Abstract
Peptides are of great therapeutic potential as vaccines and drugs. Knowledge of physicochemical descriptors, including the partition coefficient logP, is useful for the development of predictive Quantitative Structure-Activity Relationships (QSARs). We have investigated the accuracy of available programs for the prediction of logP values for peptides with known experimental values obtained from the literature. Eight prediction programs were tested, of which seven programs were fragment-based methods: XLogP, LogKow, PLogP, ACDLogP, AlogP, Interactive Analysis's LogP and MlogP; and one program used a whole molecule approach: QikProp. The predictive accuracy of the programs was assessed using r2 values, with ALogP being the most effective (r 2 = 0.822) and MLogP the least (r2 = 0.090). We also examined three distinct types of peptide structure: blocked, unblocked, and cyclic. For each study (all peptides, blocked, unblocked and cyclic peptides) the performance of programs rated from best to worse is as follows: all peptides – ALogP, QikProp, PLogP, XLogP, IALogP, LogKow, ACDLogP, and MlogP; blocked peptides PLogP, XLogP, ACDLogP, IALogP, LogKow, QikProp, ALogP, and MLogP; unblocked peptides QikProp, IALogP, ALogP, ACDLogP, MLogP, XLogP, LogKow and PLogP; cyclic peptides LogKow, ALogP, XLogP, MLogP, QikProp, ACDLogP, IALogP. In summary, all programs gave better predictions for blocked peptides, while, in general, logP values for cyclic peptides were under-predicted and those of unblocked peptides were over-predicted
Keywords: partition coefficient, logP, peptides, octanol, biphasic system, QSAR
Background
Peptides play a pre-eminent role in biological systems. However, as complex biological molecules, their physical properties have not received the attention that they deserve. In particular, their study by QSAR lags someway behind that of organic small molecules. QSAR has, traditionally, focussed on experimentally determined partition coefficients as a principal descriptor of lipophilicty or hydrophobicity. The partition coefficient is the ratio between the concentration of a chemical substance in two phases: typically one aqueous and one an organic solvent. Experimental measurement involves dissolving a compound within a biphasic system and determining its molar concentration in each layer:
P = [drug]organic / [drug]aqueous
The organic solvent used is usually 1-octanol. The partition coefficient can range over 12 orders of magnitude, and is usually quoted as a logarithm: logP. It is generally assumed that the log P of the neutral species is 2-5 log units greater than that of the ionized form, and that this is sufficiently large that the partitioning of the charged molecule into the organic phase can be neglected.
Despite problems with properly measuring logP values, they represent a potentially vital source of descriptors for QSAR studies of peptides. However, the experimental measurement of logP values is expensive, time consuming, and labour intensive. Accurate methods for the prediction of peptide logP values would thus be most useful. During the past three decades, many methods for predicting logP have been reported. At present, the most widely accepted method is a fragmental or additive approach, where a molecule is dissected into fragments (functional groups or atoms) and its logP value is obtained by summing the contributions of each fragment. ‘Correction factors’ are also introduced to rectify the calculated logP value when special substructures occur in the molecule.
Fragment-based methods are the most common. In fragment-based methods, a complex compound is divided into a series of small, simple fragments, such that each atom contained within the compound is present in one, and only one, fragment. The logP value for each fragment is known. Additive methods can also be based upon adding the logP values of each atom within the compound, rather than from a series of fragments. This alleviates the problem of missing fragments. An example of an atom based prediction method is XLOGP developed by Wang. [1,2] Other approaches are based upon the use of topological indices and quantum mechanics.
There have been various studies carried out on the logP prediction for peptides. Maybe the most convincing approach was undertaken by Akamatsu and co-workers [3], which investigate the hydrophobicity for peptides by carefully measuring the partition coefficients of a wide variety of peptides. After studying these data with linear regression analysis, they obtained different regression models for different kinds of peptides, resulting in a good correlation between observed and predicted logP values. Various physicochemical parameters are used in their models, including structural effects, β-turn formation corrections, N- and C-terminal effects, etc. This work and subsequent work by Akamatsu's group was incorporated into a logP prediction program known as PlogP. [4 ] Here, a training set included 219 blocked and unblocked peptides, varying between 2 and 5 amino acids in length. The model was further tested with 10 more peptides.
Various studies have compared the performance of different logP prediction programs. However, no study focussing on peptides has been reported. In this paper we look at prediction of logP values for peptides. It obviously focuses on a different aspect of this prediction problem compared to the prediction of properties of small molecules, which is the more typical focus for workers in the field. Our main motivation is to better understand basic physico-chemical properties in the design of peptide vaccines. Here we take a data-set of experimentallydetermined peptide logPs and use this to compare eight publicly and commercially available programs, based on 7 fragment and 1 whole-molecule based methods for logP prediction.
Methodology
Data-set
A set of peptides with known experimental logP values was compiled from the primary literature, through exhaustive, semimanual searching of a variety of different databases: PubMed [ http://www.ncbi.nlm.nih.gov/entrez/query.fcgi] , Web of Science [ wos.mimas.ac.uk], Medline [medline.cos.com], and ScienceDirect ( http://www.sciencedirect.com/). Both keyword and author searches, as well as retrospective searching, and citation matching of key authors, particularly those describing the development of an assay system, were used to identify papers detailing quantitative experimentally-derived values. The availability of measured LogP values for peptides was limited. The dataset consisted of 340 peptides, varying from 2 to 16 amino acids in length, and included 141 blocked peptides, 158 unblocked peptides, and 41 cyclic peptides.
Software Analysed
Seven fragment-based (XLogP [1 ,2], LogKow [www.syrres.com], PLogP [4], ACDLogP [ www.acdlabs.com], ALogP [www.vcclab.org], IALogP [www.logp.com] and MLogP [ www.tripos.com]) and one whole molecule approaches (QikProp [ www.schrodinger.com]) were studied in our analysis. These were downloaded or accessed on-line during June-August 2003. Methods implemented pre-defined general models for logP calculation, a peptide specific logP model, and a type of inhouse trainable model for peptide logP prediction. We used software either via internet servers or as versions installed locally. As the input requirements of each program were different, various representations of the structures were created: amino acid sequences for use with PlogP; SMILES strings [5] for ALogP, LogKow and IAlogP; 2D SYBYL ‘mol2’ files for XLOGP [www.tripos.com]; 3D structures for QikProp and MlogP. 3D Structures were generated using Corina. [6]
Results and Discussion
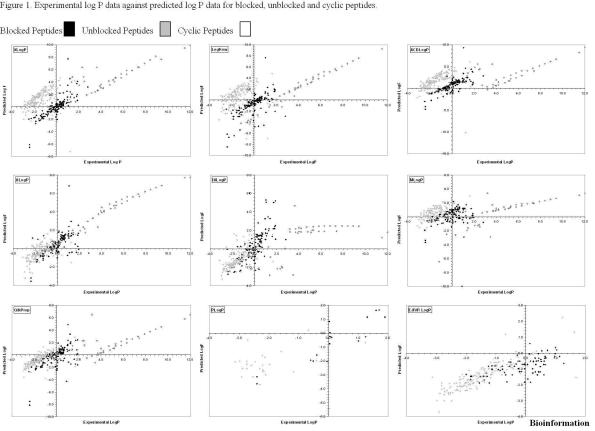
Peptides within the dataset varied widely, with large ranges of physical size and formal charge. There appeared to be no statistically significant relationship between the length of peptide and their respective logP values. Table 1 summarises results for all logP predictions. Figure 1 shows plots of experimental versus predicted logP. A list of the 379 peptide structures comprising the dataset, together with results from the various methods, is recorded in the online supplementary material (URL: http://www.jenner.ac.uk/Bioinformatics/peptide_structures.htm) .Overall, it was the fragment based method ALogP that performed the best (r 2 = 0.819). It predicted values for blocked peptides with very high accuracy (r2 = 0.822). The whole molecule method, QikProp, seems comparable to the fragment-based methods and shows a similar overall performance to PLOGP, IALogP and XLogP. QikProp is a 3D structure based method. Between -3 and 1 log units, predicted logP is well correlated with experiment. For peptides with experimental values of greater than 4 log units, QikProp's accuracy decreases and it predicts unblocked peptides poorly (r2 = 0.560). PLOGP is parameterised for peptides, has been trained on some of the dataset, and can not predict values for cyclic or chemically modified peptides. Thus the program was tested with only 44 peptides (17 blocked and 27 unblocked). Statistically the results from both types of peptides were poorly correlated, r2 = 0.482. Unblocked peptides are predicted poorly (r2 = 0.009) but blocked peptides (r2 = 0.800) are very well predicted. PLOGP was trained on peptides with five or fewer amino acids, and predictions of shorter peptides are slightly better. For XlogP, results for the whole data set were poor (r2 = 0.428), while results for the blocked and cyclic peptides (r2 = 0.665, 0.665 respectively) were reasonable; the unblocked peptides (r2 = 0.158), however, showed a much weaker correlation. IALogP produces the worst predictions for the cyclic peptides (r2 = 0.399). The program seems best at predicting values in the range -3 to 2 log units, although it over-predicted higher valued peptides. This group of four programs all show equal performance and are ranked second to ALogP, although the score for ALogP is somewhat better. Fragment based methods are, however, easier to use than QikProp, do not require training, and do not require any prior knowledge apart from the peptide structures. For LogKow, statistics for the whole dataset are poor (r2 = 0.277). Unblocked and blocked peptides were predicted unsuccessfully (r2 = 0.063 and r2 = 0.389), yet predicted cyclic peptide values very well (r2 = 0.970). For ACDLogP, blocked peptides are predicted with reasonable accuracy (r2 = 0.587) albeit predicting slightly higher than the experimental logP values. The results of the cyclic peptide (r2 = 0.462) are particularly interesting, showing a split into two distinct groups: one under-predicted, the other over-predicted. The least effective program in general was MLogP. The overall results were poor (r2 = 0.090). MLogP shows the poorest correlation for blocked and unblocked peptides, r2 = 0.060 and 0.170 respectively, although predictions are better for the cyclic peptides (r 2 = 0.661).
Table 1. Statistical Results.
| Program | No. Peptides | No. Blocked Peptides | No. Unblocked Peptides | No. Cyclic Peptides | Total | Blocked | Unblocked | Cyclic | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| r2 | RMSE | r2 | RMSE | r2 | RMSE | r2 | RMSE | |||||
| XLogP | 335 | 140 | 157 | 38 | 0.428 | 2.253 | 0.665 | 1.009 | 0.158 | 3.043 | 0.665 | 1.648 |
| LogKow | 339 | 141 | 158 | 40 | 0.277 | 2.315 | 0.389 | 1.709 | 0.063 | 2.781 | 0.970 | 2.141 |
| ACDLogP | 336 | 140 | 156 | 40 | 0.232 | 2.663 | 0.587 | 1.278 | 0.166 | 3.443 | 0.462 | 2.734 |
| AlogP | 335 | 138 | 157 | 40 | 0.822 | 1.211 | 0.382 | 0.673 | 0.394 | 0.897 | 0.946 | 0.457 |
| IALogP | 339 | 141 | 157 | 41 | 0.422 | 1.772 | 0.497 | 1.209 | 0.409 | 0.869 | 0.399 | 4.272 |
| MLogP | 338 | 140 | 158 | 41 | 0.090 | 2.351 | 0.060 | 1.402 | 0.170 | 2.272 | 0.661 | 4.411 |
| QikProp | 327 | 134 | 154 | 39 | 0.502 | 1.665 | 0.384 | 1.285 | 0.560 | 1.081 | 0.535 | 3.643 |
| PLogP | 44 | 17 | 27 | 0.482 | 1.267 | 0.800 | 1.040 | 0.009 | 1.391 | |||
Figure 1.
Experimental log P data against predicted log P for blocked, unblocked and cyclic peptides
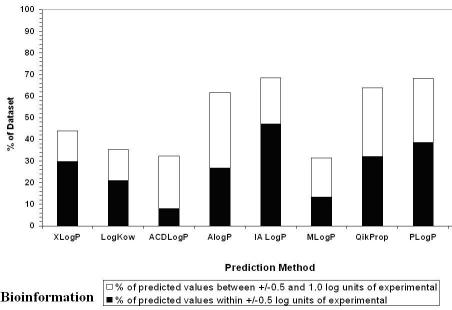
We have also calculated the percentage of predictions within +/-0.5 and between +/-0.5 and 1.0 log unit respectively of the experimental value. See Figure 2. The best accuracy within +/-0.5 log units is the IALogP method (47%). This is followed by PLogP (39%), QikProp (32%), XLogP (30%), ALogP (27%), LogKow (21%), MLogP (13%) and ACDLogP (8%). The best accuracy from between +/-0.5 and 1.0 log units is the ALogP method (35%) . This is followed by QikProp (32%), PLogP (30%), ACDLogLogP (24%), IALogP (21%), MLogP (18%), XLogP and LogKow (both at 14%).
Figure 2.
The percentage of values predicted within +/-0.5 and between +/-0.5 and 1 log unit, respectively of the experimental value
Comparing blocked, unblocked and cyclic peptides, we see that blocked peptides performed well and unblocked peptides performed worst. Unblocked peptides will be zwitterionic. The difficulties with some of these prediction methods are due to internal constraints: peptides over a certain length or those with bulky termini could not be predicted with certain programs. Far fewer cyclic peptides were studied and were usually under-predicted. Certain peptides were consistent outliers, such as the poly-lysine peptide and peptide 352A, a blocked acylated dipeptide. These may result from gross experimental error, as accurate values were not encountered for any method.
However, 1-octanol is, in reality, a poor choice of organic phase. It is “wet”, since it contains much dissolved water, and does not effectively separate hydrophobic from other interactions. Its relevance to biological systems is open to question, and many have suggested that measuring the partition into other organic phases, such as phospholipids bilayers or micelles, may prove a more rewarding avenue for seeking biologically-relevant measures of peptide hydrophobicity.
Conclusion
Fragment-based methods are sensitive to the composition but not the sequence of peptides, and any future peptide-specific logP studies should account for this. Accuracy could be improved using consensus scoring where multiple predictions are combined, by averaging or weighting, to generate better estimates. However, available methods, though inadequate, particularly for long peptides, perform better than might be imagined naively: fragmental methods are sufficient. There is little, if any, need to develop new peptide-specific treatments of the problem, such as PlogP [4], merely a need to improve fragment-based techniques and validate their use with peptides.
Our interest in this problem stems from our desire for effective measures of hydrophobicity for use in peptide QSAR studies. [7] There is a clear paucity of quality data for partition coefficients, necessitating an unsatisfying study. The dearth of reported experimental studies prevents us from obtaining a dataset of sufficient size. The peptides we found are short and have heavily biased sequence compositions. Data are both sparse and tendentious in terms of length and sequence properties. Longer peptides are of most interest, yet they are underrepresented here. The average peptide length was three amino acids, as it becomes increasingly difficult to measure logP values experimentally as peptides grow longer. As most biologically-important peptides are much longer than three amino acids, the data set is likely to compromise successful QSAR analysis. Such problems would be resolved with a properly designed training set. Our potential ability to combine in vitro and in silico analysis would allow us to improve both the scope and power of our predictions, in a way impossible using solely literature data.
Acknowledgments
The Jenner Institute (University of Oxford), formerly the Edward Jenner Institute for Vaccine Research, wishes to thank its past sponsors: GlaxoSmithKline, the Medical Research Council, the Biotechnology and Biological Sciences Research Council, and the UK Department of Health.
Footnotes
Citation:Thompson et al., Bioinformation 1(7): 237-241 (2006)
References
- 1.Wang R, et al. J Chem Inf Comp Sci. 1997;37:615. [Google Scholar]
- 2.Wang R. User manual for XLOGP v2.0. China: Peking University; 1999. [Google Scholar]
- 3.Akamatsu M, Fujita T. J Pharm Sci. 1992;81:164. doi: 10.1002/jps.2600810213. [DOI] [PubMed] [Google Scholar]
- 4.Peng T, et al. J Mol Model. 1999;5:189. [Google Scholar]
- 5.Weininger D. J Chem Inf Comp Sci. 1988;28:31. [Google Scholar]
- 6.Sadowski J. J Gasteiger Chem Rev. 1993;93:2567. [Google Scholar]
- 7.Guan P, et al. J Med Chem. 2005;48:7418. doi: 10.1021/jm0505258. [DOI] [PubMed] [Google Scholar]