Abstract
The current challenge in synthetic vaccine design is the development of a methodology to identify and test short antigen peptides as potential T-cell epitopes. Recently, we described a HLA-peptide binding model (using structural properties) capable of predicting peptides binding to any HLA allele. Consequently, we have developed a web server named T-EPITOPE DESIGNER to facilitate HLA-peptide binding prediction. The prediction server is based on a model that defines peptide binding pockets using information gleaned from X-ray crystal structures of HLA-peptide complexes, followed by the estimation of peptide binding to binding pockets. Thus, the prediction server enables the calculation of peptide binding to HLA alleles. This model is superior to many existing methods because of its potential application to any given HLA allele whose sequence is clearly defined. The web server finds potential application in T cell epitope vaccine design.
Availability
Keywords: HLA, peptide, binding, prediction, immunity, T-cell, epitope, design, vaccine
Background
Peptide vaccines are cocktails of short peptides (8-20 residues long) capable of eliciting T-cell mediated immune responses [1] upon binding to allele specific HLA (human leukocyte antigen) molecules. These peptides are referred as T-cell epitopes. The host HLA molecules are highly polymorphic with more than 1500 HLA alleles identified in the population. [2– 3] Moreover, HLA allelic variants are present at different frequencies among different ethnic groups. There are also two types of HLA molecules, HLA class I (HLAI) and HLA class II (HLAII). HLAI molecules bind peptides of length 8-10 residues HLAII bind peptides of length 10-20 residues. Given the polymorphims of HLA molecules, a critical issue in the design of T-cell epitope vaccines is the identification of peptides with proven ability to bind to many HLA alleles. Experimental testing of peptide binding to most or all HLA molecules using competitive binding assay is arduous, time consuming and expensive. For example, a 200 residues long surface antigen can ideally produce 192 overlapping peptides of length 9 residues. Testing the binding ability of these short peptides to more than 1500 HLA molecules (current status) requires cloning, expression, and purification of these molecules followed by binding assay. Hence, the selection of peptide by MHC molecules is highly specific and combinatorial. Alternatively, screening of peptide binding to HLA molecules using prediction models is economically advantageous. Thus, several validated mathematical models for the prediction of binding peptides to multiple MHC molecules are currently available.
A number of prediction servers are available over the World Wide Web. These servers facilitate prediction for few alleles of either HLA class I or HLA class II. Servers like CTLPred [4], ProPred1 [5], MAPPP [6 ], nHLAPred [7], BIMAS [8 ], LPPEP [9], SVMHC [10 ], NetMHC [11], MHCPred [12], Epitope binding [13 ], MMPRED [14] and PREDEP [ 15] facilitate prediction for class I molecules. Some servers like PREDICT [16], SYFPEITI [17 ] and RANKPEP [18] facilitate prediction for both class I and class II alleles. Servers like ProPred [19], Epipredict [ 20], HLADR4Pred [21], MHC2Pred [22] and MHCThread [23 ] facilitate prediction for class II alleles. We have implemented a web server named T-epitope Designer for MHC-peptide binding predictions. This implementation is based on a simple, efficient, and robust model described recently. [24] The model uses the definition of virtual binding pockets, mapping of virtual pockets to position specific peptide residue anchors and estimation of peptide residuevirtual binding pocket compatibility. T-epitope Designer provides a user friendly web interface for user to predict MHC binding peptides in protein sequence. Customized prediction can be performed by selecting allele specific and threshold scores. Here, we present the design methodology and features of this web server.
Methodology
T-epitope Designer is implemented based on a MHC-peptide prediction model described recently [24]. This model is built upon a selected dataset of 29 human MHC-peptide structures obtained from Protein Data Bank (PDB). The model uses the definition of virtual binding pockets, mapping of virtual pockets to position specific peptide residue anchors and estimation of peptide residue―virtual binding pocket compatibility using Q matrix developed elsewhere. [25] It has been shown that this model produces good prediction efficiencies (average 60%) with good sensitivity (˜50% - 73%) and specificity (52% - 58%). [24] Moreover, this prediction model is superior to many existing methods because of its potential application to any given MHC allele whose sequence is clearly defined.
User Interface
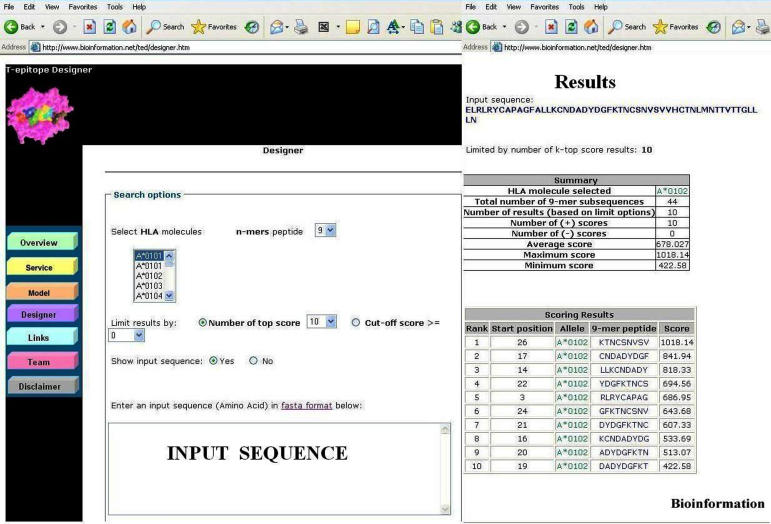
Figure 1 shows the user interface for MHC-peptide binding prediction facility at the T-epitope designer web server. There are four major options and one entry point, reflecting the different search options available for customization: Four major options
Select HLA molecules: This field provides a list of HLA alleles for user selection and customization.
Limit results by number of top score: This option enables the user to filter the number of results by specifying the required number of top MHC-peptide binding scores. The top ten peptides are returned for a selection of 10.
Limit results by number of cut-off score: This option enables the user to filter the number search results by specifying the threshold scores. Only peptides with positive scores are returned for a selection of greater than 0.
Show input sequence: This option enables the user to display the input protein sequence in the result page.
Figure 1.
A snapshot of T-EPITOPE prediction server. The input window for sequence submission and results page with prediction data are shown above
The web server reads an input sequence (in FASTA format) and then computes the binding scores between the selected allele and all the 9 residues subsequences. We choose length 9 because our previous study demonstrates MHC-peptide structures with bound peptides9 residues long are best represented in the dataset. [26] Predicted MHC binding peptides candidates are summarized and presented via the web interface in tabular form.
Prediction results
The binding score is used as a metric for MHC-peptide binding. [24] Search results are available through the web interface with the MHC-peptide binding scores given in a commonly used tabular format (Figure 1). The predicted peptides are presented in two tables, summary table and scoring results table respectively. The summary table gives details on user specified parameters like the selected allele, the total number of 9-mer subsequences and statistics of the binding scores (e.g. the number of positive/negative binding scores, maximum binding scores, etc). The scoring results table ranks the predicted peptides in descending order in terms of the binding scores.
Conclusion
Statistical and 3D structure based procedures are available for the prediction of MHC-peptide binding prediction. Data driven statistical methods are generally available for limited HLA alleles. [27] However, structure based methods can be adopted for any given MHC molecule as long as the sequence of the MHC molecules is known. It should also be noted that structural procedures utilizing structure prediction by energy minimization followed by protein-ligand interaction calculations are computationally intensive. Moreover, robust protein-ligand interaction functions are not currently available. Thus, T-epitope Designer server provides a web service that circumvents these problems by using a model that is simple, fast and robust.
Caveats
T-epitope Designer server uses a prediction method based on the definition of virtual binding pockets, mapping of virtual pockets to position specific peptide residue anchors and estimation of peptide residuevirtual binding pocket compatibility. Virtual pockets are defined using information gleaned from eight unique MHC alleles and the mapping of virtual pocket to position specific residue anchors is done using 29 MHC-peptides structures taken from PDB. The peptide residuevirtual binding pocket compatibility is estimated using the Q matrix described elsewhere. [25 ] The average Positive Predictive Value (PPV) of the model is 89%, whereas the average Negative Predictive Value (NPV) is only 18%. [24] The low NPV can be improved using (1) redefinition of virtual pockets as and when more MHC-peptide structures are available; (2) a modified Q matrix; (3) validation of the model with a dataset containing more non-binders; (4) performance of blindfold prediction and validation. We plan to improve the NPV of the model by addressing the above issues. Despite the limitations set by the model, the prediction method is novel, generic, and simple.
Footnotes
Citation:Kangueane & Sakharkar, Bioinformation 1(1): 21-24 (2005)
References
- 1.Sette A, et al. Tissue Antigens. 2002;59:443. doi: 10.1034/j.1399-0039.2002.590601.x. [DOI] [PubMed] [Google Scholar]
- 2. http://www.ebi.ac.uk/imgt/
- 3.Reche PA, Reinherz EL. J Mol Biol. 2003;331:623. doi: 10.1016/s0022-2836(03)00750-2. [DOI] [PubMed] [Google Scholar]
- 4. http://www.imtech.res.in/raghava/ctlpred/
- 5. http://www.imtech.res.in/raghava/propred1/
- 6. http://www.mpiib-berlin.mpg.de/MAPPP/binding.html.
- 7. http://www.imtech.res.in/raghava/nhlapred/
- 8. http://thr.cit.nih.gov/molbio/hla_bind/
- 9. http://zlab.bu.edu/zhiping/lppep.html.
- 10. http://www-bs.informatik.uni-tuebingen.de/Services/SVMHC.
- 11. http://www.cbs.dtu.dk/services/NetMHC/
- 12. http://www.jenner.ac.uk/MHCPred/
- 13. http://hlaligand.ouhsc.edu/prediction.htm.
- 14. http://www.imtech.res.in/raghava/mmbpred/
- 15. http://margalit.huji.ac.il/Teppred/mhc-bind/index.html.
- 16. http://research.i2r.a-star.edu.sg/predict-demo/
- 17. http://www.syfpeithi.de/
- 18. http://mif.dfci.harvard.edu/Tools/rankpep.html.
- 19. http://www.imtech.res.in/raghava/propred/
- 20. http://www.epipredict.de/Prediction/prediction.html.
- 21. http://www.imtech.res.in/raghava/hladr4pred/
- 22. http://www.imtech.res.in/raghava/mhc2pred/
- 23. http://www.csd.abdn.ac.uk/~gjlk/MHC-Thread/
- 24.Zhao B, et al. Human Immunology. 2003;64:1123. doi: 10.1016/j.humimm.2003.08.343. [DOI] [PubMed] [Google Scholar]
- 25.Venkatarajan MS, Braun W. J Mol Model. 2001;7:445. Link. [Google Scholar]
- 26.Kangueane P, et al. Human Immunology. 2001;62:539. doi: 10.1016/s0198-8859(01)00219-1. [DOI] [PubMed] [Google Scholar]
- 27. http://www.bioinformation.net/ted/links.htm.