Abstract
Simple sequence repeats (SSRs) are becoming standard DNA markers for plant genome analysis and are being used as markers in marker assisted breeding. And hence because of its great significance we have initiated this study to analyze complete genome of Arabidopsis thaliana for the prevalence of mono-, di-, tri-, tetra-, penta- and hexa- mer repeats in the coding and non-coding regions of the chromosome and to map their exact position on the sequence. We have developed a program that can search a repeat of any length, its exact position on the chromosome and also its frequency of occurrence in the genome. Analysis of the results reveal that maximum number of repeats were found in chromosome 1 followed by chromosome 2 and 4 whereas, chromosome 3 and 5 contain relatively less number of these repeats. Among the SSRs, hexamers and dimers were more predominant in the chromosomes. Overall data showed that Chromosome 5 has minimum number of repeats. The abundance or rarity of various simple repeats in different chromosomes is not explained by nucleotide composition of sequence or potential repeated motifs to form alternative DNA structures. This suggests that in addition to nucleotide composition of repeat motifs, characteristic DNA replication / repair / recombination machinery might play an important role in genesis of repeats. The positional information is given at www.geocities.com/amubioinfo/ARD. This positional information can help Arabidopsis researchers to identify new polymorphisms in chromosomal regions of interest based on the SSRs that map in the area.
Keywords: Simple sequence repeats, Arabidopsis thaliana, polymorphism, loci, coding, non-coding
Background
Simple sequence repeats (SSRs) are becoming standard DNA markers for plant genome analysis and are being used as markers in marker assisted breeding. Simple sequence repeats or microsatellite repeats are defined as regions within DNA sequences where short sequences (1-6 bp; monomers to hexamers) are repeated in tandem array [1 ] these stretches of DNA, which consist of only one, or a few tandemly repeated nucleotides, for example, a DNA stretch of GTGTGTGTGTGT would be referred to as (GT)6. These types of simple sequence have been shown to be repetitive and interspersed in many eukaryotic genomes [2] and are very polymorphic due to the high mutation rate affecting the number of repeat units. SSRs have several advantages over other molecular markers for example, microsatellites allow the identification of many alleles at a single locus; they are evenly distributed all over the genome. [3] SSRs have long been known to be distributed throughout the genomes of eukaryotes and to be highly polymorphic [4– 5], such repetitions occur primarily due to slipped-strand mispairing and subsequent error(s) during DNA replication, repair or recombination. [6].These loci mutate by insertions or deletions of one or a few repeat units, and the mutation rates generally increase with an increase in the length of repeat tracks. [7] SSRs are a ubiquitous class of repetitive DNA that is widely used in genetic analyses, there is accumulating evidence that SSRs serve a functional role, affecting gene expression, and that polymorphism of SSR tracts may be important in the evolution of gene regulation [ 8–9] such repetitive tracts has been described in all eukaryotes analyzed and is thought to result from the mutational effects of replication slippage. [10] A high level of SSR informativeness has been demonstrated for a variety of plant species and this has prompted the initiation of SSR discovery programs for the majority of agronomically important crops. [11–12] However, to date, a number of limitations have existed with SSR discovery in plants, including a lack of DNA sequence in databases, a perceived low abundance of SSRs (compared to mammals), and differences in the most common types of repeat found. In the present study we have screened the entire genome of Arabidopsis thaliana to study the occurrence of microsatellite sequences (mono- to hexa-mer repeat). Zhang et al., 2004 [13] also did same type of work but in our study we have performed detailed analysis of each repeat and also given the exact location of each repeat in the genome that is very useful information for Arabidopsis researchers.
Methodology
All the chromosome and coding region sequences were downloaded in FASTA format from Genbank ftp site [14] on 12th March 2004 that has been used for generating SSR data. The SSR data presented here includes both strands of the DNA sequence. The SSRs from 1 - 6, that is, from monomer to hexamer repeats were analyzed in the complete chromosome sequence and in the coding region of all the chromosomes of A thaliana. All the possible 501 repeat types were analyzed for their abundance in the entire genome. [ 15] The perfect repeats of 12 bp or more were analyzed because SSRs are often disrupted by single base substitutions. [16] Thus, for a 12 bp SSR, one occurrence may comprise a repeat of 12 monomers, or six dimers, or four trimers, or three tetramers and pentamers (except in Chr. 5 where pentamers were searched for two times) or two hexamers.
A perl (Practical Extraction and Report Language) programme was developed for scanning the entire genome to find out the abundance and distribution of these repeats. Perl software was downloaded and installed from the site www.perl.org. [17 ] We have developed a programme that can search repeats of any length, their exact position on chromosome and also their frequency of occurrence in the sequence. The individual chromosomes were broken into sub-sequences of one million base pairs (mbps) then the repeats were searched in these sub-sequences.
Results and Discussion
We have analyzed the distribution of perfect SSRs spanning 12 bp or more in the complete A thaliana genome. For this analysis, we calculated the average frequency (times/mbps) of the particular repeat that occurs in the genome on the per million base pairs basis in the chromosomes, we have also calculated the frequency in the coding regions of all the chromosomes and also we have taken out the exact location of the individual motif on the chromosome.
All the chromosomes of A. thaliana showed a significant variation in number of monomer repeats (Table 1). Among the two types of monomer repeats, (A)n was most abundant in all the chromosomes compared to (C)n. The maximum average frequency of (A)n was found in chromosome 1, 54.5 times/mbps (per million basepairs) while the least occurrence of this repeat type was recorded in chromosome 5 (8 times/mbps) (Figure 1a). The frequency of (C)n was comparatively less in all the chromosomes. Its highest occurrence was found in chromosome 1 and 4 while it was not found in chromosome 3 and 5 (Figure 1. a & b).
Table 1. Total of average frequency of SSRs in A. thaliana chromosomes and total frequency in coding region.
| SSRs | Chr.1 | Coding | Chr.2 | Coding | Chr.3 | Coding | Chr.4 | Coding | Chr.5 | Coding |
| Monomer | 55.5 | 3 | 43.7 | 87 | 28.5 | 35 | 52.0 | 101 | 8.0 | 4 |
| Dimer | 57.5 | 9 | 74.2 | 145 | 24.5 | 24 | 66.5 | 136 | 8.0 | 8 |
| Trimer | 58.2 | 4 | 46.7 | 104 | 33.0 | 32 | 55.5 | 114 | 5.0 | 3 |
| Tetramer | 24.8 | 16 | 20.2 | 41 | 13.5 | 12 | 23.0 | 51 | 2.5 | 2 |
| Pentamer | 238.3 | 3 | 214.0 | 15 | 128.5 | 4 | 259.2 | 15 | 25.0 | 0 |
| Hexamer | 99.0 | 134 | 101.0 | 208 | 58.0 | 52 | 110.5 | 202 | 10.0 | 12 |
Chr. = Chromosome
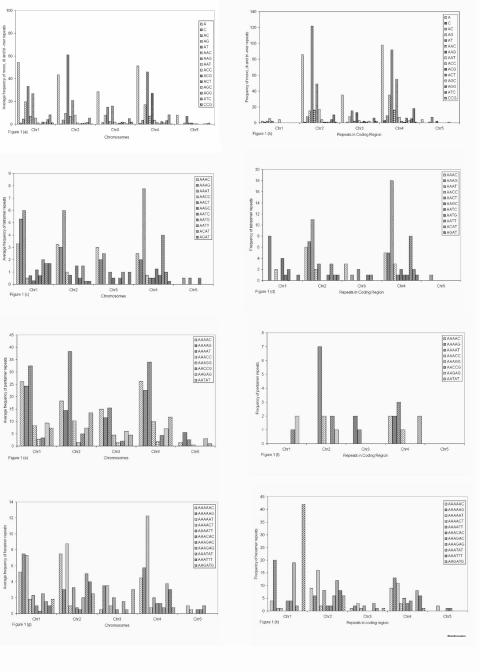
Figure 1.
Legend for Figure 1 : (a). Occurrence of mono-, di- and tri- mer repeats in all the five chromosomes of A. thaliana. (b). Occurrence of mono-, di- and tri- mer repeats in the coding region of all the five chromosomes. (c). Occurrence of tetramer repeats in all the five chromosomes. (d). Occurrence of tetramer repeats in the coding region of all the five chromosomes. (e). Occurrence of pentamer repeats in all the five chromosomes. (f). Occurrence of pentamer repeats in the coding region of all the five chromosomes. (g). Occurrence of hexamer repeats in all the five chromosomes. (h). Occurrence of hexamer repeats in the coding region of all the five chromosomes. Each bar indicates one type of repeat. Each bar has different filling that is shown in key box.
Among the dinucleotide repeats AC, AG, AT and CG, AC was most abundant in chromosome 1 (4.7 times/mbps) followed by chromosome 4, 2 and 3 respectively, this repeat was not found in chromosome 5 (Figure 1. a & b). AG was most abundant in chromosome 1 (19.8 times/mbps) followed by chromosome 4 and 2 while in chromosome 5 the average frequency of this repeat type was only 1 time/mbps. AT was most abundantly found in chromosome 2 (61 times/mbps) followed by chromosome 4, 1 and 3 (Figure 1. a & b). CG was not at all found in any of the A. thaliana chromosome. Zhang et al., 2004 [13] reported that simple repetitions comprising only pyrimidines were extremely rare in all of the fractions investigated.
Analysis of all trinucleotide repeats AAC, AAG, AAT, ACC, ACG, ACT, AGC, AGG, ATC, CCG revealed that AAG was most abundant type of repeat in the entire genome (Figure 1. a & b). Repeats that were circular permutations and reverse complements of each other were grouped together as one type.
Among the tetranucleotide repeats, the most predominate in all the chromosomes were AAAC, AAAG and AAAT. Chromosome 5 is an exception where occurrence of tetra nucleotide was found less in number (Figure 1. c & d).
Pentanucleotide repeats were found to be rare in overall genome. In chromosome 1 AAAAT was most abundant (32.5 times/mbps) followed by AAAAC, AAAAG, AAGAG and AAACC. All these repeats except AAGAG were found in non-coding region of chromosome 1. In chromosome 2 also AAAAT (38.25 times/mbps) was most abundant. Analysis of chromosome 3 revealed that AAAAT (15.5 times/mbps). In chromosome 4 also AAAAT (34 times/mbps) was most abundant. In chromosome 5 pentanucleotide repeats were found to be rare in comparison to the other chromosomes. AAAAG (5.5 times/mbps) was most abundant. The most striking feature that we observed was that none of the pentanucleotide repeats was present in the coding region of the chromosome (Figure 1. e & f).
Hexanucleotide repeats were fairly present in all the chromosomes of A thaliana. In chromosome 1, AAAAAG (7.5 times/mbps) was most abundant followed by AAAAAT, AAAAAC, AAAATT, AAAACT, AAAATC and AAAATG. The most abundant repeat in chromosome 2 was AAAAAT (8.75 times/mbps). Average frequency of these repeats was relatively less in chromosome 3, some of them were found to present 3.0 to 3.5 times/mbps while others 1.5 to 2 times/mbps only (Figure 1. g & h). Screening of chromosome 4 revealed that hexanucleotide repeats were present in good numbers and mostly the repeats were present equally in coding and non-coding part. AAAAAT (12.25 times/mbps) was the most abundant repeat followed by AAAAAG. The average frequency of these hexanucleotide repeats ranged between 0.5 - 1 times/mbps in chromosome 5 (Figure 1. g & h).
We observed that the length distributions of all SSRs indicated that the number of repeats decrease with repeat length i.e., from monomer to hexamer. Mukund et al. [18] reported in 2001 that the frequency of repeats decrease exponentially with repeat length.
The exact location of each repeat on the chromosomes was mapped. The data representing the positions of individual repeats is given at the site www.geocities.com/amubioinfo/ARD in the data files. This data on the positional information of SSRs have been checked in the AthaMap database [19] and the repeats are found at the exact location as given in the table.
Conclusion
In the present study we have examined the abundance of microsatellites with repeated unit lengths of 1-6 base pairs, that is, taking mono-, di-, tri-, tetra-, penta- and hexamers as the six classes of repeats. SSRs were well distributed throughout the genome. The overall number of each class of repeat is variable across the genome (Table 1). There is a wide range of variation in the abundance of a particular repeat type in each chromosome (Figure 1). The locations of all of the SSRs reported in this study are available in the data file at www.geocities.com/amubioinfo/ARD. This information could be useful for the selection of a wide range of microsatellite loci for studying their location and sequence-dependent evolution. Researchers can easily identify the nearest gene or the location of transcription factor binding sites around the SSR. [19] They can be used as markers for the fine analysis of recombination events along individual chromosomes. This positional information can also help Arabidopsis researchers to identify new polymorphisms in chromosomal regions of interest based on the SSRs that map in the area. SSRs are an important tool for comparative mapping because of their high polymorphism and transportability. [20 ]
Acknowledgments
The authors are grateful to Professor M Saleemuddin for providing facilities to carryout this work and his morale support throughout this project. We also thanks to the Staff of the Distribution information sub-center for their technical help. Department of Biotechnology, Ministry of Science and Technology, Government of India is acknowledged for the financial support.
Footnotes
Citation:Anwar & Khan, Bioinformation 1(2): 64-68 (2005)
References
- 1. www.ccmb.res.in/publications/newpub/paps/pap308.html.
- 2.Tautz D, Renz M. Nucleic Acids Res. 1984;12:4127. doi: 10.1093/nar/12.10.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gianfranceschi L, et al. Theor Appl Genet. 1998;96:1069. [Google Scholar]
- 4.Tautz D. Nucleic Acids Res. 1989;17:6463. doi: 10.1093/nar/17.16.6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weber JL. Genomics. 1990;7:524. doi: 10.1016/0888-7543(90)90195-z. [DOI] [PubMed] [Google Scholar]
- 6.Levinson G, Gutman GA. Mol Biol Evol. 1987;4:203. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 7.Wierdl M, et al. Genetics. 1997;146:769. doi: 10.1093/genetics/146.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenberg SM, et al. Science. 1994;265:405. doi: 10.1126/science.8023163. [DOI] [PubMed] [Google Scholar]
- 9.Moxon ER, Wills C. Sci Am. 1999;280:94. doi: 10.1038/scientificamerican0199-94. [DOI] [PubMed] [Google Scholar]
- 10.Li YC, et al. Mol Ecol. 2002;11:2453. doi: 10.1046/j.1365-294x.2002.01643.x. [DOI] [PubMed] [Google Scholar]
- 11.Weising K, et al. Nucleic Acids Res. 1989;17:10128. doi: 10.1093/nar/17.23.10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milbourne D, et al. Mol Gen Genet. 1998;259:233. doi: 10.1007/s004380050809. [DOI] [PubMed] [Google Scholar]
- 13.Zhang L, et al. Bioinformatics. 2004;20:1081. [Google Scholar]
- 14. ftp://ftp.ncbi.nlm.nih.gov/genomes/Arabidopsis_thaliana.
- 15.Jurka J, Pethiyagoda C. J Mol Evol. 1995;40:120. doi: 10.1007/BF00167107. [DOI] [PubMed] [Google Scholar]
- 16.Subramanian S, et al. Genome Biol. 2003;4:R13. doi: 10.1186/gb-2003-4-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. www.perl.org.
- 18.Mukund V, et al. Mol Biol Evol. 2001;18:1161. [Google Scholar]
- 19.Steffens NO, et al. Nucleic Acids Res. 2004:D368. doi: 10.1093/nar/gkh017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung S, et al. Funct Integr Genomics. 2005;5:136. doi: 10.1007/s10142-005-0139-0. [DOI] [PubMed] [Google Scholar]