Abstract
In this paper, a revision for the existing method of locating exons by genomic signal processing technique employing four binary indicator sequences is presented. The existing method relies on the pronounced period three peaks observed in the Fourier power spectrum of the exon regions which are absent in non-coding regions. The authors have abandoned the four sequences all together and adopted a single ‘EIIP indicator sequence’ which is formed by substituting the electron-ion interaction pseudopotentials (EIIP) of the nucleotides A, G, C and T in the DNA sequence, reducing the computational overhead by 75%. The power spectrum of this sequence reveals period three peaks for exon regions. Also a number of exons have been identified which exhibit period three peaks when mapped to ‘EIIP indicator sequence’ and which do not show the same when the binary indicator sequences are employed. We could get better discrimination between exon areas and non-coding areas of a number of genomes when the sequences are mapped to EIIP indicator sequences and the power spectra of the same are taken in a sliding Kaiser window, compared to the existing method using a rectangular window which utilizes binary indicator sequences.
Keywords: fourier, locating exons, gene finding, electron-ion interaction pseudopotential (EIIP)
Background
The pivotal problem of gene identification in eukaryotes is distinguishing exons, from introns and intergenic regions. A number of coding measures like single and polynucleotide bias differences, spectral differences etc which exist between these regions have been utilized for this purpose in various gene finding algorithms. But simultaneous improvement of sensitivity and selectivity of these algorithms is still a challenge and so the hunt for new coding measures is to be continued.
The existing method of locating exons by genomic signal processing technique employing four binary indicator sequences, one for each nucleotide, depends on the period three peaks observed in the power spectrum of the exon regions and which do not exist in non-coding regions. The method may be summarized as given below. For a DNA string x[n] of N characters (with an alphabet A, G, C & T) let us define four binary indicator sequences uA[n], uG[n], uC[n] & uT[n]. [1] Each indicator sequence has a 1 if the corresponding base exists at the position n, otherwise a zero.
For example if, :as given in the PDF file linked below
This coding measure has been utilized in the program ‘Genescan’ [4] by evaluating the N/3 component of the Fourier power spectrum of the binary indicator sequences through a sliding window and looking for the peaks, whose strength (against the average strength of the power spectrum in the region) surpass a threshold which indicate the presence of exons. Also an optimization technique [1] has been devised for locating exons employing binary indicator sequences. Another notable work is where an anti-notch filter [5] is used to locate the exons by employing the four binary indicator sequences. A recent work reported [6], employs the Cumulative Categorical Periodogram (CCP) for the same end, but giving troughs at N/3 whereas the binary indicator sequences exhibit peaks.
Methodology
The authors propose a novel coding measure scheme by replacing the four binary indicator sequences by just one sequence which we call as ‘EIIP indicator sequence’. The energy of delocalized electrons in amino acids and nucleotides has been calculated as the Electron-ion interaction pseudopotential (EIIP). [7] The EIIP values of amino acids have already been used in Resonant Recognition Models (RRM) to substitute for the corresponding amino acids in protein sequences, whose Discrete Fourier Transforms are taken to extract the information contents. [7] The Fourier cross spectra of a group of related proteins reveal a sharp peak at a frequency which is termed as the ‘characteristic frequency’ of that group of proteins as they are found to represent a particular biological function and selectively interact with targets of the corresponding ‘characteristic frequency’ (resonant recognition). [7] This has been used to identify ‘hot spots’ in proteins and for peptide design which are very useful in drug discovery. In the present work, the authors have made use of the EIIP values of the nucleotides rather than those of aminoacids for locating exons. The EIIP values for the nucleotides are given in Table 1
Table 1. Electron Ion Interaction pseudo potentials of nucleotides.
| Nucleotide | EIIP |
|---|---|
| A | 0.1260 |
| G | 0.0806 |
| C | 0.1340 |
| T | 0.1335 |
If we substitute the EIIP values for A, G, C & T in a DNA string x[n], we get a numerical sequence which represents the distribution of the free electrons' energies along the DNA sequence. This sequence is named as the ‘EIIP indicator sequence’, xe[n]. For example, if x[n] = A A T G C A T C A, then using the values from Table 1, xe[n] = [0.1260 0.1260 0.1335 0.0806 0.1340 0.1260 0.1335 0.1340 0.1260].
When Se[k] is plotted against k, it reveals a peak at N/3 for a coding region and no such peak is observable for a noncoding region. As it is evident, the method has been simplified and computational overhead is reduced by 75% as now we have to find the Fast Fourier Transform (FFT) of only one sequence instead of the FFT of four binary sequences used in the original method. This may be used as a coding measure to detect probable coding regions in DNA sequences by examining the local signal to noise ratio of the peak within a sliding window and by selecting an appropriate threshold.
Genescan [4] takes an optimal window size of 351 and the same is adopted in the present investigation. The authors have also experimented with both reduced and increased window sizes. When reduced window size is used, peaking areas become ‘sharper’ which is advantageous for detection when exons are closer (separated by comparatively shorter introns) but the subsequent increase in noise makes the discrimination poorer. On the other hand, increasing the window size makes the peaking areas wider and thus resulting in missing of exons which are closer. Instead of a rectangular window adopted by Genescan, the authors have taken Kaiser window which suppresses the noise more effectively as it has much smaller side lobes compared to rectangular window, and the binary indicator sequences are replaced by a single EIIP indicator sequence.
Results and Discussion
The authors have checked the power spectrum of several exon segments of eukaryotic genes in a number of organisms using binary sequence indicators and the proposed EIIP indicator sequence. Mainly, two data sets are used as bench mark for this purpose. One is the dataset prepared by Burset and Guigó [8,9] and the other is HMR195 [10] prepared by Sanja Rogic. In a good number of cases both methods performed well, but there are instances were EIIP indicator sequence shows the peak at the right location (near N/3) where binary sequences fail, and a few number of instances where the opposite is true, and of course there are a number of genes where both fail which proves that there exist many exons without appreciable N/3 peak. Table 2 lists some of the exons which give period three peaks when EIIP indicator sequence is employed and where the existing binary indicator sequence method fails.
Table 2. Exons from selected genes where EIIP indicator sequence gives better N/3 peaks compared to binary indicator sequences.
| Serial No. | Accession number | Description Of gene | Length of sequence | Exon area & length (N) | Comments: (a) Using binary; (b) Using EIIP |
|---|---|---|---|---|---|
| 1 | AF019074 | EKLF, mus musculus erythroid kruppel like factor gene | 6350 | 3761-4574 (814) | Peak in (a) at 131 (not near N/3), Peak in (b) at 272 (near N/3). |
| 2 | AB009589 | Human gene for Osteomodulin | 12414 | 10624-10949 (326) | Peak in (a) at 8 (not near N/3), Peak in (b) at 110 (near N/3). |
| 3 | AF065986 | Human keratocan gene | 7659 | 6638-6810 (173) | Peak in (a) at 40 (not near N/3), Peak in (b) at 53 (near N/3). |
| 4 | AF015224 | Human mammoglobin gene | 4206 | 1713-1900 (188) | Peak in (a) at 18 (not near N/3), Peak in (b) at 63 (near N/3). |
| 5 | AB016625 | Human OCTN2 gene | 25871 | 15591-15792 (172) | Peak in (a) at 76 (not near N/3), Peak in (b) at 56 (near N/3). |
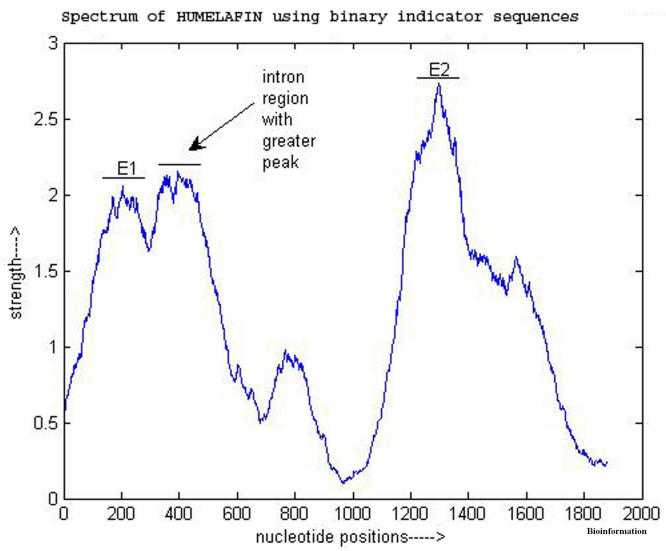
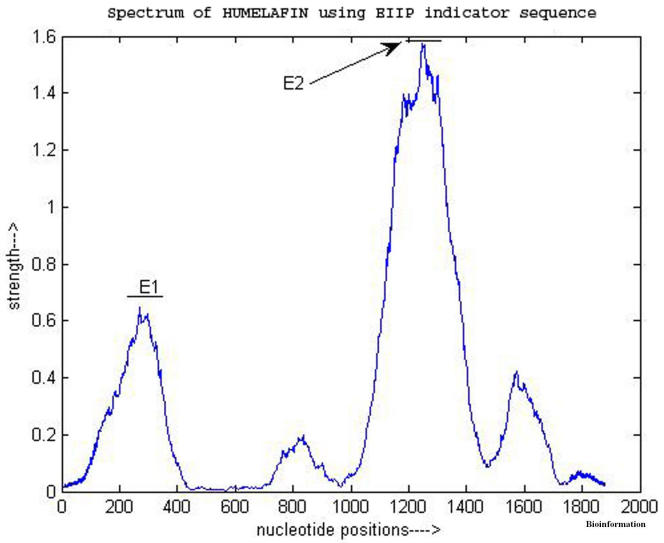
The experiments using sliding windows show that in a number of cases the EIIP indicator sequence gives a better discrimination between coding and non-coding regions. Figure 1 and Figure 2 show the power spectrum of a gene, HUMELAFIN (Acc. No. D13156, homosapiens gene for elafin), using binary indicator sequences and using EIIP indicator, respectively. HUMELAFIN has two exons, one from nucleotide positions 245 to 325, and the other from 1185 to1459. As it is evident from Figure 1, a ‘false exon’ (an intron region having greater peak than exon regions) appears when binary indicator sequences are used and the peak of first exon (at 205) is also seen shifted from the actual region. On the contrary, the use of EIIP indicator sequence has ‘removed’ the ‘false’ exon and the peak of first exon is now inside the right region as seen from Figure 2. However, the second exon peak is exhibited by both methods correctly.
Figure 1.
Power spectrum of HUMELAFIN (D13156) obtained using binary indicators
Figure 2.
Power spectrum of HUMELAFIN (D13156) obtained using EIIP indicator
Table 3 Summarizes the observations about nine genes where EIIP indicator sequence is found to be a better discriminator than the binary indicator sequences. The last two columns in Table 3 are for comparing the performances of the methods in terms of an exon-intron discrimination measure D given by, D=Lowest of theexonpeaks/Highest peakin noncoding regions
Table 3. Examples of genes whose power spectra show better discrimination between coding and non-coding regions with EIIP indicator sequence mapping than with binary indicator sequence mapping.
| No | Gene Name, Acc. No, Description | Regions (Nucleotide positions) | Highest peak (binary slide) | Highest peak (EIIP slide) | Discrimination measure D for binary slide | Discrimination measure D for EIIP slide |
|---|---|---|---|---|---|---|
| 1. | F56F11.4a,NC001135, a gene from C. elegans chromosome III | E1(929-1135) | 2.1 | 1.02 | 1.19 | 2.0 |
| E2(2528-2857) | 7.01 | 2.75 | ||||
| E3(4114-4377) | 6.0 | 2.4 | ||||
| E4(5465-5644) | 5.3 | 1.1 | ||||
| E5(7255_7605) | 3.4 | 1.25 | ||||
| Intron regions | 1.77 | 0.51 | ||||
| 2. | HUMBETGLOA L26462, human betaglobin A chain | E1(866-957) | 1.86 | 0.84 | 1.05 | 2.4 |
| E2(1088-1310) | 4.34 | 0.9 | ||||
| E3(2161-2289) | 3.0 | 1.13 | ||||
| Intron regions | 1.77 | 0.35 | ||||
| 3. | HUMCBRG, M62420, Homosapiens carbonyl reductase gene | E1(276-566) | 9.74 | 1.74 | 0.55 | 1.16 |
| E2(1112-1219) | 1.3 | 0.43 | ||||
| E3(2608-3044) | 6.67 | 1.0 | ||||
| Intron regions | 2.36 | 0.37 | ||||
| 4. | HUMELAFIN, D13156, Homo sapiens gene for elafin | E1(247-325) | 2.05 | 0.65 | 0.95 | 1.55 |
| E2(1185-1459) | 2.72 | 1.575 | ||||
| Intron regions | 2.15 | 0.42 | ||||
| 5. | GalR2, AF042784 Mus musculus galin receptor type 2 gene | E1(24-388) | 9.6 | 1.27 | 3.19 | 14.11 |
| E2(1449-2199) | 3.19 | 0.917 | ||||
| Intron regions | 1.0 | 0.09 | ||||
| 6. | PP32R1, AF00A216 Homosapiens candidate tumor suppressor gene | E(4453-5157) | 30.6 | 11.92 | 19.13 | 21.67 |
| Intergenic regions | 1.6 | 0.55 | ||||
| 7. | HMX1, AF009614, Mus musculus homeobox containing nuclear transcriptional factor gene | E1(1267-1639) | 4.76 | 1.71 | 2.14 | 2.80 |
| E2(3888-4513) | 7.09 | 3.94 | ||||
| Intron regions | 2.22 | 0.61 | ||||
| 8. | PSMB5, AB003306, Mus musculus DNA for PSMB5 | E1(1020-1217) | 3.82 | 0.95 | 1.43 | 3.17 |
| E2(2207-2513) | 2.10 | 1.05 | ||||
| E3(4543-4832) | 7.65 | 2.12 | ||||
| Intron regions | 1.47 | 0.38 | ||||
| 9. | HSODF2, X74614, Homosapiens ODF2 gene | E1(280-599) | 3.82 | 0.865 | 2.43 | 4.33 |
| E2(843-1275) | 6.725 | 1.75 | ||||
| Intron regions | 1.57 | 0.2 |
Higher the value of D better is the discrimination. If D is more than one, all exons are identified without ambiguity, D less than one indicates that at least one exon is not having enough strength to be distinguished from noncoding areas. In all the examples cited, Method using EIIP indicator sequence shows a better discrimination compared to the method using binary indicator sequences. And in two cases, (HUMCBRG and HUMELAFIN) binary indicator sequence method even fails to identify all exons.
Conclusion
Ab initio gene finding still remains a challenging and exciting field as homology searches fail to identify around 30 to 50 % genes in newly sequenced genomes and none of the existing ab inito methods (methods using Hidden Markov Models are considered to be superior) are found to have enough sensitivity and selectivity for a fail-proof prediction. The method presented in this paper which uses electron-ion interaction pseudopotentials of nucleotides in genomic signal processing method of gene finding, improves the discrimination capability of the existing method and obviously reduces the computational complexity.
The coding measure scheme using EIIP indicator sequence thus can be utilized for gene finding procedures using genomic signal processing assisted by the grammar of genes and position weight matrices (PWMs) for splice sites. Also the possibility of applying the potential of EIIP sequence to other exon prediction techniques such as Autoregressive modeling (AR), Average Magnitude Difference Function (AMDF) and Time Domain Periodogram (TDP) etc can also be explored. Thus, we hope, the fact that a physico-chemical property like EIIP has a role in the formation of protein coding regions of genomes will trigger a lot of research in related areas.
Supplementary Material
Footnotes
Citation:Nair & Sreenadhan, Bioinformation 1(6): 197-202 (2006)
References
- 1.Anastassiou D. Bioinformatics. 2000;16:12. doi: 10.1093/bioinformatics/16.12.1073. [DOI] [PubMed] [Google Scholar]
- 2.Silverman BD, Linsker R. J Theor Biol. 1986;118:3. doi: 10.1016/s0022-5193(86)80060-1. [DOI] [PubMed] [Google Scholar]
- 3.Yin C, Yau ST. J Comput Biol. 2005;12:9. doi: 10.1089/cmb.2005.12.1153. [DOI] [PubMed] [Google Scholar]
- 4.Tiwari S, et al. Comput Appl Biosci. 1997;13:3. doi: 10.1093/bioinformatics/13.3.263. [DOI] [PubMed] [Google Scholar]
- 5.Vaidyanathan PP, Yoon BJ. Journal of the Franklin Institute. 2004;341:1. [Google Scholar]
- 6.Nair AS, Mahalakshmi T. In Silico Biology. 2006;6:0019. [PubMed] [Google Scholar]
- 7.Cosic I. IEEE Trans Biomed Eng. 1994;41:12. doi: 10.1109/10.335859. [DOI] [PubMed] [Google Scholar]
- 8.Burset M, Guigó R. Genomics. 1996;34:3. doi: 10.1006/geno.1996.0298. [DOI] [PubMed] [Google Scholar]
- 9. http://genome.imim.es/datasets/genomics96.
- 10. http://www.cs.ubc.ca/~rogic/evaluation/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.