Abstract
DoD2006, an updated version of Database of Databases is an online resource maintained collectively by ProGene Biosciences and Department of Inorganic and Analytical Chemistry, Andhra University. It links to all molecular biology databases that appeared in Nucleic Acids Research 2006 database issue. DoD2006 includes 873 databases, of which, 858 are derived from Nucleic Acids Research database issue and 15 are collected from In Silico Biology, Bioinformation journals and Google Scholar search. Each database has a search option, keyword help and a brief description with direct link to the database home page. The database is freely available online at http://www.progenebio.in/DoD/index.htm
Keywords: database, molecular biology, biological databases
Background
Online availability of databases is increasing due to their utility by structural and molecular biologists. These databases are generated using a variety of programming languages and they differ in content and access methods. [1] The information explosion in biology resulted in database proliferation and this necessitates the importance of data integration, storage and retrieval. Many new databases tend to appear every year, with ever increasing information or knowledge from biology, driven by genome sequencing projects and easy internet access. [2]
On the contrary, fund crunching is a limitation for data collection, constant updates, infrastructure facilities and maintenance of databases. Biological databases with infrastructural funding are hosted by academic, governmental organizations and industry as well as non-profit organizations. [3] Collection and analysis of data from various databases is essential for computational biologists. But it is time consuming and proportional with increasing number of databases. In this context, there is a need to integrate all databases at a single interface, usually with one search engine to query all databases. Therefore, we partially address the issue by maintaining the molecular biology databases at a single interface, with links to their respective pages. In practice, there are both legal and technical impediments to cross-database communication which include digital locks, firewalls and the inability of search engines to recognize the data. [4]
Methodology
DoD2006 Content
DoD2006 is an updated version of DoD. [5] It is a cross-database interface with 858 databases collected from Nucleic Acids Research [6], Bioinformatics [7], BMC Bioinformatics [8] and other journals [9] and 15 databases collected from In Silico Biology [10], Bioinformation [11] journals and Google Scholar [12] search. The current update with 873 databases against 719 reported in 2005 [5] represents additional databases that continue to grow despite the demise of a few. [13] A small number of inaccessible databases (MitoPD, ISSD [14]) reported in 2005 are unchanged in DoD2006 so as to enable the users to know the type of database that once existed.
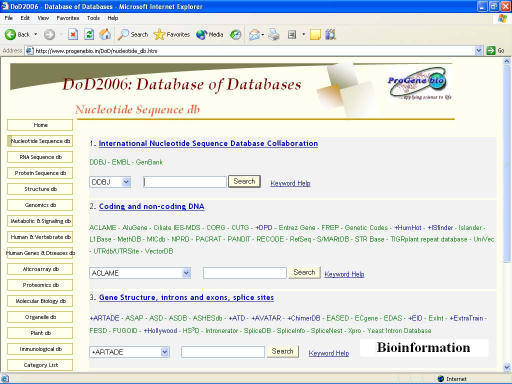
The complete list of databases is given in the website [15] and their categorization is given in Table 1. During the search, results are displayed in a separate window as this helps in further database scan without losing the home page. The screen-shot of nucleotide database is given in Figure 1. A hyperlink to ‘Keyword help’ lists keywords used to scan respective databases and the description of different links is given in our earlier publication. [5]
Table 1. List of database categories with number of database entries in each category.
| S. No. | Database Category (No. of Entries)* |
|---|---|
| 1. | Nucleotide Sequence Databases (94) |
| 2. | RNA Sequence Databases (52) |
| 3. | Protein Sequence Databases (138) |
| 4. | Structure Databases (82) |
| 5. | Genomics Databases (non-vertebrate) (198) |
| 6. | Metabolic & Signaling Pathways (68) |
| 7. | Human and other Vertebrate Genomes (87) |
| 8. | Human Genes and Diseases (100) |
| 9. | Microarray Data and other Gene Expression Databases (58) |
| 10. | Proteomics Resources (14) |
| 11. | Other Molecular Biology Databases (27) |
| 12. | Organelle Databases (22) |
| 13. | Plant Databases (72) |
| 14. | Immunological Databases (29) |
* Database entries are repeated in many cases as they fall simultaneously in more than one category
Figure 1.
Screen-shot of the DoD2006 showing sub-categories of Nucleotide Sequence Databases
Features of DoD2006
Newly appended databases in DoD2006 are distinguished from the existing ones with a ‘+’ sign and inaccessible ones are recognized by a ‘*’ sign before the name. All databases in each category are given alphabetically. The appended databases are colored blue and the remaining in green color. Those who wish to have their database to be listed can contact the authors.
DoD2006 provides an excellent and essential resource for molecular and computational biologists. Research through computational tools and software using databases has been recognized as a front-line research coupled with experimental design. Our database suits to deliver the wide biological content through the cross-database interface. Updates to DoD shall be made on yearly basis.
Footnotes
Citation:Babu et al., Bioinformation 1(6): 228-230 (2006)
References
- 1.Köhler J, et al. DDT: BIOSILICO. 2004;2:61. [Google Scholar]
- 2.Merali Z, Giles J. Nature. 2005;435:1010. doi: 10.1038/4351010a. [DOI] [PubMed] [Google Scholar]
- 3.Ellis LB, Kalumbi D. Nat Biotechnol. 1998;16:1323. doi: 10.1038/4296. [DOI] [PubMed] [Google Scholar]
- 4.Konforti B. Nature Structural & Molecular Biology. 2005;12:633. doi: 10.1038/nsmb0904-802. [DOI] [PubMed] [Google Scholar]
- 5.Babu PA, et al. In Silico Biol. 2005;5:605. [PubMed] [Google Scholar]
- 6. http://nar.oxfordjournals.org/
- 7. http://bioinformatics.oxfordjournals.org/
- 8. http://www.biomedcentral.com/bmcbioinformatics/
- 9.Galperin MY. Nucleic Acids Res. 2006;34:D3. doi: 10.1093/nar/gkj162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. http://www.bioinfo.de/isb/
- 11. http://www.bioinformation.net/
- 12. http://scholar.google.com/
- 13. http://www.infobiogen.fr/
- 14.Adzhubei IA, Adzhubei AA. Nucleic Acids Res. 1999;27:268. doi: 10.1093/nar/27.1.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. http://www.progenebio.in/DoD/index.htm.