Abstract
Physico-chemical properties of amino acids can be used to study protein sequence profiles, folding and function. We collated 242 properties for the 20 naturally occurring amino acids and created a dataset. The dataset is available as a database named APDbase( Amino acid Physico-chemical properties Data base). The database can be queried using either key words describing physico-chemical properties or pre-assigned database index number. The database contains corresponding references for each property value and facilitates deposition of new property values for processing and inclusion in the database.
Availability
The database is available for free at http://www.rfdn.org/bioinfo/APDbase.php
Keywords: Physico-chemical properties, homology, amino acids, folding, function
Background
Proteins (composed of amino acids) constitute a major group of biological macromolecules that are key for the function of a living system. Protein evolution involves selection of sequences having functional advantage over random mutants. [ 1] Closely evolved proteins resemble the parent in their sequence, structure and function which are referred to as homologs.[ 2] Often multiple sequences (strings formed by different combinations of the 20 amino acid alphabets) have the same structure and function. This introduces functional redundancy to the sequence pool. Functional redundancy can be overcome by developing metrics for sequence comparison. Comparison of protein sequences should involve a metric for the 20 amino acids. Naturally occurring amino acids can be grouped based on their similarity of physico-chemical properties. In order to understand the conservation of residues in a protein sequence, it may be important to qualitatively and quantitatively measure the differences among residues. A collection of physico-chemical properties of amino acids will be helpful to study macroscopic properties of proteins (such as aggregation), perform sequence comparison or understand conservation of functionally important residues in a protein family (physico-chemical signatures).
Methodology
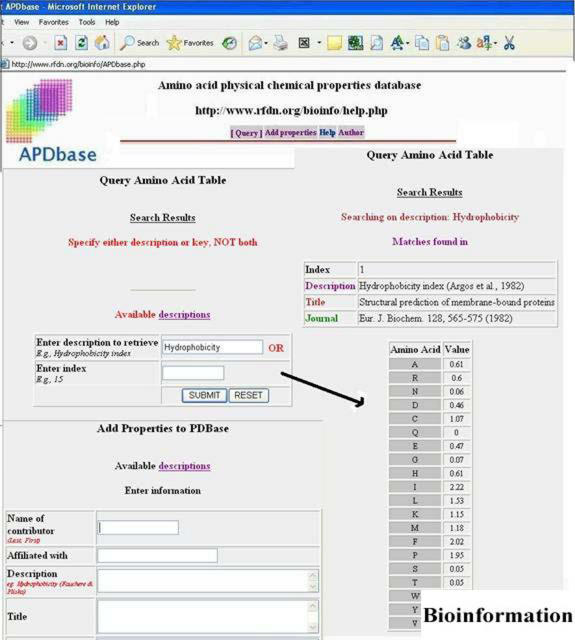
The site utilizes open-source software running on LINUX ® platform to deliver the content. Flat files that contain indexed properties, author details and journal citations were created after curating the AAINDEX [ 3] database in DBGet (Japan) and ProtScale in Swiss Expasy [ 4]. We excluded properties that have missing values for any of the twenty amino acids and those that are less relevant to the study of protein sequence, structure and function. A search interface is implemented in PHP driven by Zend engine. [ 5] Keyword search like hydrophobicity, charge, aromaticity and several others produces all listed properties in the dataset along with the journal citation. In order to facilitate future expansion, we have forms that the users can fill out to add new properties that will be audited for completion, accuracy and relevance by domain experts periodically. Numerical redundancy will be automatically avoided before incorporating into the dataset. A screen capture of the web-site is shown in Figure 1.
Figure 1.
A Screen capture of APDbase showing query and property value deposition interface is shown. This database can be queried using either amino acid property keyword or database index number. A search result for hydrophobicity is shown here as a sample
Utility
Protein sequence analysis, macroscopic property prediction, property motif identification and epitope prediction are of great interest to the biological community. Bioinformatics tools that perform such tasks are increasingly incorporating physico-chemical property based metric to increase their performance and to derive knowledge based rules.[ 6 8] APDbase was previously available as PDbase. [9] During 1999-2004 other groups have used this database for developing methods related to biological sequence analysis. [6] The previous database had 237 properties. The current dataset includes additional 5 properties that are PCP-descriptors derived elsewhere. [10]
Limitations
The dataset described here is a comprehensive sample of available properties for 20 naturally occurring amino acids. Nevertheless the dataset is not complete and requires regular updates. The dataset can be updated using property values derived from statistical analysis and experimental observations.
Future Development
We are planning to incorporate a graphics tool that can cluster available properties into a dendrogram for visual inspection. This will help to visualize relatedness among clusters. The tool can be applied to generate physico-chemical profile for sequences of interest.
Conclusion
APDbase is a dataset of physico-chemical properties of amino acids that will be converted as a tool to study protein sequence profiles.
Acknowledgments
The authors acknowledge Prof. Werner Braun (University of Texas Medical Branch, Galveston, Texas, USA) for active discussions in selecting the properties and the Roskamp Foundation for hosting the database.
Footnotes
Citation:Mathura & Kolippakkam, Bioinformation 1(1): 2-4 (2005)
References
- 1.Krylov DM, et al. Genome Res. 2003;13:2229. doi: 10.1101/gr.1589103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chothia C, et al. Embo J. 1986;5:823. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. http://www.genome.jp/dbget/dbget.links.html.
- 4. http://www.expasy.org/tools/protscale.html.
- 5. http://www.php.net .
- 6.Mathura VS, et al. Bioinformatics. 2003;19:1381. doi: 10.1093/bioinformatics/btg164. [DOI] [PubMed] [Google Scholar]
- 7.Nishikawa K, et al. J Biochem. 1983;94:997. doi: 10.1093/oxfordjournals.jbchem.a134443. [DOI] [PubMed] [Google Scholar]
- 8.Hobohm U, et al. J Mol Biol. 1995;251:390. doi: 10.1006/jmbi.1995.0442. [DOI] [PubMed] [Google Scholar]
- 9.Ganapathiraju MK, et al. IEEE signal processing magazine. 2004;21:78. [Google Scholar]
- 10.Venkatarajan MS, et al. J Mol Model. 2001;7:445. [Google Scholar]