Abstract
We have identified three C/D-box small nucleolar RNAs (snoRNAs) and one H/ACA-box snoRNA in mouse and human. In mice, all four snoRNAs (MBII-13, MBII-52, MBII-85, and MBI-36) are exclusively expressed in the brain, unlike all other known snoRNAs. Two of the human RNA orthologues (HBII-52 and HBI-36) share this expression pattern, and the remainder, HBII-13 and HBII-85, are prevalently expressed in that tissue. In mice and humans, the brain-specific H/ACA box snoRNA (MBI-36 and HBI-36, respectively) is intron-encoded in the brain-specific serotonin 2C receptor gene. The three human C/D box snoRNAs map to chromosome 15q11–q13, within a region implicated in the Prader–Willi syndrome (PWS), which is a neurogenetic disease resulting from a deficiency of paternal gene expression. Unlike other C/D box snoRNAs, two snoRNAs, HBII-52 and HBII-85, are encoded in a tandemly repeated array of 47 or 24 units, respectively. In mouse the homologue of HBII-52 is processed from intronic portions of the tandem repeats. Interestingly, these snoRNAs were absent from the cortex of a patient with PWS and from a PWS mouse model, demonstrating their paternal imprinting status and pointing to their potential role in the etiology of PWS. Despite displaying hallmarks of the two families of ubiquitous snoRNAs that guide 2′-O-ribose methylation and pseudouridylation of rRNA, respectively, they lack any telltale rRNA complementarity. Instead, brain-specific C/D box snoRNA HBII-52 has an 18-nt phylogenetically conserved complementarity to a critical segment of serotonin 2C receptor mRNA, pointing to a potential role in the processing of this mRNA.
The biogenesis of eukaryotic ribosomes involves a complex rRNA processing pathway mostly taking place in a specialized subnuclear compartment, the nucleolus. Pre-rRNA maturation includes, in addition to a series of endonucleolytic and exonucleolytic cleavages, the covalent modification of a definite subset of rRNA nucleotides, essentially by 2′-O-ribose methylation and pseudouridylation. Each of these modifications is found at about 100 sites per vertebrate ribosome (1). Although these modifications are phylogenetically conserved and restricted to the most highly conserved and functionally important regions of rRNA, their function remains largely unknown. Spliceosomal small nuclear RNAs (snRNAs) also contain a number of conserved 2′-O-methylated nucleotides and pseudouridines, confined to snRNA sequences critical for splicing, i.e., involved in contacts with pre-mRNA or other snRNAs (2). Remarkably, nucleotide modifications in the 5′ terminal region of U2 snRNA are required for assembly of a functional U2 sn-ribonucleoprotein particle (3).
The nucleolus contains a large number of small, metabolically stable RNAs, termed small nucleolar RNAs (snoRNAs) that fall into two major classes, the C/D box and H/ACA box snoRNAs, designated after common sequence motifs involved in the assembly of sno-ribonucleoprotein particles. Although each class includes a small number of snoRNAs required for definite pre-rRNA cleavages and essential for viability, the vast majority of these snoRNAs function in the posttranscriptional modification of rRNA nucleotides and are dispensable for growth. It is well documented that each 2′-O-ribose methylation or pseudouridylation in eukaryotic rRNA is selected by a cognate snoRNA belonging to either the C/D box or H/ACA box family, respectively, in both cases through formation of specific base pairing that spans the rRNA modification site (4–8). C/D box snoRNAs contain two short conserved sequence motifs, box C (5′-RUGAUGA-3′, where R is any purine) and box D (5′-CUGA-3′), always located only a few nucleotides away from the 5′ and 3′ ends, respectively, and generally brought into close proximity by base pairing of the four or five terminal nucleotides. This characteristic 5′–3′ terminal stem–box C/D structure plays a critical role in the control of snoRNA biogenesis and nucleolar localization. C/D box snoRNAs also contain, immediately upstream from box D or from another CUGA motif (box D′) in their 5′ half, 10- to 21-nt regions complementary to rRNA, thereby spanning sites of 2′-O-methylation. In the corresponding snoRNA⋅rRNA duplex, the 2′-O-ribose methylation is directed to the rRNA nucleotide paired to the fifth snoRNA nucleotide upstream from box D or box D′. C/D box snoRNAs, which also exhibit in the central region of their sequence a C′ box with up to two deviations from the consensus C box, are all immunoprecipitated by antibodies to Nop1/fibrillarin, a nucleolar protein likely to correspond to the common rRNA 2′-O-methylase (9). Recently, the range of action of C/D box methylation guide snoRNAs has been found to go much beyond the field of ribosome biogenesis, with the detection of several specimens predicted to direct 2′-O-ribose methylations in U6 snRNA, through the same kind of RNA duplex, and the function of one of them was demonstrated in Xenopus oocytes (10, 11). In vertebrates, known C/D or H/ACA box snoRNAs are ubiquitously expressed: they are encoded within introns of housekeeping genes, belonging to the 5′ terminal oligopyrimidine (5′TOP) class of genes (12, 13) and produced by processing of the pre-mRNA intron (14–17). In this study, we report the detection of tissue-specific snoRNAs clearly belonging to the two major classes of modification-guide snoRNAs. Remarkably, these specimens, which also display unique genomic organization and expression, are devoid of rRNA or snRNA complementarity and might target other cellular RNAs. Indeed, indirect evidence points to the possibility that one of the brain-specific snoRNAs targets a brain-specific mRNA for 2′-O-ribose methylation.
Materials and Methods
Construction of a cDNA Library Encoding Small Non-mRNAs from Mouse Brain.
We prepared total RNA from mouse brain by the TRIzol method (GIBCO/BRL) and fractionated RNAs on a denaturing 8% polyacrylamide gel [7 M urea/1× TBE buffer (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3)]. We excised gel bands containing RNAs of 50–500 nt, passively eluted the RNAs, and then ethanol-precipitated the RNAs. Subsequently, we C-tailed RNAs by using CTP and poly(A) polymerase. We reverse-transcribed RNAs into cDNAs and cloned cDNAs into pSPORT-1 vector by using the GIBCO/BRL system. We analyzed cloned cDNAs by cycle sequencing on the Applied Biosystems Prism 377 (Perkin–Elmer). Novel RNA sequences were confirmed by BLAST searches. Subsequently, in an initial screen for tissue specificity, we performed Northern blot analysis (see below) on brain, liver, and testis tissue extracts.
Oligonucleotides.
We selected all primers with the LASERGENE primer selection program. Oligonucleotides were synthesized by MWG Biotech (Ebersberg, Germany) or Y. De Preval (Laboratoire de Biologie Moléculaire Eukaryote, Toulouse, France). For Northern blot hybridization to detect MBII or HBII snoRNAs, 5.8S rRNA, and U21 and U40 RNAs (as controls), we used the following probes: MBII-13, 5′-CTTCAGAGTAATCATTTTGAGCATCA-3′; MBII-52, 5′-CCTCAGCGTAATCCTATTGAGCATGAA-3′; MBII-85, 5′-TTCCGATGAGAGTGGCGGTACAGA-3′; MBI-36, 5′-TGTACCCACCTGCATGCGGGAGTAGCCCAC-3′; HBII-13, 5′-AGTAATCACGTTGAGCTTTACTTCATCAT-3′; HBII-52, 5′-CCTCAGCGTAATCCTATTGAGCATGA-3′; HBII-85, 5′-AGAGTTTTCACTCATTTTGTTCAGC-3′; HBI-36, 5′-TATGTACCCAGCTGCATGCAGGAGTAG-3′; U21, 5′-TGCCATCAGTCCCGTCTTGAAAC-3′; U40, 5′-AGTCAGACATTGATACTGATTCAGG-3′; and 5.8S rRNA, 5′-TCCTGCAATTCACATTAATTCTCGCAGCTAGC-3′.
For Southern blot hybridization, in addition to HBII/MBII probes (see above), we used probe IPW1, 5′-CCTGCAGATAAGAAATGCTTAAGTGAG-3′, for detection of the human IPW gene.
Northern Blot Analysis.
We separated total RNAs from various tissues on 8% denaturing polyacrylamide gels (7 M urea/1× TBE buffer) or 1.2% agarose/formaldehyde gels and transferred RNAs onto nylon membranes (Qiabrane Nylon Plus, Qiagen Hilden, Germany) by using the Bio-Rad semidry blotting apparatus (Trans-blot SD, Bio-Rad). After immobilizing of RNAs using the Stratagene cross-linker, we prehybridized nylon membranes for 15 min in 1 M sodium phosphate buffer, pH 6.2/7% SDS. Oligonucleotides complementary to respective RNA species were end-labeled with [32P]ATP and T4 polynucleotide kinase and hybridization was carried out at 58°C in 1 M sodium phosphate buffer, pH 6.2/7% SDS for 12 h. We washed blots twice at room temperature for 15 min in 2× SSPE buffer (20 mM sodium phosphate, pH 7.4/0.3 M NaCl/2 mM EDTA)/0.1% SDS and then for 1 min at 58°C in 0.1× SSPE/0.5% SDS. Membranes were exposed to Kodak MS-1 film for 1–12 h for autoradiography.
Southern Blot Analysis.
We digested aliquots (2 μg) of whole-blood DNA and genomic clones [yeast artificial chromosome and bacteriophage P1-derived artificial chromosome (PAC) clones] with EcoRI or PstI. We separated the fragments by gel electrophoresis, transferred the DNA to Biodyne A nylon membranes (Pall) or GeneScreen Plus membranes (NEN), and hybridized the blots with radioactively labeled probes. We used primer IPW1 (see above) for detection of the IPW gene and oligonucleotides MBII 52 or HBII 52 and MBII 85 or HBII 85 (see above) for detection of snoRNA genes.
Sources of Genomic Clones.
We isolated PACs from the mouse library RCPI 21 (Roswell Park Cancer Institute, Buffalo, NY, generated by Kazutoyo Osoegawa and Pieter de Jong) or from the human library RCPI 6 (Roswell Park Cancer Institute generated by Pieter de Jong). We obtained filters and clones from the Resource Centre of the German Human Genome Project at the Max-Planck-Institute for Molecular Genetics in Berlin.
Filter Hybridization and Isolation of Clones.
We end-labeled oligonucleotides as described above and subsequently hybridized oligonucleotides to DNA arrays of mouse or human PACs spotted on filters (see above). We performed hybridizations in 0.5 M sodium phosphate, pH 7.2/7% SDS/1 mM EDTA at 53°C for 12 h. We washed filters twice at room temperature for 15 min in 40 mM sodium phosphate buffer, pH 7.2/0.1% SDS and exposed filters to Kodak MS-1 film overnight.
DNA Sequencing and Sequence Analysis.
We sequenced cDNA clones encoding small non-mRNAs or subclones of PACs by using appropriate primers and the BigDye terminator cycle sequencing reaction kit (Applied Biosystems). We analyzed sequences on an Applied Biosystems Prism 377 sequencer using the LASERGENE sequence analysis program package. We sequenced PAC clones A17157 (insert size, 160 kb) and P0950 (insert size, 75 kb) in collaboration with MWG Biotech (Ebersberg, Germany).
Processing of MBII-52 snoRNA from Intronic Sequences.
Transfection of the construct pCMV/MBII-52 into mouse cell line L929 and processing assays were as described (7).
Immunoprecipitation.
We performed immunoprecipitation of U2, U3, MBII-13, MBII-52, and MBII-85 RNAs as described (18).
Results and Discussion
In the course of a systematic search for small non-mRNAs that could be specifically expressed in brain, we have isolated several clones of four types, MBI-36, MBII-13, MBII-52, and MBII-85, from a mouse adult brain cDNA library. The latter two are by far the most abundant in this library, amounting to 37 and 56 clones, respectively, of the 2,000 clones analyzed. The four clone types exhibit structural hallmarks of either H/ACA (MBI-36) or C/D box (MBII-13, MBII-52, and MBII-85) snoRNAs (Fig. 1A). However, none of these snoRNA-like sequences contains at the expected location an antisense element that could direct a pseudouridylation or ribose methylation in rRNA or snRNAs.
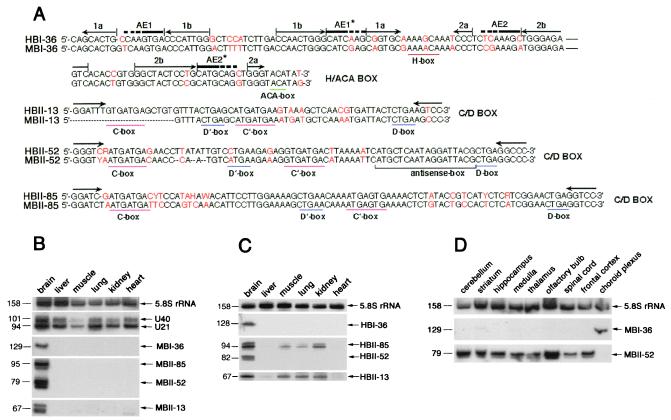
Figure 1.
Sequence alignments between the four human and mouse snoRNAs (A) and Northern blot analysis showing their tissue-specific expression (B-D). (A) Comparison between sequences of human and mouse snoRNAs with nucleotide substitutions indicated in red. Conserved box motifs, H and ACA for H/ACA box snoRNA and C, C′, D, and D′ for C/D box snoRNAs are underlined. For HBI-36/MBI-36 canonical location (8) of the two potential, bipartite antisense elements, AE1 and AE2, are shown by a thick overline (with dots corresponding to potential extensions of each element). Stem portions of the two-hairpin structure typical of H/ACA box snoRNA (8) are overlined by pairs of arrows in opposite orientation (hairpins 1 and 2 each contain a large internal loop). For C/D box snoRNAs, the 5-nt inverted sequence allowing formation of a typical 5′–3′ terminal stem–box C/D structure is shown by horizontal arrows. For mouse and human C/D box snoRNAs MBII-52/HBII-52 or MBII-85/HBII-85, respectively, the sequences shown are the consensus of the repeated genomic copies. For MBII-13 the 5′ terminal nucleotides of the snoRNA were not determined. In MBII-52/HBII-52, the conserved 18-nt antisense element complementary to an edited segment of the serotonin receptor 5-HT2C mRNA is also underlined (refs. 19 and 35–37; see Fig. 3.). (B) Northern blot analysis showing brain-specific expression of the four novel snoRNAs in mouse. (C) Northern blot analysis showing tissue-specific expression of the four novel snoRNAs in human. (D) Northern blot analysis showing expression of the rat homologues of MBI-36 and MBII-52 in different rat brain areas. On longer exposures, weak expression of MBI-36 snoRNA could also be observed in striatum, medulla, thalamus, olfactory bulb, and spinal cord. Probes for 5.8S rRNA and ubiquitous C/D box snoRNAs U21 and U40 provided internal controls. As frequently observed with other snoRNAs, some of the snoRNAs were revealed as doublets (or larger bands) reflecting the presence of some terminal heterogeneity. Sizes (in nucleotides) of the RNAs are indicated on the left.
For the two abundant types of clones, the different cDNA sequences generally differ from each other by a few base changes resulting in 17 variants of MBII-52 and 15 variants of MBII-85, suggesting that these small RNAs might be transcribed from multiple gene copies (see below). The predicted snoRNAs have been detected by Northern blot analysis (Fig. 1B) and primer extensions (data not shown). As expected, the three C/D box snoRNAs and the H/ACA snoRNA are immunoprecipitated from a mouse brain cellular extract by antibodies to the cognate sno-ribonucleoproteins fibrillarin and Gar1, respectively (data not shown). However, in striking contrast to previously described snoRNAs, they are detected in brain only (Fig. 1B). We also detected their human orthologues, designated HBI-36, HBII-13, HBII-52, and HBII-85 (Fig. 1A, see also below). Likewise, HBI-36 and HBII-52 snoRNAs are exclusively expressed in human brain (Fig. 1C). As for HBII-13 and HBII-85, although predominantly found in brain, they are also detected—at reduced levels—in muscle, kidney, and lung. We next determined the distribution of these snoRNAs in various brain areas, choosing rat as a more convenient biological system (Fig. 1D). The three C/D box snoRNAs accumulate at roughly similar levels in all rat brain areas (Fig. 1D and data not shown) except that in choroid plexus where they were not detected. Strikingly, brain-specific H/ACA snoRNA MBI-36 displayed a reverse distribution: it was detected mainly in the choroid plexus (Fig. 1D).
By a BLASTN database search, we discovered the conserved human orthologue of MBI-36 (87% sequence similarity), designated as HBI-36 (Fig. 1A). In addition, we were able to determine its genomic location on chromosome X, within the second intron of a gene predominantly expressed in choroid plexus epithelial cells, encoding the serotonin receptor 2C (19). Southern blot analysis demonstrated that MBI-36 is present in a single copy gene per haploid genome (data not shown). Thus, MBI-36 is an intronic snoRNA host gene that is nonubiquitous and does not belong to the 5′TOP gene family (12, 13). Ubiquitous, intronic, modification-guide snoRNAs in vertebrates are generally hosted by genes coding for proteins directly involved in ribosome biogenesis or function, suggesting this peculiar gene organization could provide the basis for a coordinate production of functionally linked gene products (8–17). In this context, it is tempting to speculate that the presence of brain-specific H/ACA box snoRNA HBI-36 within an intron of the serotonin 2C receptor might reflect some functional link between HBI-36 snoRNA and its host gene.
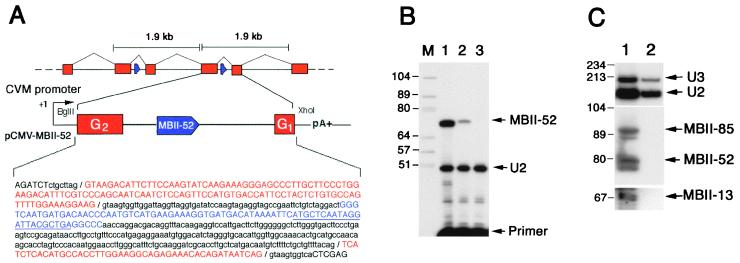
To isolate the gene encoding C/D box snoRNA MBII-52, we screened a mouse PAC library with a MBII-52 probe and identified a 1.9-kb PstI restriction fragment in three overlapping murine PAC clones (B16410, I10450, and B13499). Sequence analysis of five subclones from PAC I10450 revealed that they all contained slightly divergent copies (sequence similarity 95%) of the MBII-52-containing fragment, indicating the presence of multiple tandem copies of a 1.9-kb repeat unit (Fig. 2A). By a BLASTN database search with one of the cloned repeats, we identified 89% sequence similarity between two short separate domains of the query sequence and two adjacent parts (nucleotides 1–47 and 49–144) of exon G of the Ipw (imprinted in Prader–Willi syndrome) gene in mouse chromosome region 7C (20). Although exon G was reported to be a single exon, our data show this segment to be tandemly repeated and interrupted by a sequence that displays hallmarks of a bona fide intron that, in turn, contains a single copy of MBII-52 (Fig. 2A). The region of the repeat spanning the snoRNA-containing intron and two flanking exons, taken from a randomly selected unit, was transiently expressed under the control of a strong viral promoter (Fig. 2A) in mouse L929 cells, which do not express the endogenous snoRNA gene. Faithfully processed MBII-52 snoRNA was readily detected in transfected cells (Fig. 2B). Moreover, predicted spliced exons G2/G1 were detected by sequence analysis of reverse transcription–PCR products (data not shown), ruling out the possibility that MBII-52 is processed by a splicing-independent pathway. The human IPW gene is highly divergent from mouse (see below), preventing the identification of the human equivalent of mouse exons G. Accordingly, we do not know whether HBII-52 is encoded in an IPW gene intron. MBII-52 and the three other snoRNAs were not immunoprecipitated by an anti-trimethyl cap antibody, strongly suggesting their 5′ ends do not mark a transcription initiation site but result from processing of longer transcripts (Fig. 2C).
Figure 2.
Genomic organization of the repeated MBII-52 snoRNA genes in mouse (A), processing of the MBII-52 snoRNA from one repeat unit (B), and immunoprecipitation of MBII-13, MBII-52, and MBII-85 snoRNAs with an anti-trimethyl cap antibody (C). A) Schematic organization of the tandemly repeated units containing mouse snoRNA MBII-52-genes and the construct used for expressing the MBII-52 snoRNA locus in mouse cells. A BglII–XhoI DNA fragment of the mouse 1.9-kb repeat unit was inserted into the eukaryotic vector pRCEN downstream from a cytomegalovirus (CMV) promoter (not drawn to scale). In the sequence of the insert, the Ipw bipartite exon G is indicated in red, the intron is in lowercase type, and the MBII-52 snoRNA (blue) is in uppercase type (the underlining denotes the location of the primer used for the reverse transcription, see B). (B) Transient expression of MBII-52 snoRNA in transfected mouse L929 cells. The snoRNA was assayed by primer extension performed on total RNA from mouse L929 cells transfected with the construct depicted in A (lane 2) or an unrelated DNA (lane 3). Splicing of exons G1 onto G2 has been experimentally checked in this system (data not shown). Lane 1 shows control detection of MBII-52 performed on total brain RNA (from rat). The U2 snRNA content of each sample was assayed in parallel through reverse transcription with another appropriate primer to provide a control for gel loading. Lane M contains DNA marker (size in nucleotides). (C) Lack of immunoprecipitation of MBII-13, MBII-52, and MBII-85 snoRNAs with an anti-trimethyl cap antibody. Lanes: 1, input (total RNA from mouse brain); 2, RNA fraction precipitated by the R1131 antibody. The three snoRNAs were assayed by Northern blot hybridization with specific oligonucleotide probes, and positive controls were 5′ trimethyl-capped U2 snRNA and U3 snoRNA probed with appropriate oligonucleotides. Markers sizes in nucleotides are indicated to the left.
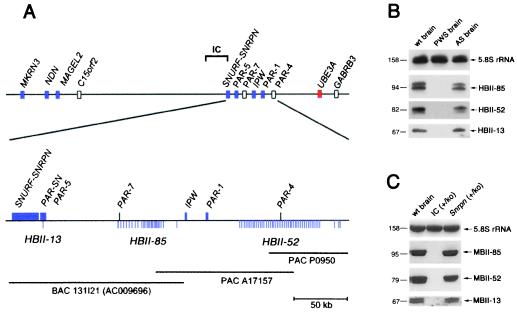
To identify the genomic location of the human orthologue of MBII-52, we screened a human PAC library with a probe derived from the MBII-52 gene. Four clones (O19203, A17157, H17223, and P0950) were identified. Similar to the situation in mouse (see above), we identified HBII-52 and the human IPW transcript on the same clone, A17157, which maps to chromosome 15q11–13 (21). This region is involved in two diverse genetic defects, PWS and Angelman syndrome. PWS is a neurogenetic disease resulting from a deficiency of paternal gene expression and associated with significant developmental, behavioral, and mental problems (22). Surprisingly, clone A17157 was also positive for an MBII-85 probe, indicating that at least two of the snoRNA genes mapped to the PWS region. By complete sequence analysis of the overlapping PAC clones A17157 and P0950 spanning a region of 220 kb, we identified 47 copies of HBII-52. With some exceptions, they were regularly spaced within a stretch of 99 kb (Fig. 3A) and embedded within 1.9-kb units substantially different from each other (average sequence divergence from the consensus is 22% over the entire repeat unit but only 3% over the snoRNA coding region). Except for the snoRNA coding region, the human 1.9-kb units are completely different from the sequenced mouse MBII-52 containing 1.9-kb repeat units. Interestingly, part of repeat unit number 23, including a copy of the HBII-52 gene, is identical to the published sequence (343 bp) of a previously identified transcribed sequence called PAR-4 (23). This suggests that PAR-4 is derived from the HBII-52 locus.
Figure 3.
Chromosomal location of the human HBII-13, HBII-52, and HBII-85 snoRNA genes (A) and their lack of expression, assayed by Northern blot analysis, in a PWS patient (B) or in a PWS mouse model (C). (A Upper) Overview of the proximal chromosomal 15q region containing genes for PWS and Angelman syndrome (not drawn to scale). Blue boxes, paternally expressed genes (italic type)/transcribed sequences; red boxes, maternally expressed genes; open boxes, biallelically expressed genes or genes with unknown imprint status. (Lower) Fine mapping of the three snoRNA genes with regard to known genes or expressed sequences distal to SNRPN. Each vertical line below the map represents one snoRNA gene. Horizontal lines indicate the PAC and bacterial artificial chromosome clones. (B) Lack of expression of HBII-13, HBII-52, and HBII-85 snoRNAs in the brain of a PWS patient. RNA samples were taken from the cortex of a human control sample (wt brain), a PWS patient (PWS brain), and an Angelman syndrome patient (AS brain). (C) Lack of expression of MBII-13, MBII-52, and MBII-85 snoRNAs in a PWS mouse model (25). Total brain RNA samples of a wild-type mouse (wt brain), a paternal imprinting center (IC) deletion strain (IC+/ko), and a phenotypically normal mouse with a Snrpn deletion (Snrpn+/ko) were analyzed.
On examination of the nucleotide sequence proximal to the IPW gene (PAC clone A17157 and bacterial artificial chromosome clone RP11-131I21, GenBank accession no. AC009696), we identified 24 copies of the human homologue of the mouse MBII-85 gene (HBII-85) within a region of 55 kb. These copies were less evenly spaced than the HBII-52 genes (Fig. 3A). Copies of the HBII-85 snoRNA coding region are substantially less conserved than HBII-52 genes, exhibiting on the average 20% sequence divergence among each other (Fig. 1A). Part of repeat number 1 contains a sequence derived from a previously identified transcribed sequence, designated as PAR-7 (23). Surprisingly, by a BLASTN databank search with the MBII-13 sequence, we identified a single copy of the human homologue HBII-13 within the overlap of the cDNA clones PAR-SN (24) and PAR-5 (23) close to SNRPN on chromosome 15q12 (Fig. 3A), thus showing that all three brain-specific box C/D snoRNAs are located within the same region on chromosome 15 involved in PWS. It can be speculated that the previously reported transcribed sequences PAR-4, PAR-7, and PARSN/PAR-5 might be derived from precursor products in the processing pathway of HBII-13, HBII-52, and HBII-85 snoRNAs. We made a noteworthy observation in light of the lack of sequence conservation of intervening non-RNA coding segments between human and mouse and the redundancy of coding regions for HBII-52 and HBII-85 snoRNAs: almost the entire (central) repeat units for each snoRNA are totally void of repetitive elements (e.g., SINEs, LINEs, LTRs, etc.) amounting to 72 and 46 kb, respectively.
In view of their location in an imprinted domain on human chromosome 15, we examined whether HBII-13, HBII-52, and HBII-85 snoRNAs show paternal-only expression, as observed for other genes in this region (22). We detected all three snoRNAs in cortex RNA from a normal control brain and from a patient with Angelman syndrome, who had a maternal deletion of 15q11–q13, but not from a patient with PWS who had a paternal deletion of 15q11–q13 (Fig. 3B).
We also tested whether a PWS mouse model with a paternal deletion of the imprinting center on chromosome 7 would be devoid of expressing the novel C/D box snoRNA genes. It had previously been shown that this deletion resulted in a lack of transcription of the paternally expressed genes Zfp127, Ndn, and Ipw (25). As a control, we used both normal mice and mice with a paternal deletion of the Snrpn ORF (25), which are known to be phenotypically normal. Consistent with the finding in the PWS patient, we did not observe expression of MBII-13, MBII-52, and MBII-85 snoRNAs in the PWS mouse model but did in control animals (Fig. 3C). These results demonstrate that snoRNA genes can be subject to genomic imprinting and expressed from the paternal allele only.
We note with interest that a paternal deletion from Snrpn to Ube3a, which includes all three C/D box snoRNA genes, causes hypotonia, growth retardation, and partial lethality in mice, providing evidence for a gene or genes contributing to PWA (26). HBII-13 and HBII-85 are positional candidates for these genes but HBII-52 is not because a paternally derived deletion including Ipw and Gabrb3 in mice (Fig. 3A) and a similar deletion with a breakpoint distal to IPW and inside GABRB3 in humans (27–29) are without any phenotypic effect when transmitted through the male germ line. It has been suggested that the non-protein-coding transcripts between SNRPN and UBE3A (PAR-SN, PAR-5, PAR-7, IPW, PAR-1, and PAR-4) might exhibit a regulatory function (22). From our findings, these genes may host snoRNAs that could well be the only functional entities of their transcripts as reported for some ubiquitous box C/D snoRNAs (30). Among the three brain-specific C/D box snoRNAs whose genes are located in the same region on human chromosome 15q11–q13, HBII-52 and HBII-85 are particularly intriguing because they are expressed from tandemly amplified genes, a feature unique so far among snoRNAs. These are also among the most abundant snoRNAs in the brain.
The unexpected finding of brain-specific snoRNAs, three of which are subject to genomic imprinting in mice and humans, raises the tantalizing issue of their possible function. Apparently unable to guide any hypothetical brain-specific ribose methylations or pseudouridylation in rRNA because of the absence of appropriate antisense elements, they could nevertheless function in still unknown, developmentally regulated aspects of rRNA processing and/or ribosome assembly. Alternatively, they could direct nucleotide modification of other types of cellular RNAs. Indeed, C/D box snoRNAs able to direct ribose methylations of snRNA U6 (10, 11) or other snRNAs (A.H., J.-P.B., and J.B., unpublished results) have been identified recently. However, like two ubiquitous methylation guide snoRNAs devoid of rRNA complementarity (31), the three brain-specific C/D box snoRNAs do not show any complementarity of at least 9 bp to a known snRNA or a non-protein-coding RNA known to transiently localize to the nucleolus, such as RNase P, signal-recognition particle RNA, or telomerase RNA (32).
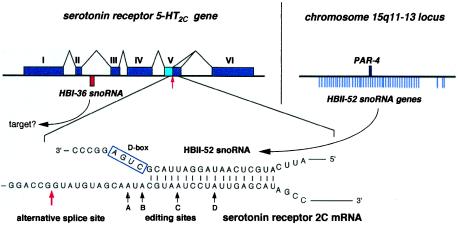
Some pre-mRNAs also transiently localize to the nucleolus (33), and the general view that mRNAs are essentially devoid of nucleotide modifications has been recently challenged by the detection of inosine at tissue-specific levels, particularly elevated in brain (34). However, the presence of ribose-methylated nucleotides in mRNA-coding regions has not been reported so far. Not surprisingly, the three C/D box snoRNAs contain, at the location of an antisense element, complementarities of at least 14–15 nt to a mRNA or an expressed sequence tag in sequence databases, most of which in all likelihood are matches by chance and without biological significance. One particular complementarity, however, seems unlikely to be fortuitous and deserves further scrutiny, because of its outstanding length, its perfect conservation in mice and humans, and the identity of the mRNA involved. HBII-52 exhibits, at the appropriate position (Fig. 1A), an 18-nt complementarity to the mRNA encoded, curiously, by the host gene of HBI-36, i.e., the gene encoding serotonin receptor 2C (Fig. 4). Moreover, in the mRNA-coding region (19, 35), this complementarity matches a conserved region critical because it is subject to both alternative splicing (36) and adenosine-to-inosine editing at four vicinal sites (37). Remarkably, the mRNA nucleotide potentially targeted for ribose methylation by MBII-52 corresponds precisely to one of the four edited adenosines, site C (Fig. 4). Ribose methylation of this adenosine should dramatically hamper its deamination to inosine (38). However, we have not been able so far to demonstrate such a role for MBII-52 in a negative control of the serotonin 5-HT2C receptor mRNA editing, but this may be due to technical limitations (data not shown). Alternatively, MBII-52 snoRNA might be involved in the regulation of alternative splicing of 5-HT2C heterogeneous nuclear RNA. A splice donor site leading to a variant form of the receptor (36) is located only 13 nt upstream of the site of potential snoRNA pairing in the mRNA sequence (Fig. 4). Interestingly, the alternatively spliced mRNA form is mostly enriched in the choroid plexus (36), the only brain area devoid of MBII-52 snoRNA (Fig. 1D).
Figure 4.
Structure of the serotonin receptor 5-HT2Cgene containing the intron-encoded HBI-36 gene (Upper Left) and potential base-paired interaction between the serotonin receptor 5-HT2C mRNA and C/D box snoRNA HBII-52 (Lower). (Upper Left) The six exons of the 5-HT2C gene including the alternative splice site in exon V are indicated with the location of the HBI-36 snoRNA gene within the second intron shown by a red bar (most introns of the 5-HT2C gene are extremely large; not drawn to scale). (Upper Right) Location of 47 copies of the HBII-52 snoRNA genes with respect to the PAR-4 gene in the PWS locus on chromosome 15 (see Fig. 3A) is indicated on the right. (Lower) Potential base pairing of HBII-52 snoRNA with the editing sites/alternative splice site of exon V. The corresponding guide duplex should direct ribose methylation to the nucleotide paired to the fifth nucleotide upstream from box D (4–7), i.e., the adenosine at position C. The adenosine at position C is one out of four sites of adenosine-to-inosine editing within the serotonin receptor mRNA (ref. 37; the four sites of edition in 5-HT2C receptor mRNA are denoted by solid arrows and labeled A–D). The alternative splicing site present in the serotonin receptor 5-HT2C mRNA (36) is indicated by a red arrow.
The detection of additional tissue-specific specimens snoRNAs, in brain or other tissues, and a thorough comparative analysis of their potential targets in the complete human and mouse genomes might provide further insight into the range of functions fulfilled by the unexpectedly large families of snoRNAs in mammals.
Acknowledgments
We thank P. Veyrac (Laboratoire de Neurobiologie de L'Apprentissage, Orsay, France), S. Mazan (Laboratoire de Biologie Cellulaire 4, Orsay), and J. M. Zajac (Institut de Pharmacologie et de Biologie Structurale) for assistance in rat brain dissections. A.H. thanks C. Sorg for his steady interest and encouragement during this work. This work was supported by an Interdisziplinäres Zentrum für Klinische Forschung grant (Teilprojekt F3, Münster) to A.H., the German Human Genome Project through the Bundesministerium für Bildung, Wissenschaft, Forschung, und Technologie Grant 01KW9616 to J.B., Deutsche Forschungsgemeinschaft Grant Ho949/12–3 to B.H., and a grant from the Association pour la Recherche sur le Cancer and laboratory funds from the Centre National de la Recherche Scientifique and Université Paul-Sabatier, Toulouse, to J.P.B.
Abbreviations
- snoRNA
small nucleolar RNA
- PWS
Prader–Willi syndrome
- snRNA
small nuclear RNA
- PAC
bacteriophage P1-derived artificial chromosome
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. for PAC clones A17157 and P0950, AF250841).
See commentary on page 14035.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.250426397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.250426397
References
- 1.Maden B E H. Prog Nucleic Acid Res Mol Biol. 1990;39:241–303. doi: 10.1016/s0079-6603(08)60629-7. [DOI] [PubMed] [Google Scholar]
- 2.Massenet S, Mougin A, Branlant C. In: Modification and Editing of RNA: The Alteration of RNA Structure and Function. Grosjean H, Benne R, editors. Washington, DC: Am. Soc. Microbiol.; 1998. pp. 201–228. [Google Scholar]
- 3.Yu Y T, Shu M D, Steitz J A. EMBO J. 1998;17:5783–5795. doi: 10.1093/emboj/17.19.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith C M, Steitz J A. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- 5.Bachellerie J P, Cavaillé J. Trends Biochem Sci. 1997;22:257–261. doi: 10.1016/s0968-0004(97)01057-8. [DOI] [PubMed] [Google Scholar]
- 6.Kiss-Laszlo Z, Henry Y, Bachellerie J P, Caizergues-Ferrer M, Kiss T. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 7.Cavaillé J, Nicoloso M, Bachellerie J P. Nature (London) 1996;383:732–735. doi: 10.1038/383732a0. [DOI] [PubMed] [Google Scholar]
- 8.Ganot P, Bortolin M-L, Kiss T. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 9.Wang H, Boisvert D, Kim K K, Kim R, Kim S H. EMBO J. 2000;19:317–323. doi: 10.1093/emboj/19.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tycowski T H, You Z H, Graham P J, Steitz J A. Mol Cell. 1998;2:629–638. doi: 10.1016/s1097-2765(00)80161-6. [DOI] [PubMed] [Google Scholar]
- 11.Ganot P, Jady B E, Bortolin M-L, Darzacq X, Kiss T. Mol Cell Biol. 1999;19:6906–6917. doi: 10.1128/mcb.19.10.6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith C M, Steitz J A. Mol Cell Biol. 1998;18:6897–6909. doi: 10.1128/mcb.18.12.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pelczar P, Filipowicz W. Mol Cell Biol. 1998;18:4509–4518. doi: 10.1128/mcb.18.8.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maxwell E S, Fournier M J. Annu Rev Biochem. 1995;35:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 15.Fragapane P, Prislei S, Michienzi A, Caffarelli E, Bozzoni I A. EMBO J. 1993;12:2921–2928. doi: 10.1002/j.1460-2075.1993.tb05954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tycowski K T, Shu M D, Steitz J A. Genes Dev. 1993;7:1176–1190. doi: 10.1101/gad.7.7a.1176. [DOI] [PubMed] [Google Scholar]
- 17.Cavaillé J, Bachellerie J P. Biochimie (Paris) 1996;78:443–456. doi: 10.1016/0300-9084(96)84751-1. [DOI] [PubMed] [Google Scholar]
- 18.Lerner M R, Hardin J A, Steitz J A. Science. 1981;211:400–402. doi: 10.1126/science.6164096. [DOI] [PubMed] [Google Scholar]
- 19.Xie E, Zhu L, Zhao L, Chang L S. Genomics. 1996;35:551–561. doi: 10.1006/geno.1996.0397. [DOI] [PubMed] [Google Scholar]
- 20.Wevrick R, Francke U. Hum Mol Genet. 1997;5:325–332. doi: 10.1093/hmg/6.2.325. [DOI] [PubMed] [Google Scholar]
- 21.Wevrick R, Kerns J A, Francke U. Hum Mol Genet. 1994;3:1877–1882. doi: 10.1093/hmg/3.10.1877. [DOI] [PubMed] [Google Scholar]
- 22.Nicholls R D, Saitoh S, Horsthemke B. Trends Genet. 1998;14:194–200. doi: 10.1016/s0168-9525(98)01432-2. [DOI] [PubMed] [Google Scholar]
- 23.Sutcliffe J S, Nakao M, Christian S, Orstavik K H, Tommerup N, Ledbetter D H, Beaudet A L. Nat Genet. 1994;8:52–58. doi: 10.1038/ng0994-52. [DOI] [PubMed] [Google Scholar]
- 24.Ning Y, Roschke A, Christian S L, Lesser J, Sutcliffe J S, Ledbetter D H. Genome Res. 1996;6:742–746. doi: 10.1101/gr.6.8.742. [DOI] [PubMed] [Google Scholar]
- 25.Yang T, Adamson T E, Resnick J L, Leff S, Wevrick R, Francke U, Jenkins N A, Copeland N G, Brannan C I. Nat Genet. 1998;19:25–31. doi: 10.1038/ng0598-25. [DOI] [PubMed] [Google Scholar]
- 26.Tsai T F, Jiang Y H, Bressler J, Armstrong D, Beaudet A L. Hum Mol Genet. 1999;8:1357–1364. doi: 10.1093/hmg/8.8.1357. [DOI] [PubMed] [Google Scholar]
- 27.Nicholls R D. Nat Genet. 1999;23:132–134. doi: 10.1038/13758. [DOI] [PubMed] [Google Scholar]
- 28.Saitoh S, Kubota T, Ohta T, Jinno Y, Niikawa N, Sugimoto T, Wagstaff J, Lalande M. Lancet. 1992;339:366–367. doi: 10.1016/0140-6736(92)91686-3. [DOI] [PubMed] [Google Scholar]
- 29.Greger V, Woolf E, Lalande M. Hum Mol Genet. 1993;2:921–924. doi: 10.1093/hmg/2.7.921. [DOI] [PubMed] [Google Scholar]
- 30.Tycowski K T, Shu M D, Steitz J A. Nature (London) 1996;379:464–466. doi: 10.1038/379464a0. [DOI] [PubMed] [Google Scholar]
- 31.Jady B E, Kiss T. Nucleic Acids Res. 2000;28:1348–1354. doi: 10.1093/nar/28.6.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pederson T. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bond V C, Wold B. Mol Cell Biol. 1993;13:3221–3230. doi: 10.1128/mcb.13.6.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paul M S, Bass B L. EMBO J. 1998;17:1120–1127. doi: 10.1093/emboj/17.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu L, Nguyen H, Le H, Bloem L J, Kozak C A, Hoffman B J, Snutch T P, Lester H A, Davidson N, Lübbert H. Mol Brain Res. 1991;11:143–149. doi: 10.1016/0169-328x(91)90116-f. [DOI] [PubMed] [Google Scholar]
- 36.Canton H, Emeson R B, Barker E L, Backstrom J R, Lu J T, Chang M S, Sanders Bush E. Mol Pharmacol. 1996;50:799–807. [PubMed] [Google Scholar]
- 37.Burns C M, Chu H, Rueter S M, Hutchinson L K, Canton H, Sanders Bush E, Emeson R B. Nature (London) 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 38.Yi-Brunozzi H Y, Easterwood L M, Kamilar G M, Beal P A. Nucleic Acids Res. 1999;27:2912–2917. doi: 10.1093/nar/27.14.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]