ESEEM spectroscopy. J. Am. Chem. Soc. 2006;128:16764–16770. doi: 10.1021/ja057035p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim N.-K., Murali A., DeRose V.J. A distance ruler for RNA using EPR and site-directed spin labeling. Chem. Biol. 2004;11:939–948. doi: 10.1016/j.chembiol.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 17.Macosko J.C., Pio M.S., Tinoco I., Shin Y.K. A novel 5 displacement spin-labeling technique for electron paramagnetic resonance spectroscopy of RNA. RNA. 1999;5:1158–1166. doi: 10.1017/s1355838299990830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin P.Z., Dieckmann T. Application of NMR and EPR methods to the study of RNA. Curr. Opin. Struct. Biol. 2004;14:350–359. doi: 10.1016/j.sbi.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Qin P.Z., Butcher S.E., Feigon J., Hubbell W.L. Quantitative analysis of the isolated GAAA tetraloop/receptor interaction in solution: a site-directed spin labeling study. Biochemistry. 2001;40:6929–6936. doi: 10.1021/bi010294g. [DOI] [PubMed] [Google Scholar]
- 20.Edwards T.E., Okonogi T.M., Robinson B.H., Sigurdsson S.T. Site-specific incorporation of nitroxide spin-labels into internal sites of the TAR RNA; structure-dependent dynamics of RNA by EPR spectroscopy. J. Am. Chem. Soc. 2001;123:1527–1528. doi: 10.1021/ja005649i. [DOI] [PubMed] [Google Scholar]
- 21.Piton N., Schiemann O., Mu Y., Stock G., Prisner T.F., Engels J.W. Synthesis of spin labeled RNAs for long range distance measurements by PELDOR. Nucleosides Nucleotides Nucleic Acids. 2005;24:771–775. doi: 10.1081/ncn-200060139. [DOI] [PubMed] [Google Scholar]
- 22.Qin P.Z., Hideg K., Feigon J., Hubbell W.L. Monitoring RNA base structure and dynamics using site-directed spin labeling. Biochemistry. 2003;42:6772–6783. doi: 10.1021/bi027222p. [DOI] [PubMed] [Google Scholar]
- 23.Ramos A., Varani G. A new method to detect long-range protein-RNA contacts: NMR detection of electron-proton relaxation induced by nitroxide spin-labeled RNA. J. Am. Chem. Soc. 1998;120:10992–10993. [Google Scholar]
- 24.Milov A.D., Salikhov K.M., Shchirov M.D. Application of the double resonance method to electron spin echo in a study of the spatial distribution of paramagnetic centers in solids. Soviet. Phys. Sol. Stat. 1981;23:565–569. [Google Scholar]
- 25.Larsen R.G., Singel D.J. Double electron-electron resonance spin-echo modulation: spectroscopic measurement of electron spin pair separations in orientationally disordered solids. J. Chem. Phys. 1993;98:5134–5146. [Google Scholar]
- 26.Martin R.E., Pannier M., Diederich F., Gramlich V., Hubrich M., Spiess H.W. Determination of end-to-end distances in a series of TEMPO diradicals of up to 2.8 nm length with a new four-pulse double electron electron resonance experiment. Angew. Chem. Int. Ed. 1998;37:2834–2837. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2833::AID-ANIE2833>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 27.Borbat P.P., Freed J.H. Multiple-quantum ESR and distance measurements. Chem. Phys. Lett. 1999;313:145–154. [Google Scholar]
- 28.Jeschke G., Bender A., Paulsen H., Zimmermann H., Godt A. Sensivity enhancement in pulse EPR distance measurements. J. Magn. Reson. 2004;169:1–12. doi: 10.1016/j.jmr.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 29.Berliner LJ, Eaton SS, Eaton GR, editors. Biological Magnetic Resonance. Vol. 19. New York: Kluwer Academic/Plenum Publisher; 2000. [Google Scholar]
- 30.Bowman M.K., Maryasov A.G., Kim N., DeRose V.J. Visualization of distance distribution from pulsed double electron-electron resonance data. Appl. Magn. Reson. 2004;26:23–39. [Google Scholar]
- 31.Schiemann O., Weber A., Edwards T.E., Prisner T.F., Sigurdsson S.T. Nanometer distance measurements on RNA Using PELDOR. J. Am. Chem. Soc. 2003;125:3434–3435. doi: 10.1021/ja0274610. [DOI] [PubMed] [Google Scholar]
- 32.Borbat P.P., Davis J.H., Butcher S.E., Freed J.H. Measurement of large distances in biomolecules using double-quantum filtered refocused electron spin-echoes. J. Am. Chem. Soc. 2004;126:7746–7747. doi: 10.1021/ja049372o. [DOI] [PubMed] [Google Scholar]
- 33.Schiemann O., Piton N., Mu Y., Stock G., Engels J.W., Prisner T. A PELDOR-based nanometer distance ruler for oligonucleotides. J. Am. Chem. Soc. 2004;126(18):5722–5729. doi: 10.1021/ja0393877. [DOI] [PubMed] [Google Scholar]
- 34.Rist M., Amann N., Wagenknecht H.-A. Preparation of 1-Ethynylpyrene-modified DNA via sonogashira-type solid-phase couplings and characterization of the fluorescence properties for electron-transfer studies. Eur. J. Org. Chem. . 2003:2498–2504. [Google Scholar]
- 35.Malakhov A.D., Skorobogatyi M.V., Prokhorenko I.A., Gontarev S.V., Kozhich D.T., Stetsenko D.A., Stepanova I.A., Shenkarev Z.O., Berlin Y.A., Korshun V.A. 1-(Phenylethynyl)pyrene and 9,10-Bis(phenylethynyl)anthracene, useful fluorescent dyes for DNA labeling: excimer formation and energy transfer. Eur. J. Org. Chem. . 2004:1298–1307. [Google Scholar]
- 36.Hofmann T., Zweig K., Engels J.W. A new synthetic approach for the synthesis of N2-modified guanosines. Synthesis. 2005;11:1797–1800. [Google Scholar]
- 37.Beaucage S.L., Iyer R.P. The synthesis of modified oligonucleotides by the phosphoramidite approach and their applications. Tetrahedron. 1993;49:6123–6194. [Google Scholar]
- 38.Ogilvie K.K., Beaucage S.L., Schifman A.L., Theriault N.Y., Sadana K.L. The synthesis of oligoribonucleotides. II. The use of silyl protecting groups in nucleoside and nucleotide chemistry. VII. Can. J. Chem. 1978;56:2768–2780. [Google Scholar]
- 39.Beránek J., Drašar P. Selective O-acylation. Preparation of 1-(2,3,5-Tri-O-acetyl-beta-D-arabinopentofuranosyl)cytosine hydrochloride. Analogues of nucleosides XII. Coll. Czech. Chem. Comm. 1977;42:366–369. [Google Scholar]
- 40.Bobek M., Kavai I., Sharma R.A., Grill S., Dutschman, Cheng Y.C. Acetylenic nucleosides. 4. 1-(beta-D-Arabinofuranosyl)-5-ethynylcytosine. Improved synthesis and evaluation of biochemical and antiviral properties. J. Med. Chem. 1987;30:2154–2157. doi: 10.1021/jm00394a039. [DOI] [PubMed] [Google Scholar]
- 41.Robins M.J., Uznánski B. Nucleic acid related compounds. 33. Conversions of adenosine and guanosine to 2,6-dichloro, 2-amino-6-chloro, and derived purine nucleosides. Can. J. Chem. 1981;59:2601–2607. [Google Scholar]
- 42.Matsuda A., Shinozaki T., Yamaguchi T., Homma H., Nomoto R., Miyasaka T., Watanabe Y., Abiru T. Nucleosides and nucleotides. 103. 2-Alkynyladenosines: a novel class of selective adenosine A2 receptor agonists with potent antihypertensive effects. J. Med. Chem. 1992;35:241–252. doi: 10.1021/jm00080a007. [DOI] [PubMed] [Google Scholar]
- 43.Nair V., Young D.A. A new synthesis of isoguanosine. J. Org. Chem. 1985;50:406–408. [Google Scholar]
- 44.Seela F., Hersmann K., Grasby J.A., Gait M.J. 7-Deazaadenosine: oligoribonucleotide building block synthesis and autocatalytic hydrolysis of base-modified hammerhead ribozymes. Helv. Chim. Acta. 1993;76:1809–1819. [Google Scholar]
- 45.Gannett P.M., Darian E., Powell J., Johnson E.M. II, Mundoma C., Greenbaum N.L., Ramsey C.M., Dalal N.S., Budil D.E. Probing triplex formation by EPR spectroscopy using a newly synthesized spin label for oligonucleotides. Nucleic Acids Res. 2002;30:5328–5337. doi: 10.1093/nar/gkf634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rozantsev E.G., Sholle V.D. Synthesis and reactions of stable nitroxyl radicals II. Reactions. Synthesis. 1971:, 401–414. [Google Scholar]
- 47.Scaringe SA. 2000. Orthoester protecting groups. Patent US6111086. [Google Scholar]
- 48.Scaringe S.A. RNA oligonucleotide synthesis via 5′-silyl-2′-orthoester chemistry. Methods. 2001;23:206–217. doi: 10.1006/meth.2000.1132. [DOI] [PubMed] [Google Scholar]
- 49.Scaringe S.A., Wincott F.E., Caruthers M.H. Novel RNA synthesis method using 5′-O-silyl-2′-O-orthoester protecting groups. J. Am. Chem. Soc. 1998;120:11820–11821. [Google Scholar]
- 50.Scaringe S.A., Kitchen D., Kaiser R., Marshall W.S. Preparation of 5′-silyl-2′-orthoester ribonucleosides for use in oligoribonucleotide synthesis. Curr. Prot. Nucl. Acid Chem., . 2004:2.10.1–2.10.15. doi: 10.1002/0471142700.nc0210s16. [DOI] [PubMed] [Google Scholar]
- 51.Markiewicz W.T. Tetraisopropyldisiloxane-1,3-diyl, a group for simultaneous protection of 3′- and 5′-hydroxy functions of nucleosides. J. Chem. Res. Synop. 1979:, 181–197. [Google Scholar]
- 52.Chiang Y.-W., Borbat P.P., Freed J.H. The determination of pair distance distributions by pulsed EPR using Tikhonov regularization. J. Magn. Reson. 2005;172:279–295. doi: 10.1016/j.jmr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 53.Chiang Y.-W., Borbat P.P., Freed J.H. Maximum entropy: a complement to Tikhonov regularization for determination of pair distance distributions by pulsed EPR. J. Magn. Reson. 2005;177:184–196. doi: 10.1016/j.jmr.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 54.Jeschke G., Panek G., Godt A., Bender A., Paulsen H. Data analysis procedures for pulse ELDOR measurements of broad distance distributions. Appl. Magn. Reson. 2004;26:223–244. [Google Scholar]
- 55.Cai Q., Kusnetzow A.K., Hubbell W.L., Haworth I.S., Gacho G.P.C., Eps N.V., Hideg K., Chambers E.J., Qin P.Z. Site-directed spin labeling measurements of nanometer distances in nucleic acids using a sequence-independet nitroxide probe. Nucleic Acids Res. 2006;34:4722–4730. doi: 10.1093/nar/gkl546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Batey R.T., Rambo R.P., Doudna J.A. Tertiary motifs in RNA structure and folding. Angew. Chem. Int. Ed. 1999;38:2326–2343. doi: 10.1002/(sici)1521-3773(19990816)38:16<2326::aid-anie2326>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 57.Auffinger P., Westhof E. RNA solvation: a molecular dynamics simulation perspective. Biopolymers. 2000;56:266–274. doi: 10.1002/1097-0282(2000)56:4<266::AID-BIP10027>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 58.Beveridge D.L., McConnell K.J., Nucleic acids: theory and computer simulation, Y2K. Curr. Opin. Struct. Biol. 2000;10:182–196. doi: 10.1016/s0959-440x(00)00076-2. [DOI] [PubMed] [Google Scholar]
- 59.Cheatham T.E., III Simulation and modeling of nucleic acid structure, dynamics and interactions. Curr. Opin. Struct. Biol. 2004;14:360–367. doi: 10.1016/j.sbi.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 60.Norberg J., Nilsson L. Molecular dynamics applied to nucleic acids. Acc. Chem. Res. 2002;35:465–472. doi: 10.1021/ar010026a. [DOI] [PubMed] [Google Scholar]
- 61.Williams D.J., Hall K.B. Experimental and theoretical studies of the effects of deoxyribose substitutions on the stability of the UUCG tetraloop. J. Mol. Biol. 2000;297:251–265. doi: 10.1006/jmbi.2000.3547. [DOI] [PubMed] [Google Scholar]
- 62.Zacharias M. Simulation of the structure and dynamics of nonhelical RNA motifs. Curr. Opin. Struct. Biol. 2000;10:311–317. doi: 10.1016/s0959-440x(00)00089-0. [DOI] [PubMed] [Google Scholar]
- 63.Lu X.-J., Olson W. K. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lindahl E., Hess B., van der Spoel D. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Mod. 2001;7:306. [Google Scholar]
- 65.Cheatham T.E., Cieplak P., Kollman P.A. A modified version of the Cornell et al. force field with improved sugar pucker phases and helical repeat. J. Biomol. Struct. Dyn. 1999;16:845. doi: 10.1080/07391102.1999.10508297. [DOI] [PubMed] [Google Scholar]
- 66.Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman, J.R. Montgomery J.A., Vreven T., Kudin K.N., Burant J.C., et al. Gaussian03. Wallingford CT: Gaussian Inc.; 2004. [Google Scholar]
- 67.Case D.A., Pearlman D.A., Caldwell .J.W., Cheatham T.E. III, Ross W.S., Simmerling C.L., Darden T.A., Merz K.M., Stanton R.V., Cheng A.L., et al. AMBER 6. San Francisco, CA: University of California; 1999. [Google Scholar]
- 68.Darian E, Gannett P.M. Application of molecular dynamics simulations to spin-labeled oligonucleotides. J. Biomolec. Struct. Dyn. 2005;22:579. doi: 10.1080/07391102.2005.10507028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.