Abstract
LEA (late embryogenesis abundant) proteins are associated with tolerance to water stress resulting from desiccation and cold shock. Although various functions have been proposed to LEA proteins, their precise role is not fully defined. In silico analysis of the amino acid sequence of two LEA proteins (early methionine-labeled Vigna, EMV) from the tropical legume crop, Vigna radiata identified a 20 residues motif ‘GGQTRKQQLGSEGYHEMGRK’ characteristic to group 1 LEA proteins. Structural analyses hypothesize these proteins to function like DNA/RNA binding proteins in protecting macromolecules/ membrane stabilization at the time of dehydration process.
Keywords: LEA proteins, Vigna radiate, 20-mer motif, function assignments, DNA/RNA binding proteins
Background
Some proteins are highly expressed during late stage(s) of seed development and are referred to as LEA (late embryogenesis abundant) proteins. These proteins are found in a wide range of plant species and are suggested to involve in desiccation tolerance based on their accumulation and physicochemical properties. [1] LEA-type proteins fall into a number of families with diverse structures and functions, that differ in the arrangement and number of conserved motifs. These proteins are also hydrophilic in nature, and are transcriptionally regulated in response to ABA. [2] Prediction of secondary structures suggests that these proteins exist as largely unfolded molecules in their native state although a few members do exist as dimers or tetramers. [3] The precise function of LEA-type proteins is largely unknown. However, their considerable synthesis during the late stage of embryogenesis, induction by stress and other biophysical characteristics, such as hydrophilicity, random coils and repeating motifs permit prediction of some of their possible functions. LEA-type proteins are reported to act as water-binding molecules, in ion sequestration and in macromolecule and membrane stabilization. [2,4]
LEA proteins are ubiquitous among photosynthetic organisms and have been reported in mono- and dicot plants as well as in nematodes, yeast, bacteria and cyanobacteria. [5] These proteins are encoded by multigene families with different number of conserved residues motifs arranged in tandem as reported in cotton, maize, barley, Arabidopsis, mung bean, soybean etc. [ 6] Our earlier work showed the occurrence of early-methionine (Em)-labeled proteins in the mung bean (Vigna radiata) axes, referred as EMV proteins, the first ever report of such proteins in the Fabaceae family. [7] The cDNAs encoding these proteins were isolated, characterized and found to show certain level of similarity with other Em/ LEA proteins.
The results of an in silico analyses of these proteins based on their deduced amino acid sequences prove that these belong to group 1 LEA protein with possible DNA/RNA binding function. Such a property may facilitate hydrogen bonding of these proteins with essentially any macromolecule or membrane thus protecting the internal structures of the cell from being damaged due to altered physiological conditions.
Methodology
Datasets
The Em protein sequences of Vigna radiate, (EMV) - EMV1 and EMV2 (NCBI GenBank accession numbers U31210 and U31211; UniProt acc. Nos. Q41684 and Q41685) and other sequences examined in this study were retrieved from the public databases, http://www.ncbi.nlm.nih.gov [8] and http://www.ebi.ac.uk. [9] Structurally homologous subsets of the experimentally determined 3D structures of the EMV proteins were retrieved from PDB (Protein Data Bank) and SCOP (Structural Classification of Proteins) databases.
Similarity Search and Pattern Recognition
BLAST searches of the Swiss-Prot TrEMBL and Uniprot-curated databases were performed using WU-BLAST2.0 algorithm developed by Washington University [10] in order to identify local alignments for the mung bean (Vigna radiate) proteins. Further analysis of sequences was performed with tools available in the ExPasy Server (http://www.expasy.org/tools). [11] The Interpro analysis, based on the PROSITE and Pfam databases, was done to identify sequence patterns associated with the protein family and to determine the modular architecture of these proteins. Physicochemical properties of the selected proteins were determined using the ProtParam tools. Hydropathy plots of the deduced proteins were determined based on Kyte and Doolittle values. [12]
Homology Modeling and Protein Function-Prediction
The secondary structure analysis was performed using the PELE program of the SDSC Biology workbench (http://workbench.sdsc.edu/ [13]); intrinsic disorders in the peptide sequences were identified by GLOBPLOT analysis based on Linding's values. [14] Tertiary structure of the Vigna radiata LEA proteins was modeled by submitting the deduced amino acid sequences of EMV proteins to the Computational Biology Service Unit, Cornell Theory Center, Cornell University, USA. The protein models were generated by aligning to the structural homologues in the fold recognition program of LOOPP v3.0 (Learning, Observing and Outputting Protein Patterns) server. [15] The quality of the protein models was assessed using PROCHECK. The visual displays of the models were performed by SwissPDB viewer. [11] Function assignments were made based on the structural homologues identified for the test EMV proteins.
Results and Discussion
Sequence homologues and presence of 20 residues long conserved motifs
WU-BLAST2 analysis of EMV proteins revealed the highest sequence identity to a group 1 LEA protein from the black locust, Robinia pseudoacacia (87 %; UniProt: P93509) followed by several other group 1 LEA sequences including Arabidopsis thaliana (83 %; UniProt: Q42489), water-stress protectant protein from Gossypium hirsutum (80 %; UniProt: Q03791), Em-like protein from Daucus carota (78 %; UniProt: Q5KTS7) and Quercus robur (79 %; UniProt: Q7XBA7). Analysis of EMV2 protein sequence showed maximum sequence identity (87 %) to Glycine max Em protein (UniProt: P93165) followed by Em protein from Robinia pseudoacacia (80 %; UniProt: P93510), LEA protein from Arachis hypogaea (83 %; UniProt: Q4U4M1) and Arabidopsis thaliana Em like protein GEA6 (81 %; EM6). In addition, sequences with similarity to EMV proteins were detected in the moss, Physcomitrella patens and in Bacillus subtilis, specific for the gsiB gene encoding a stress-related protein identified based on the glucose starvation-inducibility. [16] The other matches include hypothetical proteins from different organisms with less significant E-values.
The occurrence of a 20 residues long repeat in group 1 LEA proteins (Table 1) identified a 20-mer motif ‘GGQTRKQQLGSEGYHEMGRK’ at positions 44-63 and 64-83 in EMV1 and at position 44-63 in EMV2, characteristic in plants and other organisms indicating that these proteins belong to group1 LEA family based on the revised classification system proposed by Wise [17]. This remarkable conservation points to an important role of LEA proteins in stress adaptation.
Table 1. Characteristics of group1 LEA proteins from Vigna radiata and other sources.
| Species | Identity | E-Value | No. of amino acids | No. of 20-mer repeats | Consensus at amino acida |
pI | % negative aa residues | % positive aa residues | Hydro pathicityb | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | |||||||||
| V. radiata | Q41684 | 4e-29c | 112 | 2 | G | G | Q | T | R | K | Q | Q | L | G | S | E | G | Y | H | E | M | G | T | K | 8.86 | 17.0 | 18.8 | -1.421 |
| Q41685 | 5e-14c | 99 | 1 | G | G | QE | TA | R | K | E | Q | L | G | T | E | G | Y | K | E | M | G | RH | K | 6.62 | 19.2 | 19.2 | -1.497 | |
| R. pseudoacacia | P93509 | 2e-24c | 112 | 2 | G | G | H | T | R | K | Q | Q | L | G | S | E | G | Y | H | E | M | G | TR | K | 6.21 | 17.9 | 16.1 | -1.455 |
| Q. robur | Q7xBA7 | 2e-20c | 113 | 2 | G | G | Q | T | R | K | E | Q | L | G | H | E | G | Y | Q | E | MI | G | SR | K | 6.85 | 16.8 | 16.8 | -1.345 |
| A. hypogaea | Q45W85 | 6e-14c | 91 | 1 | G | G | Q | T | R | R | E | Q | L | G | T | E | G | Y | Q | E | M | G | R | K | 5.28 | 19.8 | 16.5 | -1.355 |
| G. max | P93165 | 1e-13c | 105 | 1 | G | G | Q | T | R | K | E | Q | L | G | T | E | G | Y | Q | E | M | G | R | K | 5.51 | 18.1 | 16.2 | -1.617 |
| S. cereale | Q9LD94 | 1e-11c | 93 | 1 | G | G | Q | T | R | R | E | Q | M | G | E | E | G | Y | S | E | M | G | R | K | 5.29 | 19.4 | 17.2 | -1.317 |
| A. thaliana | Q42489 | 2e-22c | 112 | 2 | G | G | Q | T | R | K | E | Q | L | G | H | E | G | Y | Q | E | I | G | H | K | 5.59 | 20.5 | 16.1 | -1.359 |
| Q07187 | 2e-40d | 152 | 4 | G | G | EQ | AT | R | K | E | Q | L | G | H | E | G | Y | Q | E | I | G | HR | K | 5.75 | 20.4 | 15.8 | -1.468 | |
| H. vulgare | Q02400 | 4e-41d | 133 | 3 | G | G | Q | T | R | K | Q | Q | L | G | S | E | G | Y | H | E | M | G | T | K | 5.38 | 21.8 | 17.3 | -1.479 |
| Q05191 | 2e-37d | 153 | 4 | G | G | EQ | T | R | K | E | Q | L | G | E | E | G | Y | R | E | M | G | HR | K | 5.58 | 21.6 | 18.3 | -1.551 | |
| T. aestivum | P42755 | 3e-31d | 93 | 1 | G | G | E | T | R | K | E | Q | L | G | E | E | G | Y | R | E | M | G | R | K | 5.14 | 21.5 | 17.2 | -1.265 |
| P08000 | 4e-29d | 93 | 1 | G | G | Q | T | R | R | E | Q | M | G | E | E | G | Y | S | E | M | G | R | K | 5.50 | 19.4 | 17.2 | -1.372 | |
| D. carota | P17639 | 5e-31d | 92 | 1 | G | G | Q | T | R | K | E | Q | L | G | G | E | G | Y | H | E | M | G | R | K | 6.74 | 17.4 | 17.4 | -1.365 |
| Z. mays | P46517 | 7e-30d | 91 | 1 | G | G | Q | T | R | R | E | Q | L | G | Q | Q | G | Y | S | E | M | G | K | K | 6.61 | 17.6 | 17.6 | -1.265 |
| O. sativa | P46520 | 3e-28d | 95 | 1 | G | G | Q | T | R | K | E | Q | M | G | E | E | G | Y | R | E | M | G | R | K | 5.57 | 17.9 | 16.8 | -1.369 |
| G. hirsutum | P09443 | 2e-26d | 102 | 1 | G | G | E | T | R | K | Q | Q | L | G | T | E | G | Y | Q | E | M | G | R | K | 5.49 | 17.6 | 15.7 | -1.265 |
| P. glauca | Q40864 | 4e-26d | 91 | 1 | G | G | Q | T | R | K | E | Q | I | G | S | E | G | Y | Q | E | M | G | R | K | 5.89 | 18.7 | 17.6 | -1.460 |
| R. sativus | P11573 | 4e-23d | 83 | 1 | G | G | N | T | R | K | E | Q | L | G | S | E | G | Y | Q | Q | M | G | R | K | 6.59 | 18.1 | 18.1 | -1.625 |
| H. annuus | P46514 | 2e-21d | 92 | 1 | G | G | Q | T | R | K | D | Q | L | G | T | E | G | Y | K | E | M | G | K | K | 5.36 | 21.7 | 19.6 | -1.688 |
| B. subtilis | P26907 | 7e-06d | 122 | 5 | G | G | E | AT | T | S | K | N | H | D | K | E | F | Y | Q | E | I | G | QE | K | 5.32 | 22.1 | 16.4 | -1.798 |
| P. patens | Q4FE77 | 1e-07c | 88 | 1 | G | G | Q | T | R | A | E | Q | L | G | H | E | G | Y | T | E | M | G | K | K | 4.80 | 17.0 | 11.4 | -0.977 |
aThe consensus sequence for each motif represents the most frequent amino acid at each position. When more than one amino acid occurs frequently, additional amino acids are listed if their frequency is >25%. Amino acids shown in the smaller font indicates less frequent amino acids. b Grand average of hydropathicity. c BLASTP analysis of Q41684 against the UniProtKB database. d BLASTP analysis of Q41684 against the Swiss-Prot /TrEMBL database.
EMV proteins are hydrophilic and belong to pfam00477 Cluster
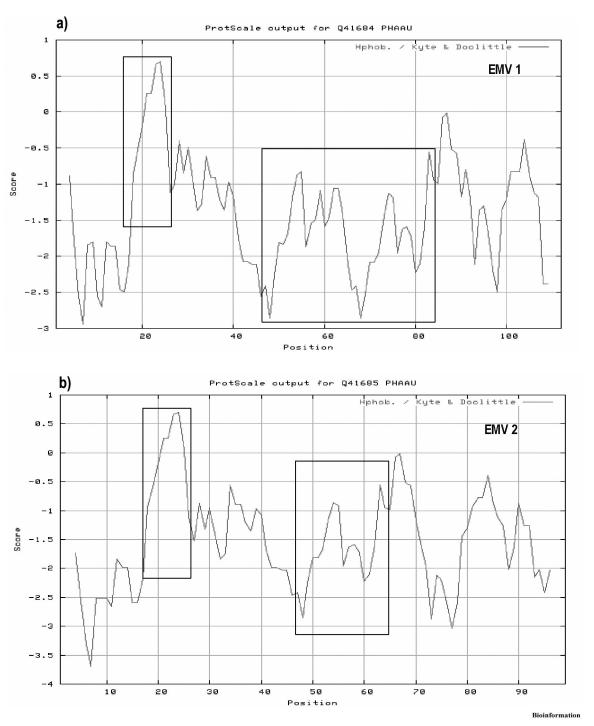
The accumulation of hydrophilic transcripts was demonstrated in E. coli and S. cerevisiae as well as from nematode and moss. Hydropathy plots revealed that EMV proteins are highly hydrophilic with over 95 % residues falling in the hydrophilic regions with negative scores (Figure 1). The grand average hydropathicity values of EMV1 (-1.421) and EMV2 (-1.497) suggest that these proteins are highly hydrated in aqueous environment. Profile search for Vigna LEA proteins revealed 78 % identity to LEA19 protein of Gossypium hirsutum; thus, classifying this protein into a pfam 00477 cluster (LEA-5 domain) with a small hydrophilic plant signature motif (GETWPGGT) at the residues 18-26 in both EMV1 and EMV2.
Figure 1.
Hydropathy analysis of predicted proteins EMV1 and EMV2 based on Kyte and Doolittle values, using a seven-residue window. Those values below zero are negative and obviously hydrophilic. Areas corresponding to highly conserved blocks are highlighted in boxes. (a) Hydropathy plot of EMV1 with regions from positions 18 to 26 indicate hydrophilic plant seed protein signature and positions 44 to 83 indicate two 20-mer repeat motifs arranged consecutively. (b) Hydropathy plot of EMV2 with regions from positions 18 to 26 indicate hydrophilic plant seed protein signature and positions 44 to 63 indicate a 20-mer repeat motif.
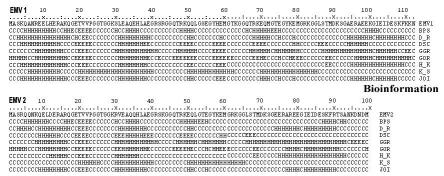
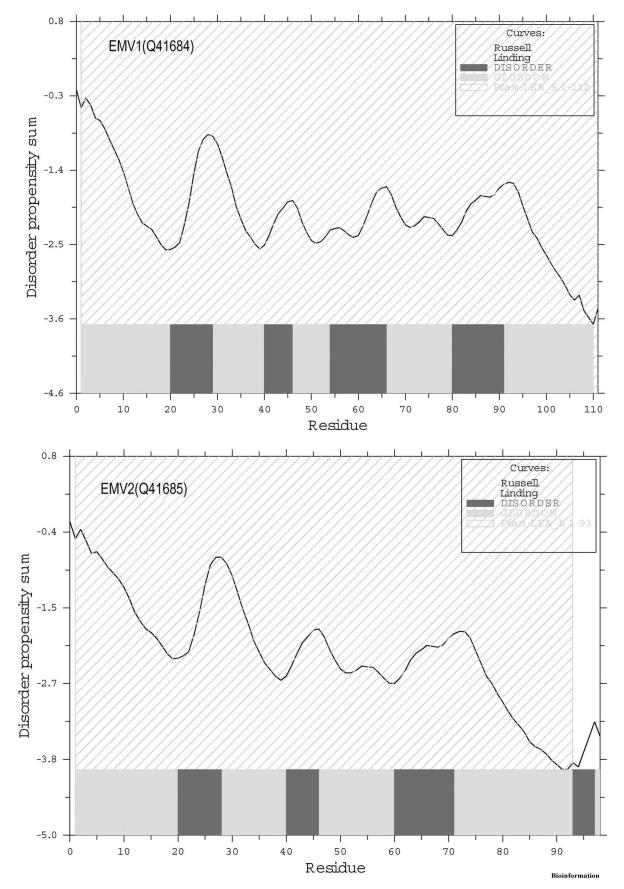
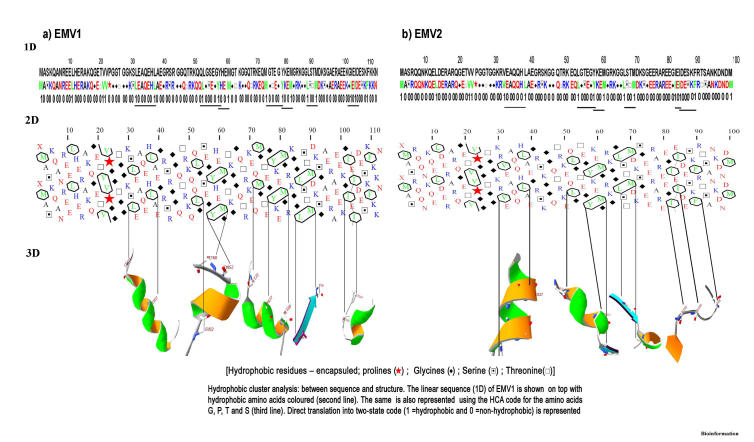
The structure prediction program and intrinsic disorder prediction suggest that EMV proteins maybe largely or entirely unstructured in solution. Firstly, the consensus identified for EMV proteins by structure prediction programs revealed predominantly random coil structure with two small regions of β sheets and five distinct helical blocks (Figure 2). Secondly, the GLOBPLOT analysis (Fig. 3) revealed intrinsic disorders in EMV1 (residue positions 20-29, 40-46, 49-66 and 80-91) and EMV2 (residues 20-28, 40-46, 60-71 and 93-97). Secondary structures were also observed as hydrophobic clusters and corresponded with the 1D and 3D representations (Figure 4). Low hydrophobic levels of proteins with relatively high overall charge are associated with a lack of compactness in proteins under physiological conditions resulting in a natively unfolded structure [18]
Figure 2.
Secondary structure analysis of the predicted EMV proteins performed with PELE program available on the SDSC Biology Workbench (http://workbench.sdsc.edu [13]). Seven different structure predictions are shown, with the most likely structural feature at each residue indicated by H (α-helices), E (β-sheets) or C (random coils). The programs used are denoted BPS, D_R, DSC, GGR, GOR, H_K and K_S. The “winner-takes-all” joint prediction was given by the JOI program.
Figure 3.
GLOBPLOT analysis of EMV proteins indicating the regions of intrinsic disorders and conserved LEA_5 domain along the length of sequence.
Figure 4.
Correspondence between 1D sequence, 2D HCA plot and 3D organization of secondary structures of EMV proteins.
Polypeptide chain flexibility and conserved double glycine residues
The internal hydrophilic motif of EMV proteins is flanked by the conserved double glycine residues with approximately 20 amino acid intervals giving a pattern in which the entire sequence of the mung bean LEA proteins could be viewed as consisting of 20 residues domains separated by structurally flexible double glycine residues. Variable number of hydrophilic motif suggests a higher water-binding capacity as seen by the presence of repeats that are most hydrophilic part of mung bean LEA proteins. This observation is in good agreement with the findings of [ 19] for the barley B19 LEA proteins.
Functional implications of Vigna LEA proteins
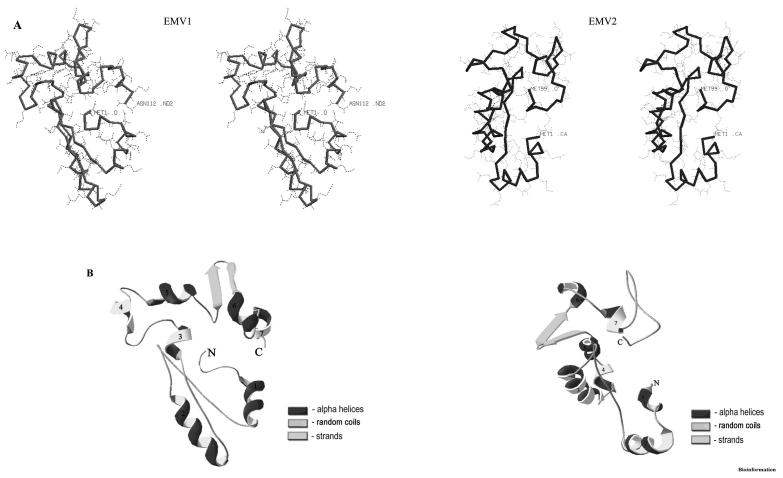
SCOP analysis showed that EMV proteins share structural homology to proteins with helical bundles of small proteins and DNA/RNA binding proteins (PDB: 2CCY, 1IG6 and 1EXE). Ab initio predictions indicate that 32.14% of the EMV1 and 32.32% of EMV2 protein attain helical conformation as represented by helical blocks in the 3D hypothetical models (Figure 5). Our earlier observation [7] of low molecular weight protein from dry mungbean embryonic axes showing aggregation of 12 kDa monomeric polypeptide into tetramer with apparent molecular weight of 50 kDa in the gel filtration studies corroborates the above finding. [9] Their highly conserved nature and stress induced structural transition suggest EMV proteins to resemble LEA proteins and consequently share similar hypothetical functions.
Figure 5.
Predicted structure of the EMV proteins. A) Backbone stereo view of EMV proteins (EMV 1- residues 1-112; EMV2- residues 1-99). B) Ribbon view of the representative EMV proteins structure (closest to average). Numbers represents the order of helices, and the N and C terminal of the protein are labeled. The structures were generated with the molecular graphics program Swiss PDB viewer.
Homology models validated by PROCHECK essentially satisfy the stereo-chemical parameters with well-refined structures at similar resolutions. [20] The distribution of residues in the most favored regions of the Ramachandran plot for EMV1 and EMV2 is 87.6 % and 93 %, respectively (Table 2). Thus, EMV1 is ‘fairly good’ while that of EMV2 is a ‘good’ hypothetical protein model. The homology model of mung bean LEA proteins, thus generated in this study, could aid in determining the mechanistic function of this important class of proteins.
Table 2. Ramachandran plot statistics of EMV proteins.
| Quadrangular regions of plot | Scattered residues | |||
|---|---|---|---|---|
| EMV 1 | EMV 2 | |||
| Number | Percentage | Number | Percentage | |
| Most favoured regions [A, B, L] | 78 | 87.6 | 76 | 93.8 |
| Additional allowed regions [a, b, l, p] | 8 | 9.0 | 3 | 3.7 |
| Generously allowed regions [~a, ~b, ~l, ~p] | 3 | 3.4 | 2 | 2.5 |
| Disallowed regions [XX] | 0 | 0.0 | 0 | 0.0 |
| Non glycine & non proline residues | 89 | 100.0 | 81 | 100.0 |
| End-residues (excluding Gly and Pro) | 2 | 2 | ||
| Glycine residues | 20 | 15 | ||
| Proline residues | 1 | 1 | ||
| Total number of residues | 112 | 99 | ||
Conclusions
Vigna radiata EMV proteins are classified under group 1 LEA proteins based on their extreme hydrophilicity and predominantly random-coiled arrangement of the residues along with the adoption of helical conformation as revealed by ab initio secondary structure predictions. Function assignments of these two LEA proteins suggest that they are involved in DNA/RNA binding action like other group 1 LEA proteins. Such a property may facilitate hydrogen bonding of these EMV proteins with essentially any macromolecule or membrane thus protecting the internal structures of the cell from being damaged due to altered physiological conditions. EMV proteins with the consistent spatial arrangements hence point to the possibility that they have a functional role in the plant's response to dehydration.
Acknowledgments
S.R is grateful to CSIR (Council of Scientific and Industrial Research) New Delhi, India for the Research Fellowship. Authors thank Dr. L. Arul for critical reading of the manuscript. We acknowledge the crews of NCBI, EBI, MRC Lab-UK and SIB for making computational biology data/tools publicly available.
Footnotes
Citation:Rajesh & Manickam, Bioinformation 1(4): 133-138 (2006)
References
- 1.Dure L, et al. Plant Mol Biol. 1989;12:475. doi: 10.1007/BF00036962. [DOI] [PubMed] [Google Scholar]
- 2.Ingram G, Bartels D. Annu Rev Plant Physiol Plant Mol Biol. 1993;47:377. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- 3.Ceccardi TL, et al. Protein Express Purif. 1994;5:266. doi: 10.1006/prep.1994.1040. [DOI] [PubMed] [Google Scholar]
- 4.Thomashow F. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:571. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- 5.Dure L. Protein Pept Lett. 2001;8:115. [Google Scholar]
- 6.Ramanjulu S, Bartels D. Plant Cell Environ. 2002;25:141. doi: 10.1046/j.0016-8025.2001.00764.x. [DOI] [PubMed] [Google Scholar]
- 7.Manickam A, et al. Physiol Plant. 1996;97:524. [Google Scholar]
- 8. http://www.ncbi.nlm.nih.gov.
- 9. http://www.ebi.ac.uk.
- 10.Lopez R, et al. Nucl Acids Res. 2003;31:3795. doi: 10.1093/nar/gkg573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. http://www.expasy.org/tools.
- 12.Kyte J, Doolittle RF. J Mol Biol. 1982;157:105. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 13. http://workbench.sdsc.edu/
- 14.Linding R, et al. Nucl Acids Res. 2003;31:3701. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teodorescu O, et al. Proteins Str Fun Genet. 2004;54:41. doi: 10.1002/prot.10474. [DOI] [PubMed] [Google Scholar]
- 16.Stacy R, Aalen R. Planta. 1998;206:476. doi: 10.1007/s004250050424. [DOI] [PubMed] [Google Scholar]
- 17.Wise M. BMC Bioinformatics. 2003;4:52. doi: 10.1186/1471-2105-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaboriaud C, et al. FEBS Lett. 1987;224:149. doi: 10.1016/0014-5793(87)80439-8. [DOI] [PubMed] [Google Scholar]
- 19.Espelund M, et al. Plant J. 1992;2:241. [PubMed] [Google Scholar]
- 20.Morris L, et al. Proteins. 1992;12:345. doi: 10.1002/prot.340120407. [DOI] [PubMed] [Google Scholar]