Abstract
Mycolyl-transferases are a family of proteins that are specifically present in the CMN (Corynebacterium, Mycobacterium and Nocardia) genera and are responsible for the synthesis of cell wall components. We modeled the three-dimensional structures of mycolyl-transfersases from Corynebacterium and Nocardia using homology modeling methods based on the crystal structures of mycolyl-transferases from M. tuberculosis. Comparison of the models revealed significant differences in their substrate binding site. Some mycolyl-transferases identified by the following Gene Ids: Nfa25110, Nfa45560, Nfa7210, Nfa38260, Nfa32420, Nfa23770, Nfa43800, Nfa30260, Dip0365, Ncgl0987, Ce1488, Ncgl0885, Ce0984, Ncgl2101, Ncgl0336, Ce0356 are associated with a relatively larger substrate binding site and amino acid residue mutations (D40N, R43D/G, S236N/A) are likely to affect binding to trehalose.
Keywords: CMN, Mycobacterium, Corynebacterium, Nocardia, mycolyl-transferases, homology modeling
Background
The CMN group constitutes the organisms of the genera Corynebacterium, Mycobacterium and Nocardia, which are grouped together on the basis of factors that include complex cell wall components, presence/type of mycolic acids, adjuvant activity, presence of cord factor, sulfo-lipids, iron-chelating compounds, polyphosphate, and serological cross-reactivity. The cell walls of the organisms that belong to the CMN group consists of interconnected peptidoglycan and polysaccharide-mycolate complex and are characterized by the presence of mycolic acid on their surface. [1] Mycolic acids are long chain fatty acids that form a part of the unique cell envelope, responsible for the pathogenesis and survival of the organism inside the host. The mycolic acids are named according to the individual genus from which they are isolated; i.e., corynomycolic acids from Corynebacterium comprising ~22-36 carbons, mycolic/eumycolic acids from Mycobacterium comprising ~60-90 carbons and nocardiomycolic acids from Nocardia comprising ~40-60 carbons. [2 –4]
In M. tuberculosis, the mycolyl-transferases are also termed antigen 85 or Ag85 complex enzymes. [5] These correspond to three secreted proteins; Ag85A (Gene Id: Rv3804), Ag85B (Gene Id: Rv1886) and Ag85C (Gene Id: Rv0129). These proteins comprise a signal peptide at the N-terminus followed by a carboxylesterase domain. It has been demonstrated that Ag85 enzymes catalyze the transfer of mycolyl residue from one molecule of α, α' TMM (trehalose monomycolate) to another leading to the formation of α, α' TDM (trehalose dimycolate) and hence these enzymes are termed mycolyl-transferases. [6] Also, in Corynebacterium and Nocardia, orthologous proteins synthesize TDCM (trehalose dicorynomycolate) and TDNM (trehalose dinocardiomycolate), respectively. Further, this family of enzymes is specific only to the CMN group of organisms because of their unique cell envelope. Mycolyl-transferases are also termed fibronectin-binding proteins, since they are involved in binding to fibronectin and entry of the organism into host cells. [7,8] Hence, it is important to understand the structure and function of the proteins responsible for the synthesis of cell wall components in CMN.
The structures of Ag85A (PDB Ids: 1SFR) [9], Ag85B (PDB Ids: 1F0N, 1F0P) [10] and Ag85C (PDB Ids: 1DQZ, 1DQY, 1VA5) [11] were determined for both native and substrate bound forms. The structure corresponds to a α/β hydrolase fold and the catalytic triad responsible for the mycolyl-transferase activity comprise the amino acid residues S126, E230 and H262 (numbering is according to PDB Id: 1F0P). The structural comparison of the three mycolyl-transferases (PDB Ids: 1SFR, 1F0P, 1DQZ) revealed that the active sites are virtually identical indicating that these share a common function. [9] However, in contrast to the high level of similarity within the substrate-binding site and the active site, it was observed that the surface residues disparate from the active site are quite variable indicating that all three Ag85 enzymes in M. tuberculosis are needed to evade the host immune system. The genome sequencing of M. tuberculosis [12], C. glutamicum [13], C. efficiens [14], C. diphtheria [15] and Nocardia farcinica [16] is completed. The M. tuberculosis comprising 3,986 genes is the causative agent of tuberculosis that causes 3 million deaths worldwide. The C. glutamicum comprising 3,002 genes is a soil bacterium and widely used by the industry in the production of amino acids. The C. efficiens comprising 3,069 genes is a non-pathogenic bacterium. The C. diphtheria comprising 2,320 genes is the causative agent of diphtheria. The genome of N. farcinica comprising 5,674 genes is the causative agent of nocardiosis, affecting the lung, central nervous system and cutaneous tissues of humans and animals.
In our earlier work [17], we identified mycolyl-transferases in C. glutamicum and C. efficiens genomes and modeled their three dimensional structures. We reported the relative binding of corynomycolyl-transferases towards trehalose. Our findings are in accordance with the experimental data [18, 19] that reported the gene deletion mutation studies and measured the concentration of TMCM / TDCM. The genomes of N. farcincia, a representative species from Nocardia and C. diphtheria were also subsequently sequenced and we now have complete data available in the public databases on all mycolyl-transferases from species that belong to the CMN group. Therefore we have carried out sequence analysis corresponding to all mycolyl-transferases and modeled the structures of Nocardia and C. diphtheria and compared their substrate binding sites. Such comparative analysis is relevant in situations when the structural information for proteins from only one organism is available and useful inferences can be made about the structure, function and nature of the substrate binding sites for related members from other organisms.
Methodology
Sequence data
The amino acid sequences corresponding to mycolyl-transferases from M. tuberculosis; Ag85A, Ag85B and Ag85C were obtained from the EBI (European Bioinformatics Institute) [20] and are represented by the following Ids; GI: 15610940, GI: 15609023, GI: 57116693, respectively as shown in Table 1.
Table 1. Mycolyl-transferases in CMN group.
| Gene Id | GeneBank Id | Source | Protein Length | % similarity | BLASTP E-value |
|---|---|---|---|---|---|
| Rv1886c | GI:15609023 | M. tuberculosis | 325 | 100 | 9e-173 |
| Rv3804c | GI:15610940 | M. tuberculosis | 338 | 90 | 1e-146 |
| Rv0129c | GI:57116693 | M. tuberculosis | 340 | 81 | 3e-123 |
| Rv3803c | GI:57117159 | M. tuberculosis | 299 | 52 | 2e-50 |
| Nfa1830 | GI:54022147 | N. farcinica | 345 | 53 | 5e-48 |
| Nfa1810 | GI:54022145 | N. farcinica | 347 | 51 | 2e-47 |
| Nfa1820 | GI:54022146 | N. farcinica | 353 | 48 | 1e-45 |
| NCgl2777 | GI:19554065 | C. glutamicum | 657 | 50 | 2e-44 |
| Ce2709 | GI:25029265 | C. efficiens | 669 | 52 | 5e-44 |
| Nfa1840 | GI:54022148 | N. farcinica | 624 | 50 | 1e-40 |
| NCgl2779 | GI:19554067 | C. glutamicum | 341 | 50 | 2e-38 |
| Dip2193 | GI:38234734 | C. diphtheriae | 638 | 49 | 3e-3 |
| Ce2710 | GI:25029266 | C. efficiens | 360 | 51 | 9e-3 |
| Dip2194 | GI:38234735 | C. diphtheriae | 338 | 49 | 7e-35 |
| Nfa5610 | GI:54022528 | N. farcinica | 319 | 48 | 2e-33 |
| Nfa30260 | GI:54024995 | N. farcinica | 341 | 45 | 8e-28 |
| Nfa32420 | GI:54025211 | N. farcinica | 351 | 44 | 9e-27 |
| Nfa38260 | GI:54025796 | N. farcinica | 353 | 42 | 2e-26 |
| Nfa7210 | GI:54022688 | N. farcinica | 340 | 42 | 4e-26 |
| Ncgl0987 | GI:19552252 | C. glutamicum | 411 | 45 | 8e-26 |
| Nfa25110 | GI:54024480 | N. farcinica | 311 | 45 | 5e-25 |
| Ce1488 | GI:25028044 | C. efficiens | 390 | 43 | 9e-24 |
| Dip0365 | GI:38232981 | C. diphtheriae | 355 | 43 | 1e-23 |
| Nfa45560 | GI:54026529 | N. farcinica | 324 | 44 | 4e-23 |
| Ncgl0885 | GI:19552148 | C. glutamicum | 483 | 43 | 5e-23 |
| Ncgl2101 | GI:19553383 | C. glutamicum | 483 | 43 | 8e-23 |
| Nfa23770 | GI:54024346 | N. farcinica | 339 | 42 | 4e-22 |
| Nfa43800 | GI:54026351 | N. farcinica | 337 | 43 | 9e-22 |
| Dip2339 | GI:38234873 | C. diphtheriae | 406 | 44 | 3e-20 |
| Ce0356 | GI:25026912 | C. efficiens | 381 | 41 | 5e-20 |
| Ce0984 | GI:25027540 | C. efficiens | 484 | 42 | 1e-19 |
| Ncgl0336 | GI:19551592 | C. glutamicum | 365 | 42 | 8e-18 |
Database searching
The homologous proteins were identified for the Mycobacterium, Corynebacterium, and N. farcinica using BLASTP [21] with the Ag85B as the query sequence against GenBank release 153 [22]. The BLOSUM62 matrices were used and the results were sorted using E-value (expected value) with the gap costs set to existence at 11 and extension at 1.
Multiple sequence analysis
Thirty-one mycolyl-transferase sequences were aligned using the CLUSTALW program [23] available at EBI. A penalty of 10 for gap opening, 0.05 for gap extension and 8 for gap separation (default parameters) was assigned for the alignment and shown in Figure 1.
Figure 1.
Multiple sequence alignment corresponding of CMN mycolyl-transferases. Conserved amino acid residues (*), sites of insertion (inverted triangle).
Homology modeling
The three-dimensional models were constructed using MODELER [24] available in InsightII (Accelrys Inc., USA). The structures of Ag85A (PDB Id: 1SFR), Ag85B (PDB Id: 1F0N) and Ag85C (PDB Ids: 1DQZ) were used as templates for modeling. MODELER is an automated comparative modeling program designed to find the most probable structure of a protein sequence, given its alignment with related structures. The model is obtained by the optimal satisfaction of spatial restraints derived from the alignment and is expressed as probability density function for the features restrained. The optimization procedure is a variable target function method that applies conjugate gradients algorithm to position all non-hydrogen atoms. [25] In all seventeen homology models were constructed for the mycolyl-transferases from N. farcincia and C. diphtheria species.
Model evaluation
The models were evaluated using PROCHECK. [26] The RMSD (root mean square deviation) values corresponding to topologically equivalent residues between the models and corresponding crystal structures obtained via structural superposition were derived using programs in InsightII (Accelrys Inc., USA) The method of Profiles-3D that measures the compatibility of an amino acid sequence to a protein of known three-dimensional structure was used to further assess the model. [27]
Substrate docking
The trehalose substrate was docked into the binding site of all protein models using QUANTA (Accelrys Inc., USA). The enzyme-substrate complex was refined using molecular mechanics (MM) and molecular dynamics (MD) calculations in order to understand their interactions. Hydrogen atoms were added to the structures at pH 7.00 using BIOPOLYMER in Insight II. The parameter ‘capping mode off’ was chosen so that the protein ends remain uncharged with the NH2 and COOH groups. The CVFF (Consistent Valence Force Field) force field was chosen and the ‘Fix’ option was used to select the potential atom types, partial charges and formal charges for the protein-substrate complex. The docked complex was subjected to energy minimization using 3000 steps steepest descent followed by conjugate gradient until an energy gradient < 0.01 kcal/mol/A0 was achieved. The energy minimized structures were further subjected for MD simulations which were performed in the canonical ensemble (NVT) at 298° K using CVFF force field implemented in Discover-3 and equilibrated for 3000 femtoseconds with step size of 1 femtosecond.
Results and Discussion
Sequence searches identified four mycolyl-transferases each in M. tuberculosis and C. diphtheria, six in C. glutamicum, five in C. efficiens, and thirteen in N. farcinica. The details of mycolyl-transferases analysed and modeled in this work are provided in Table 1. The mycolyl-transferases corresponding to the mycobacteria species; M. tuberculosis, M. leprae and M. bovis are highly similar. Therefore, the mycolyl-transferases from M. tuberculosis H37Rv strain are used in our analysis. Also, M. tuberculosis consists a mycolyl-transferase precursor protein MPT51 (Gene Id: Rv3803) that does not possess mycolyl-transferase activity [28] and was also therefore excluded from our analysis. The multiple sequence alignment of thirty-one mycolyl-transferases is shown in Figure 1. Despite low sequence similarity shared between these proteins, we observed 16 amino acid residues are conserved. These amino acid residues are; L39, W51, P71, D81, W82, W97, F100, G124, S126, S150, D192, G214, E230, G260, H262 and W264. The alignment also indicated some proteins have an insertion sequence of variable length (between 2 and 19 amino acid residues) that precedes the catalytic E230. Further, two N. farcinia proteins (Nfa1810 and Nfa1820) comprise a 27 amino acid residue insertion sequence rich in glycine and serine present between the conserved W82 and W97 (see Figure 1).
The three-dimensional models are useful to identify the positions of these highly conserved resides and regions of insertions. Further, we can also infer the nature of the substrate binding pockets defined by interactions with ‘trehalose’. Evaluation of the three-dimensional models corresponding to corynomycolyl-transferases and nocardiomycolyl-transferases according to PROCHECK indicated more than 85% amino acid residues are in the allowed regions of the Ramachandran plot [29] suggesting that the models are of good quality. Further, according to Profiles-3D, the ‘observed’ scores for the models lie between 124-134 as ‘expected’, suggesting the compatibility of structure and sequence. Also, the RMSD of the respective structures is ~0.68Å and residues that form the catalytic site S126, E230 and H262 can be highly superimposed. The conservation of catalytic residues and their positions in the three dimensional models indicated that all corynomycolyl transferases and nocardiomycolyl transferases must also retain catalytic activity. Examination of the models on computer graphics showed that, the conserved residues L39, P71, D81, W82, W97 and F100 constitute the ‘hydrophobic tunnel’. These are needed in order to accommodate the alkyl chain of mycolic acid, indicating a functional conservation in these proteins. The invariant S126 and G260 are close to the catalytic active site comprising E230. The indole side chains of W51 and W264 are perpendicular to each other and are in proximity to G124 associated with the β5 strand. The amino acid residue D192 is away from the active site indicating that the conservation extends beyond the catalytic site in CMN mycolyl-transferases. We observed that the disulphide connectivity patterns are different. The structures of 1SFR (Ag85A) and 1F0N (Ag85B) consist a disulphide bridge connecting half-cystine residues on β5 and β6 strands. In some proteins, half-cystine residue on the α10 helix and half-cystine residue on the loop connecting β6 strand and α6 helix are involved in the disulphide bridge. The information on the disulphide connectivity pattern is provided in Table 2. Based on the structural superposition, we observed that the differences between these structures are only in the loop regions. The 27 amino acid residue insertion in Nfa1810 and Nfa1820 is located between the β5 and β6 strands that is away from the active site and we therefore predict that it may not be involved in the activity of the protein. According to the structure of 1F0P (Ag85B bound to the substrate trehalose), two substrate binding pockets are present. We observed that the variable region preceding the E230 forms an “insertion loop” close to the trehalose 1151 binding site (Figure 1). The length and amino acid composition of this insertion loop is variable and is given in Table 2. The proteins with a long insertion loop formed a larger substrate binding pocket relative to the mycolyl-transferases. The corynomycolyl-transferases and nocardiomycolyl-transferases with larger substrate binding pocket are: Nfa7210, Nfa38260, Nfa32420, Nfa23720 Nfa43800, Nfa30260, Nfa45560, Nfa25110, Nfa5610, Ce0356, Ncgl0336, Dip0365, Ncgl2101, Ncgl0885 and Ce0984. In order to get an insight into the nature of interaction between the enzymes and substrate, trehalose was docked into the substrate binding site of all modeled structures and optimized using energy minimization. The specificity pockets defined by interaction with trehalose substrate were examined and the results are presented in Table 2. While some proteins retain the nature of residues lining the specificity pockets, mutations such as D40N, R43D/G, S236N/A are observed in Nfa25110, Nfa45560, Nfa7210, Nfa38260, Nfa32420, Nfa23770, Nfa43800, Nfa30260, Dip0365, Ncgl0987, Ce1488, Ncgl0885, Ce0984, Ncgl2101, Ncgl0336 and Ce0356. In these proteins specificity may be affected. Further, we observed that proteins with large substrate binding site were also associated with specific amino acid residue mutations. Therefore, in these proteins binding to trehalose is affected. Also, we observed that proteins comprising conserved amino acid residues in the substrate binding site are not associated with an insertion loop. Therefore, such proteins may bind trehalose.
Table 2. ‘Insertion loop’ amino acid sequence, disulphide bridges and substrate binding pockets in CMN mycolyl-transferases.
| Protein | ‘Insertion loop’ amino acid sequence | Disulphide bridge | Trehalose 1151 binding residues | Trehalose 1152 binding residues | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1F0P | Cys 87- | 40D | 43R | 126S | 223N | 262H | 263S | 264W | 154D | 157Q | 159M | 231N | 232F | 235S | 236S | 239K | |
| Cys 92 | |||||||||||||||||
| Rv0129 | 38D | 41R | 124S | 221N | 260H | 261S | 262W | 152N | 155E | 157W | 229G | 230L | 233R | 234T | 237T | ||
| Rv3804 | Cys 87- | 40D | 43R | 126S | 223N | 262H | 263S | 264W | 154D | 157Q | 159M | 231G | 232F | 235T | 236S | 239K | |
| Cys 92 | |||||||||||||||||
| Ncgl2777 | AIGPA | 40D | 43R | 121S | 216G | 261H | 262S | 263W | 149D | 152S | 154G | 231V | 232I | 235M | 236T | 239T | |
| Ce2709 | ATGPA | 40D | 43R | 121S | 215G | 261H | 262A | 263W | 149D | 152S | 154G | 231L | 232I | 235M | 236T | 239T | |
| Ncgl2779 | DH | 41D | 44R | 128S | 223V | 266H | 267G | 268W | 156N | 159A | 161G | 236F | 237V | 240T | 241S | 244I | |
| Ce2710 | DH | 41D | 44R | 128S | 223T | 266H | 267S | 268W | 156T | 159A | 161G | 236A | 237V | 240A | 241T | 244A | |
| Ncgl0987 | SEKEPFYN | 41D | 44G | 125S | 219D | 267H | 268N | 269W | 153S | 156D | 158I | 240S | 241C | 244A | 245L | 248S | |
| Ce1488 | YADEPFYN | 41D | 44G | 125S | 219E | 267H | 268N | 269W | 153S | 156D | 158I | 240S | 241C | 244A | 245L | 248A | |
| Ncgl0885 | DNAPIDEDAFKNR | 41G | 44D | 124S | - | 272H | 273A | 274W | 152E | 155S | 157M | 241A | 242M | 245T | 246C | 249N | |
| Ce0984 | ENAPEDEKGLKNR | 41G | 44D | 124S | - | 272H | 273A | 274W | 152E | 155S | 157M | 241A | 242L | 245T | 246C | 249N | |
| Ncgl2101 | DNAPIDEDAFKNR | 41G | 44D | 124S | - | 272H | 273A | 274W | 152E | 155S | 157M | 241A | 242M | 245T | 246C | 249N | |
| Ncgl0336 | SPRFEGLNQQVQSIAMAET | 41N | 44D | 124S | 218D | 276H | 277S | 278W | 152A | 155S | 157L | 246A | 247A | 250K | 251C | 254D | |
| Ce0356 | SPRFNGLDQAYLSLAMTET | 41N | 44D | 124S | 218N | 276H | 277S | 278W | 152S | 155Q | 157L | 246A | 247A | 250K | 251C | 254D | |
| Nfa1810 | FG | 40D | 43R | 153S | 249N | 291H | 292N | 293W | 181N | 184A | 186G | 260V | 261L | 264A | 265N | 268A | |
| Nfa1820 | FN | 40D | 43R | 148S | 244S | 286H | 287A | 288W | 176N | 179A | 181G | 255A | 256L | 259A | 260N | 263A | |
| Nfa1830 | SPVGVFN | 39D | 42R | 124S | 218N | 264H | 265S | 266W | 152N | 155A | 157G | 234A | 235L | 238V | 239N | 242A | |
| Nfa1840 | PGVST | 41D | 44R | 122S | 217S | 263H | 264S | 265W | 150T | 153T | 155G | 233I | 234L | 237L | 238T | 241N | |
| Nfa25110 | Cys 146- | 38A | 41G | 120S | - | 252H | 253T | 254W | 148W | 151D | 153P | 222A | 223I | 226T | 227C | 230A | |
| Cys 227 | |||||||||||||||||
| Nfa45560 | APGIDGNPLDLVER | Cys 146- | 38N | 41D | 120S | 241T | 266H | 267S | 268W | 148R | 151D | 153A | 237T | 238V | 241A | 242C | 245P |
| Cys 242 | |||||||||||||||||
| Nfa7210 | GPYALPGSYGLANQ | Cys 149- | 41N | 44G | 123S | 218N | 271H | 272S | 273W | 151Q | 154D | 156V | 241A | 242G | 245Y | 246C | 249N |
| Cys 246 | |||||||||||||||||
| Nfa38260 | GPHAMPGSDGLTNQ | Cys 150- | 41A | 44G | 123S | 217N | 270H | 271S | 272W | 151Q | 154D | 156V | 240A | 241G | 244H | 245C | 248N |
| Cys 246 | |||||||||||||||||
| Nfa32420 | YLNAAPGPMGAVN- | Cys 150- | 41N | 44D | 123S | 218Y | 270H | 271Y | 272W | 151Q | 154D | 156T | 240A | 241A | 244Q | 245C | 248N |
| Cys 246 | |||||||||||||||||
| Nfa23770 | NPRLHNDDRQLLNQ | Cys 157- | 41N | 44G | 130S | 224A | 278H | 279S | 280W | 158M | 161D | 163L | 247S | 248V | 241L | 252C | 255R |
| Cys 253 | |||||||||||||||||
| Nfa43800 | AVGGDPMQLGYQ | Cys 149- | 41N | 44S | 122S | - | 267H | 268A | 269W | 150R | 153D | 155Q | 237A | 238V | 241M | 242C | 245Q |
| Cys 243 | |||||||||||||||||
| Nfa30260 | GPGIDADPLALADQ | Cys 149- | 41N | 44T | 123S | 217Q | 270H | 271S | 272W | 151P | 154D | 156R | 240A | 241V | 244D | 245C | 248E |
| Cys 245 | |||||||||||||||||
| Nfa5610 | KPQLAEN | Cys 148- | 41D | 43D | 122S | 214L | 260H | 261S | 262W | 150D | 153L | 155T | 230V | 231G | 234I | 235C | 238A |
| Cys 235 | |||||||||||||||||
| Dip0365 | SPRLAGKDPVTIFATNLIT | 39N | 42D | 122S | 216S | 274H | 275S | 276W | 150A | 153S | 155L | 244A | 245G | 248M | 249C | 252D | |
| Dip2339 | PKEDGPFT | 41D | 44T | 125S | 219G | 269H | 270S | 271W | 153S | 156N | 158S | 240R | 241C | 244E | 245L | 248S | |
| Dip2193 | ANKKG | 40D | 43R | 121S | 215G | 261H | 262D | 263W | 149D | 152S | 154G | 231V | 232I | 235M | 236T | 239T | |
| Dip2194 | ND | 41D | 44R | 125S | 220Y | 263H | 264N | 265W | 153S | 156V | 158G | 233I | 234A | 237V | 238S | 241I | |
It is often observed that, during evolution, gene duplications, rearrangements and gene loss occur in genomes due to a complex, general purpose mechanism for rapid adaptation of the organism. As a result of gene duplication, extra copies of selected genes are evolved. Duplications are important because they effectively allow at least one of the gene copies to evolve while the function of the original gene can remain intact. Many new functions arise from duplication and subsequent change of old genes. In this way, duplication of pre-existing genetic information provides the raw material from which new gene functions can evolve thereby contributing to the genetic complexity during evolution. With reference to mycolyl-transferases in the CMN genera, the presence of varying number of proteins in each organism reflects gene duplication events during evolution of these organisms. Further, we identified that the overall structure, active site and hydrophobic tunnel are identical in all proteins, with significant differences in substrate specificity pockets which may be a result of selective pressure during evolution. From this work, we propose that trehalose is the original substrate and this binding is retained only in some corynomycolyl-transferases and nocardiomycolyl-transferases. During gene duplication, mutations in the substrate binding site have occurred such that the newly evolved proteins can bind to other sugars so as to synthesize organism specific polysaccharide-mycolate cell wall component.
Further, the mycolyl-transferases Nfa1840, Ncgl2777, Ce2709 and Dip2193 comprise a 300 amino acid residue C-terminal extension as a result of gene fusion events. Brand et al., 2003 reported that deletion of Ncgl2777 gene led to a 10-fold increase in the cell volume of the organism. We reported the identification of 55 amino acid residue tandem LGFP (conserved sequence motif; leucine, glycine, phenylalanine, proline) repeats in the C-terminal region of Ncgl2777 and Ce2709 [30] and suggested that the abnormal increase in the cell volume of C. glutamicum is due to the loss of C-terminal domain corresponding to the LGFP tandem repeats that may be responsible for maintaining the integrity of the cell wall. The presence of these LGFP repeats in C-terminal region of Nfa1840 and Dip2193 imply that these are also cell surface proteins and may be important in maintaining cell wall integrity in analogous manner.
Conclusion
This work describes the comparison of the three-dimensional models for mycolyl-transferases in CMN genera. Although the sequence identities in some cases is as low as 17%, yet the overall α/β fold characteristic of mycolyl-transferases is conserved. This conservation extends to the active site comprising amino acid residues; S126, E230 and H262. However, the amino acid residues comprising the substrate binding pockets defined by interactions with trehalose vary owing to certain mutations in some mycolyl-transferases. Also, significant differences are observed in the size of the substrate binding pocket owing to the close proximity of an insertion loop between the conserved W82 and W97. The size and nature of amino acid residues corresponding to the substrate binding pockets is likely to affect mycolyl-transferase substrate specificity. These observations lead us to believe that during the course of evolution, gene duplication events followed by mutagenesis at the substrate binding pockets, may have resulted in those mycolyl-transferases that are responsible for synthesis of polysaccharide-mycolate complex in an organism specific manner.
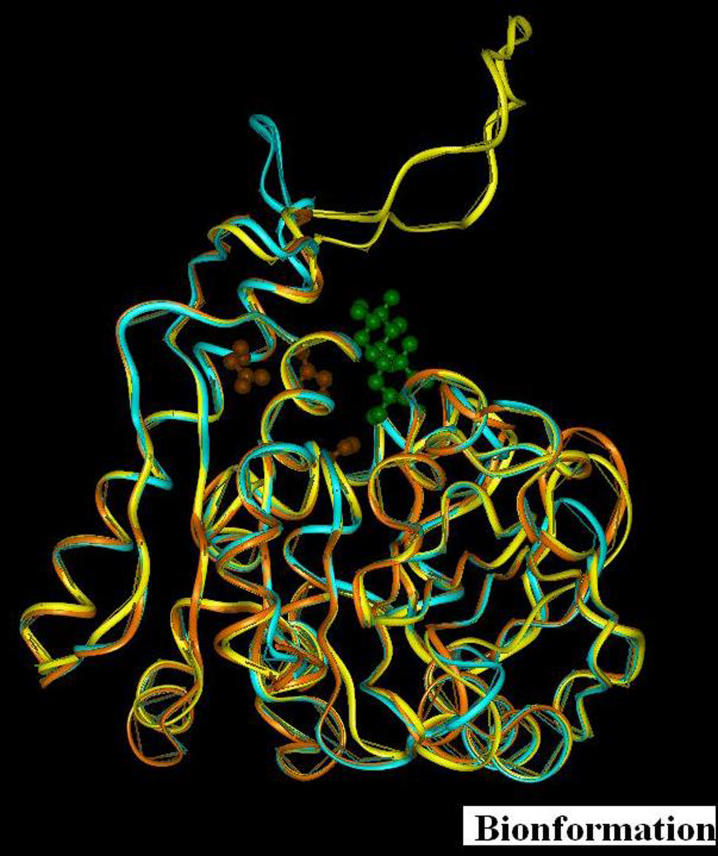
Figure 2.
The structural superposition of representative CMN mycolyl-transferases (PDB Id: 1F0P (brown), Ncgl0336 (yellow), Ncgl0987 (blue). The side chains of the active site residues S126, E230, H262 (red) and trehalose 1151 (green) are represented in ball and stick model.
Acknowledgments
HGR thanks UGC, New Delhi for a JRF fellowship. SA thanks CSIR New Delhi for a SRF fellowship. LGP thanks DBT, New Delhi for research funding. We thank referees for their valuable comments.
Footnotes
Citation:Ramulu et al., Bioinformation 1(5): 161-169 (2006))
References
- 1.Cocito C, Delville J. Eur J Epidemiol. 1985;1:202. doi: 10.1007/BF00234095. [DOI] [PubMed] [Google Scholar]
- 2.Collins MD, et al. J Gen Microbiol. 1982;128:129. doi: 10.1099/00221287-128-1-129. [DOI] [PubMed] [Google Scholar]
- 3.Daffé M, Draper P. Adv Microb Physiol. 1998;39:131. doi: 10.1016/s0065-2911(08)60016-8. [DOI] [PubMed] [Google Scholar]
- 4.Alshamaony L, et al. J Gen Microbiol. 1976;92:188. doi: 10.1099/00221287-92-1-183. [DOI] [PubMed] [Google Scholar]
- 5.Wiker HG, Harboe M. Microbiol Rev. 1992;56:648. doi: 10.1128/mr.56.4.648-661.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belisle JT, et al. Science. 1997;276:1420. doi: 10.1126/science.276.5317.1420. [DOI] [PubMed] [Google Scholar]
- 7.Abou-Zeid C, et al. Infect Immun. 1988;56:3046. doi: 10.1128/iai.56.12.3046-3051.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ratliff TL, et al. J Gen Microbiol. 1988;134:1307. doi: 10.1099/00221287-134-5-1307. [DOI] [PubMed] [Google Scholar]
- 9.Ronning R, et al. J Biol Chem. 2004;279:36771. doi: 10.1074/jbc.M400811200. [DOI] [PubMed] [Google Scholar]
- 10.Anderson H, et al. J Mol Biol. 2001;307:671. doi: 10.1006/jmbi.2001.4461. [DOI] [PubMed] [Google Scholar]
- 11.Ronning R, et al. Nat Struct Biol. 2000;7:141. doi: 10.1038/72413. [DOI] [PubMed] [Google Scholar]
- 12.Cole ST, et al. Nature. 1998;393:537. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 13.Kalinowski J, et al. J Biotechnol. 2003;104:5. doi: 10.1016/s0168-1656(03)00158-5. [DOI] [PubMed] [Google Scholar]
- 14.Kawarabayasi Y, et al. Unpublished. 2002.
- 15.CerdenoTarraga AM, et al. Nucleic Acids Res. 2003;31:6516. [Google Scholar]
- 16.Ishikawa J, et al. Proc Natl Acad Sci. 2004;101:14925. doi: 10.1073/pnas.0406410101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adindla S, et al. Int J Biol Macromol. 2004;34:181. doi: 10.1016/j.ijbiomac.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 18.Brand S, et al. Arch Microbiol. 2003;180:33. doi: 10.1007/s00203-003-0556-1. [DOI] [PubMed] [Google Scholar]
- 19.De SousaD' Auria C, et al. FEMS Microbiol Lett. 2003;224:35. [Google Scholar]
- 20. http://srs.ebi.ac.uk/
- 21.Altschul SF, et al. J Mol Biol. 1990;215:403. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22. http://www.ncbi.nlm.nih.gov/BLAST/
- 23.Higgins D, et al. Nucleic Acids Res. 1994;22:4673. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sali A, Blundell TL. J Mol Biol. 1993;234:779. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 25.Sanchez R, Sali A. Methods Mol Biol. 2000;143:97. doi: 10.1385/1-59259-368-2:97. [DOI] [PubMed] [Google Scholar]
- 26.Laskowski RA, et al. J Appl Crystallogr. 1993;26:283. [Google Scholar]
- 27.Luthy R, et al. Nature. 1992;356:83. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- 28.Kremer L, et al. Lett Appl Microbiol. 2002;34:233. doi: 10.1046/j.1472-765x.2002.01091.x. [DOI] [PubMed] [Google Scholar]
- 29.Ramachandran N, Sasisekharan V. Adv Protein Chem. 1968;23:283. doi: 10.1016/s0065-3233(08)60402-7. [DOI] [PubMed] [Google Scholar]
- 30.Adindla S, et al. Comp Funct Genomics. 2004;5:2. doi: 10.1002/cfg.358. [DOI] [PMC free article] [PubMed] [Google Scholar]