Abstract
Connective tissue growth factor (CTGF) is a downstream mediator of transforming growth factor-β1 (TGF-β1) and thus a potential target for antifibrotic treatment strategies. CTGF is up-regulated in disorders such as atherosclerosis, scleroderma, and fibrosis of kidneys and lungs. We investigated the temporospatial expression patterns of CTGF and TGF-β1 mRNA in rat livers with acute fibrogenesis (after a single dose of CCl4) and with advanced fibrosis (6 weeks after complete bile duct occlusion). Multiprobe ribonuclease protection assay revealed increasing TGF-β1 and CTGF mRNA levels 6 hours after injection of CCl4, with peak levels after 72 hours. In biliary fibrosis TGF-β1 and CTGF mRNA levels increased fourfold and sevenfold, respectively (P < 0.001). In situ hybridization combined with cell-specific markers revealed CTGF transcripts in desmin-positive cells after a single dose of carbon tetrachloride, whereas no transcripts were found in normal livers. In biliary fibrosis, however, proliferating bile duct epithelial cells were the predominant source of CTGF mRNA. We conclude that in rat liver fibrogenesis CTGF is up-regulated in close association with TGF-β1 and that, contrary to a previous report, not solely hepatic stellate cells but activated bile duct epithelial cells are the main source of this profibrogenic factor.
Connective tissue growth factor (CTGF) is a cysteine-rich polypeptide identified originally in human umbilical vein endothelial cells by its chemotactic and mitogenic effects on fibroblasts. 1 In vitro and in vivo experiments have shown that expression of CTGF in endothelial cells and fibroblasts is induced by transforming growth factor-β1 (TGF-β1), but not by other mediators such as platelet-derived growth factor, epidermal growth factor, or basic fibroblast growth factor. 2-5 CTGF shares biological properties with TGF-β1 such as triggering rat kidney (NRK) fibroblast proliferation and up-regulating mRNA expression of procollagen α1(I), fibronectin, and the fibronectin receptor, integrin α5β1. Unlike TGF-β1, however, CTGF does not stimulate anchorage-independent growth of NRK fibroblasts or inhibit growth of mink lung epithelial cells, indicating that CTGF does not share all biological activities with TGF-β1. 2,6 In this line CTGF seems to be a downstream mediator for only some actions of TGF-β1, notably promotion of fibroblast proliferation and extracellular matrix production. 7,8 Co-injection of CTGF in subcutaneous tissue of mice enhances and perpetuates the fibrotic response after injection of TGF-β1. 9 Because of its potentiating effect in organ fibrosis and because of its specific expression by and action on mesenchymal cells, CTGF is considered an attractive target for antifibrotic therapies.
Several studies demonstrated that CTGF mRNA is coordinately expressed with TGF-β1 in fibrogenic lesions such as atherosclerosis, 10 scleroderma, 11-14 and fibrosis of kidneys, 15 lungs, 16 and intestine. 17 Recently, hepatic stellate cells (HSCs) were identified as major cellular source of CTGF in liver fibrogenesis, 18 in which TGF-β1 is a prominent profibrogenic cytokine. 19 However, CTGF transcripts in activated HSC were only shown for a single case of (human) liver cirrhosis. By studying the temporospatial expression of CTGF in rats with acute and chronic hepatic fibrogenesis, we identified bile duct epithelia as a dominant source of CTGF mRNA in liver subsequent to bile duct occlusion (BDO).
Materials and Methods
Cloning of Partial Rat CTGF cDNA by Reverse Transcription-Polymerase Chain Reaction
Total liver RNA was reverse transcribed with an oligo-dT primer and Superscript II reverse transcriptase (Life Technologies, Karlsruhe, Germany) according to the manufacturer’s instructions. Rat CTGF cDNA was amplified with primers CAA CCG CAA GAT TGG AGT GT and CTC CAG TCT GCA GAA GGT ATT G according to positions 398 to 417 and 806 to 827 of the mouse CTGF sequence (fisp-12, GenBank accession number: M70642). 7,20 Polymerase chain reaction products were cloned into the EcoRV site of pZErO-1 and sequenced on both strands. The rat sequence of the CTGF amplicon has been submitted to GenBank/EMBL (accession number AJ236872). The rat protein displays 95% homology to mouse, human, and porcine CTGF.
Animal Models
Animal experiments were performed in accordance with local institutional and governmental regulations on the use of experimental animals. Wistar rats were maintained in 12-hour light/dark cycles at 23°C with a humidity of 60 ± 10%. Fifteen male rats (∼125 g; Animal Production Facilities, Schoenwalde, Germany) received a single intraperitoneal injection of CCl4 (1.25 ml/kg body weight), mixed with an equal volume of vegetable oil. Groups of three animals were sacrificed 6, 12, 24, 48, and 72 hours after the injection, and three untreated rats served as controls. In eight female rats (∼250 g) the common bile duct was injected in a retrograde manner with the sclerosant sodium amidotriazoate (0.2 ml/kg body weight, Ethibloc; Ethicon, Norderstedt, Germany) as reported previously. 21,22 Another eight rats, in whom the abdominal cavity was opened and resealed, were used as sham-operation controls. After 6 weeks, rats were sacrificed and aliquots of right and left liver lobes were snap-frozen in liquid nitrogen.
In Situ Hybridization Combined with Immunohistochemistry
Plasmids containing rat CTGF (430 bp, see above), TGF-β1 (a 570-bp fragment from position 987 to 1550 in X52498), kindly provided by Dr. X. L. Tian, Max Delbrueck Center for Molecular Medicine, Berlin, Germany) 23 and rat procollagen α1(I) (a 1.3-kb PstI/HindIII-fragment of plasmid α1R1, provided by Dr. D. Rowe, Department of Pediatrics, University of Connecticut Health Center, Farmington, CT, subcloned in pGEM (Promega, Mannheim, Germany) 24 were linearized prior to transcription. Antisense and sense (negative control) RNA probes were prepared by in vitro transcription with [35S]-UTP (NEN, Cologne, Germany) and used for in situ hybridization combined with immunohistochemistry on a single section as described before. 25-27 Briefly, 5-μm frozen sections were mounted onto 3-aminopropyl triethoxysilane-coated slides and fixed in 4% paraformaldehyde/phosphate-buffered saline (PBS) for 20 minutes, followed by washes in PBS, dehydration in graded ethanol, and air-drying. After prehybridization, slides were hybridized with antisense or sense probes at 52°C for 16 hours, followed by high stringency washing, and RNase A digestion to remove unhybridized RNA. For double labeling, sections were stained with monoclonal antibodies to desmin (DAKO, Hamburg, Germany) by the alkaline phosphatase anti-phosphatase (APAAP) method before prehybridization under RNase-free conditions. Sections were dehydrated and dipped into photo emulsion (Amersham, Braunschweig, Germany), followed by autoradiographical exposure at 4°C for 1 to 2 weeks, development, fixation, and counterstaining with hematoxylin and eosin.
RNA Isolation and Multiprobe Ribonuclease Protection Assay
RNA was extracted from snap-frozen liver tissues by the single-step method of Chomczynski and Sacchi. 28 Concentration and purity of RNA were determined by spectrophotometry at 260/280 nm and integrity of RNA was verified by visualization of the 18S and 28S rRNA bands after agarose (1.4%) electrophoresis and ethidium bromide staining.
[α-32P]-UTP (NEN)-labeled antisense riboprobes for rat CTGF (the full length of our cloned sequence), glyceraldehyde-3- phosphate dehydrogenase (GAPDH) 29 and TGF-β1 were prepared by in vitro transcription and purified by polyacrylamide gel electrophoresis. The ribonuclease protection assay (RPA) was performed as described 22 using the RPA II kit (Ambion, Austin, TX) according to the manufacturer’s instructions. In brief, 20 μg of total RNA and 30,000 cpm of each probe (20,000 cpm for the GAPDH mRNA) were hybridized in 20 μl of hybridization buffer containing 80% formamide at 45°C for 16 hours and digested with RNase T1 at 37°C for 1 hour. Protected probes were separated on a 5% polyacrylamide/8 mol/L urea gel followed by autoradiography with X-ray films (Kodak, Rochester, NY) at −70°C for 16 hours. Sizes of the protected sequences for CTGF, TGF-β1, and GAPDH were 430, 253, and 102 nucleotides, respectively.
Signal Quantification and Statistical Analysis
Autoradiographies were analyzed with the public domain program NIH Image (developed at the National Institutes of Health, Bethesda, MD) on a Power Macintosh 7500/100 computer. Target mRNA signals of CTGF and TGF-β1 were normalized to the GAPDH signal and expressed as relative abundance (arbitrary units). The differences among the various time points after injection of CCl4 were analyzed by one-way analysis of variance. The differences between sham-operated and BDO rats were compared by the unpaired t-test. P < 0.05 was regarded as statistically significant.
Results
Rat CTGF Is Highly Homologous to CTGF of Other Species
Using primers derived from the mouse CTGF mRNA sequence, we amplified a 430-bp fragment from rat liver cDNA. After excluding the 42-bp mouse primer regions, this rat sequence shared 95 and 97% identity with mouse CTGF at the mRNA and the deduced amino acid level, respectively. Amino acid identity was also 95% with human and pig, and 82% with bovine and frog CTGF.
CTGF mRNA Levels in Rats with Acute and Chronic Liver Fibrogenesis
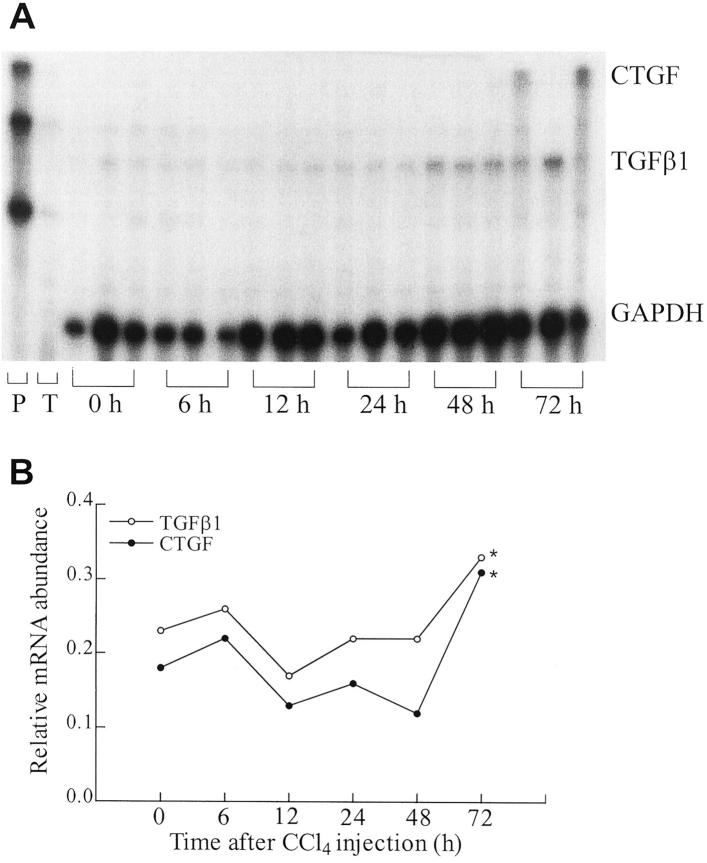
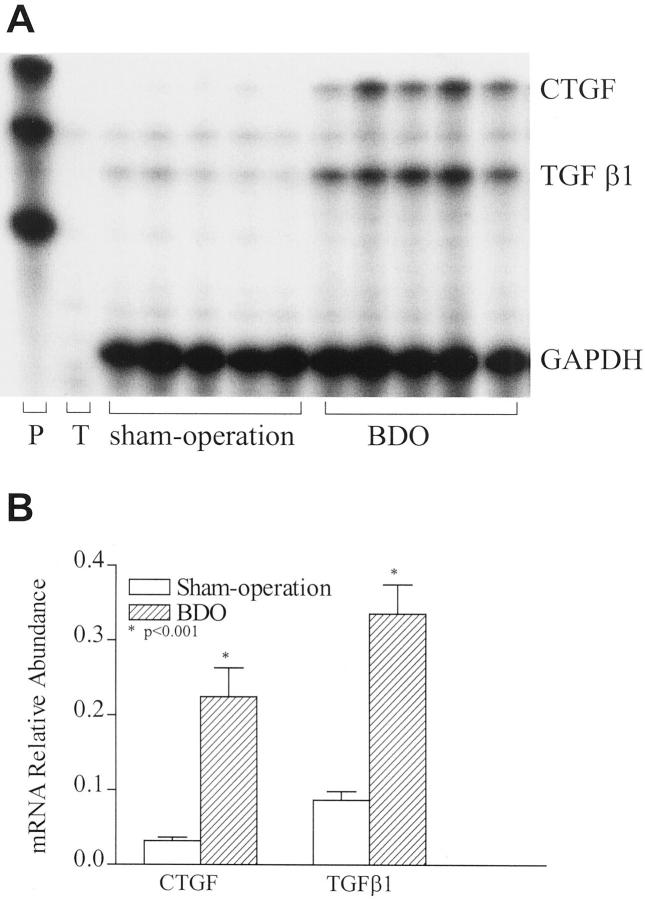
CTGF mRNA levels in normal rat livers were low. After a single injection of CCl4, mRNAs for TGF-β1 and CTGF rose slowly, but only increased significantly (∼50% greater than baseline) 72 hours after injection (Figure 1) ▶ . However, at 72 hours one out of three rats showed a low CTGF and a high TGF-β1 mRNA steady state level, indicating that at later time points expression of these two cytokines may dissociate (Figure 1A) ▶ . Furthermore, this finding underlines the well-known individual heterogeneity of the CCL4 model of fibrogenesis. In contrast, normalized mRNA levels of TGF-β1 and CTGF in livers of rats with secondary biliary cirrhosis (BDO) were fourfold and sevenfold higher than those of the sham-operated controls (P < 0.001, Figure 2 ▶ ).
Figure 1.
Coordinate expression of CTGF and TGF-β1 in acute liver fibrogenesis. A: Multiprobe ribonuclease protection assay for CTGF and TGF-β1 mRNA in rats after acute CCl4-intoxication. Twenty μg of liver RNA were hybridized with [α-32P]-labeled CTGF and TGF-β1 riboprobes. After RNase T1 digestion the protected probes were resolved through denaturing polyacrylamide gel electrophoresis and subjected to autoradiography at −70°C for 16 hours. B: P, full-length probes; T, tRNA control. Autoradiographic signals were normalized to that of GAPDH and expressed as arbitrary units. Each time point represents the mean value of three rats. *, P < 0.05 compared with all other time points.
Figure 2.
Coordinate expression of CTGF and TGF-β1 in chronic liver fibrogenesis. Multiprobe ribonuclease protection assay for CTGF and TGF-β1 mRNA from rats after BDO for 6 weeks. Twenty μg of liver RNA were hybridized with [α-32P]-labeled CTGF and TGF-β1 riboprobes and processed as described in Figure 1 ▶ . P, full-length probes; T, tRNA control. Error bars represent standard errors derived from five rats. *, P < 0.001 compared with sham-operated controls.
Cellular Sources of CTGF in Rat Liver
By combining in situ hybridization with immunostaining for cell-specific markers CTGF mRNA was barely detectable in liver sections of normal rats, except for a local up-regulation in portal and central vein endothelia and in myocytes of portal arteries (Figure 3, A and B) ▶ . After acute CCl4 intoxication signals for CTGF mRNA were increased predominantly in activated HSCs/myofibroblasts, identified by positivity for desmin (Figure 3C) ▶ . Surprisingly, CTGF mRNA was highly up-regulated and almost exclusively found over proliferating, pan-cytokeratin-positive and desmin-negative bile duct epithelia in rats after 6 weeks of BDO (Figure 3 ▶ ; D, E, F, and G). Here, signal intensity varied among cells of a single bile duct. These CTGF-expressing cells were surrounded by a rim of procollagen α1(I) expressing, desmin-positive HSCs/myofibroblasts (Figure 3H) ▶ .
Figure 3.
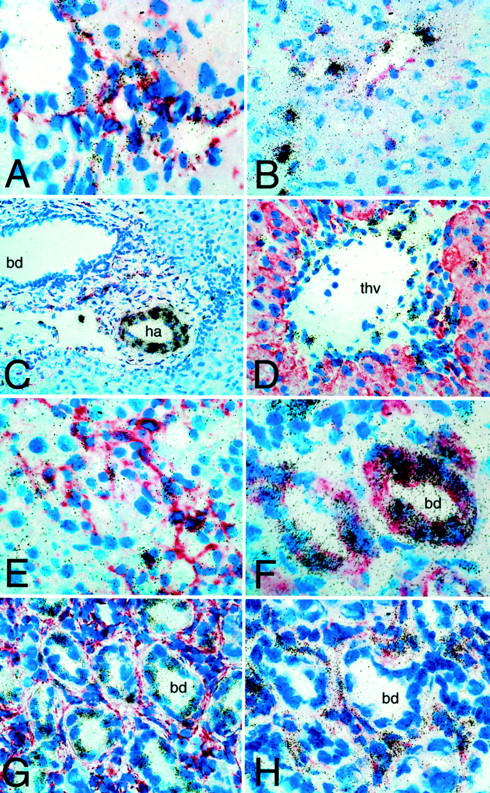
Expression patterns of CTGF and procollagen α1(I) mRNA in acute and chronic hepatic fibrogenesis. Radioactive in situ hybridization, in part in combination with immunostaining for desmin (A-D, G, and H) or pan-cytokeratin (E) on rat livers. CTGF antisense: A: Normal liver portal tract; labeling of HSCs/portal fibroblasts (original magnification, ×600). B: Six hours after CCl4, labeling of desmin-positive HSC and endothelial cells (original magnification, ×400). C: Six hours after CCl4, prominent signals in hepatic arterial (ha) myocytes in a larger portal tract, no expression over a larger bile duct (bd) (original magnification, ×200). D: Seventy-two hours after CCl4, strong labeling of single centrilobular HSC and endothelial cells of a terminal hepatic vein (thv) (original magnification, ×600). E: Seventy-two hours after CCl4, enhanced CTGF mRNA expression in most portal tract fibroblasts (original magnification, ×400). F and G: CTGF mRNA is almost exclusively expressed in desmin-negative (F: original magnification, ×900) and pan-cytokeratin-positive (G: original magnification, ×600) bile duct epithelia (bd in biliary fibrosis); note the patchy ductular CTGF mRNA expression pattern. H: Up-regulation of procollagen α1(I) mRNA in desmin-positive myofibroblasts surrounding proliferating bile ducts (bd) after 6 weeks of BDO (original magnification, ×700).
Coordinate Expression of TGF-β1 and CTGF mRNA
Although in acute fibrogenesis the time course and quantitative expression of TGF-β1 and CTGF mRNA were virtually superimposable (Figure 1) ▶ , in BDO-induced fibrosis proliferating bile duct epithelia expressed TGF-β1 mRNA at minimal levels whereas it was highly up-regulated in the surrounding HSCs/myofibroblasts (data not shown).
Discussion
CTGF is a member of the emerging CCN (connective tissue growth factor/fisp 12, Cyr61/cef10 and nov) family that comprises several immediate early gene products induced by growth factors or oncogenes. Recent data indicate that CTGF is up-regulated by and acts in concert with TGF-β1. Because mesenchymal cells have been held to be both the major source and target of CTGF, inhibition of expression or blockade of this growth factor and its yet unidentified cellular receptor are attractive targets for an antifibrogenic therapy.
By combining in situ hybridization and immunohistochemistry we identified activated HSCs as the main cellular source of CTGF in later time points of acute fibrogenesis triggered by CCl4, confirming a recent report. 18 In normal as well as CCl4-injured livers, however, CTGF transcripts were most prominent over venous endothelial cells and arterial myocytes. This is consistent with the results obtained in rats and other species, where CTGF transcripts were predominantly found in fibroblasts and endothelia of the skin, 13 gingiva, 30 kidney, 15 pancreas, 31 and lung, 16 as well as in smooth muscle cells of atherosclerotic blood vessels 10 and in hypertrophic chondrocytes of costal cartilage. 32 The basal expression of CTGF in these cell types was low, but highly up-regulated during wound healing or fibrogenesis. A single in vivo study showed some CTGF expression by renal tubular epithelial cells. 33 Our observation of a prominent expression of CTGF by proliferating bile duct epithelial cells in vivo is unexpected. However, it explains the prominent fibroductular reaction around proliferating bile duct epithelia in biliary fibrosis, as exemplified by an up-regulated procollagen I expression by periductular myofibroblast-like cells. 34 It is also in line with the TGF-β1 and TGF-β2 mRNA expression patterns of these ductular epithelial cells and periductular myofibroblasts that we found in an earlier study. 35 Furthermore, the deposition of CTGF immune-reactive material in the periductular extracellular matrix observed by Paradis and colleagues 18 mostly likely reflects the affinity of this growth factor to abundant matrix proteins such as collagens and fibronectin. 36
The observed patchy distribution of CTGF mRNA in proliferating bile duct cells does not seem to be related to proliferation, because we showed previously that the proliferation marker Ki-67 is distributed homogeneously in larger as well as proliferating biliary epithelia. 34,35
Our findings may have therapeutic implications. TGF-β1 has been regarded as a key profibrogenic cytokine that is capable of promoting mesenchymal proliferation and extracellular matrix production and of suppressing extracellular matrix degradation. 37 Thus blocking the expression or activity of TGF-β1 can effectively prevent organ fibrosis in various experimental models. 38 However, TGF-β1 is a pleiotropic, multifunctional, and context-dependent cytokine, including a strong immunosuppressive and proapoptotic effect for lymphocytes, macrophages, and hepatocytes. 39 TGF-β1 knockout mice die shortly after birth of an excessive inflammatory response, because of massive mononuclear infiltration into organs such as intestine and liver. Furthermore, depending on the context the role of TGF-β1 in carcinogenesis can be divergent, varying from inhibition to promotion of tumor formation or metastasis. 40 Therefore, complete blockade of its expression or activity is accompanied by unfavorable if not lethal side effects.
On the other hand, the effect of CTGF, which acts in conjunction with TGF-β1, seems to be limited to the mesenchymal compartment. 2 Therefore, inhibition of CTGF bioactivity is expected to be a better target for antifibrotic therapies. 41 However, as we showed in our study, activated epithelial cells have to be considered as a major source of CTGF in vivo.
In conclusion, we found that in rat biliary fibrosis bile-duct epithelial cells are the dominant cellular source of CTGF and that the expression of CTGF mRNA is coordinately up-regulated with that of TGF-β1, supporting the suggested role of CTGF as an associated if not downstream mediator of TGF-β actions.
Acknowledgments
We thank Mrs. Kathrin Thomsen-Mund for preparing tissue sections for in situ hybridization.
Footnotes
Address reprint requests to Detlef Schuppan, M.D., Ph.D., Department of Medicine I, University of Erlangen-Nuernberg, Krankenhausstrasse 12, 91054 Erlangen, Germany. E-mail: detlef.schuppan@med1.imed.uni-erlangen.de.
Supported by a grant from the Federal Ministry of Science and Research (BMBF-IZKF, University Erlangen-Nuernberg).
N.S. and J.-D.J. contributed equally to this report.
References
- 1.Bradham DM, Igarashi A, Potter RL, Grotendorst GR: Connective tissue growth factor: a cysteine-rich mitogen secreted by human vascular endothelial cells is related to the SRC-induced immediate early gene product CEF-10. J Cell Biol 1991, 114:1285-1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frazier K, Williams S, Kothapalli D, Klapper H, Grotendorst GR: Stimulation of fibroblast cell growth, matrix production, and granulation tissue formation by connective tissue growth factor. J Invest Dermatol 1996, 107:404-411 [DOI] [PubMed] [Google Scholar]
- 3.Duncan MR, Frazier KS, Abramson S, Williams S, Klapper H, Huang X, Grotendorst G: Connective tissue growth factor mediates transforming growth factor-induced collagen synthesis: down-regulation by cAMP. FASEB J 1999, 13:1774-1786 [PubMed] [Google Scholar]
- 4.Grotendorst GR, Okochi H, Hayashi N: A novel transforming growth factor beta response element controls the expression of the connective tissue growth factor gene. Cell Growth Differ 1996, 7:469-480 [PubMed] [Google Scholar]
- 5.Igarashi A, Okochi H, Bradham DM, Grotendorst GR: Regulation of connective tissue growth factor gene expression in human skin fibroblasts and during wound repair. Mol Biol Cell 1993, 4:637-645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kothapalli D, Frazier KS, Welply A, Segarini PR, Grotendorst GR: Transforming growth factor beta induces anchorage-independent growth of NRK fibroblasts via a connective tissue growth factor-dependent signaling pathway. Cell Growth Differ 1997, 8:61-68 [PubMed] [Google Scholar]
- 7.Brunner A, Chinn J, Neubauer M, Purchio AF: Identification of a gene family regulated by transforming growth factor-beta. DNA Cell Biol 1991, 10:293-300 [DOI] [PubMed] [Google Scholar]
- 8.Grotendorst GR: Connective tissue growth factor: a mediator of TGF-beta action on fibroblasts. Cytokine Growth Factor Rev 1997, 8:171-179 [DOI] [PubMed] [Google Scholar]
- 9.Mori T, Kawara S, Shinozaki N, Hayashi M, Kakinuma T, Igarashi A, Takigawa M, Nakanishi T, Takehara K: Role and interaction of connective tissue growth factor with transforming growth factor-beta in persistent fibrosis: a mouse fibrosis model. J Cell Physiol 1999, 181:153-159 [DOI] [PubMed] [Google Scholar]
- 10.Oemar BS, Werner A, Garnier JM, Do DD, Godoy N, Nauck M, Marz W, Rupp J, Pech M, Luscher TJ: Human connective tissue growth factor is expressed in advanced atherosclerotic lesions. Circulation 1997, 95:831-839 [DOI] [PubMed] [Google Scholar]
- 11.Haustein UF, Anderegg U: Pathophysiology of scleroderma: an update. J Eur Acad Dermatol Venereol 1998, 11:1-8 [PubMed] [Google Scholar]
- 12.Igarashi A, Nashiro K, Kikuchi K, Sato S, Ihn H, Grotendorst GR, Takehara K: Significant correlation between connective tissue growth factor gene expression and skin sclerosis in tissue sections from patients with systemic sclerosis. J Invest Dermatol 1995, 105:280-284 [DOI] [PubMed] [Google Scholar]
- 13.Igarashi A, Nashiro K, Kikuchi K, Sato S, Ihn H, Fujimoto M, Grotendorst GR, Takehara K: Connective tissue growth factor gene expression in tissue sections from localized scleroderma, keloid, and other fibrotic skin disorders. J Invest Dermatol 1996, 106:729-733 [DOI] [PubMed] [Google Scholar]
- 14.Kikuchi K, Kadono T, Ihn H, Sato S, Igarashi A, Nakagawa H, Tamaki K, Takehara K: Growth regulation in scleroderma fibroblasts: increased response to transforming growth factor-beta 1. J Invest Dermatol 1995, 105:128-132 [DOI] [PubMed] [Google Scholar]
- 15.Riser BL, Denichilo M, Cortes P, Baker C, Grondin JM, Yee J, Narins RG: Regulation of connective tissue growth factor activity in cultured rat mesangial cells and its expression in experimental diabetic glomerulosclerosis. J Am Soc Nephrol 2000, 11:25-38 [DOI] [PubMed] [Google Scholar]
- 16.Lasky JA, Ortiz LA, Tonthat B, Hoyle GW, Corti M, Athas G, Lungarella G, Brody A, Friedman M: Connective tissue growth factor mRNA expression is upregulated in bleomycin-induced lung fibrosis. Am J Physiol 1998, 275:L365-L371 [DOI] [PubMed] [Google Scholar]
- 17.Dammeier J, Brauchle M, Falk W, Grotendorst GR, Werner S: Connective tissue growth factor: a novel regulator of mucosal repair and fibrosis in inflammatory bowel disease? Int J Biochem Cell Biol 1998, 30:909-922 [DOI] [PubMed] [Google Scholar]
- 18.Paradis V, Dargere D, Vidaud M, De Gouville AC, Huet S, Martinez V, Gauthier JM, Ba N, Sobesky R, Ratziu V, Bedossa P: Expression of connective tissue growth factor in experimental rat and human liver fibrosis. Hepatology 1999, 30:968-976 [DOI] [PubMed] [Google Scholar]
- 19.Olaso E, Friedman SL: Molecular regulation of hepatic fibrogenesis. J Hepatol 1998, 29:836-847 [DOI] [PubMed] [Google Scholar]
- 20.Ryseck RP, Macdonald-Bravo H, Mattei MG, Bravo R: Structure, mapping, and expression of fisp-12, a growth factor-inducible gene encoding a secreted cysteine-rich protein. Cell Growth Differ 1991, 2:225-233 [PubMed] [Google Scholar]
- 21.Boigk G, Stroedter L, Herbst H, Waldschmidt J, Riecken EO, Schuppan D: Silymarin retards collagen accumulation in early and advanced biliary fibrosis secondary to complete bile duct obliteration in rats. Hepatology 1997, 26:643-649 [DOI] [PubMed] [Google Scholar]
- 22.Cho JJ, Hocher B, Herbst H, Jia JJ, Boigk G, Hahn EG, Riecken EO, Schuppan D: An oral endothelin A receptor antagonist blocks collagen synthesis and deposition in advanced rat secondary biliary fibrosis. Gastroenterology 2000, 118:1169-1178 [DOI] [PubMed] [Google Scholar]
- 23.Qian SW, Kondaiah P, Roberts AB, Sporn MB: cDNA cloning by PCR of rat transforming growth factor beta-1. Nucleic Acids Res 1990, 18:3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Genovese C, Rowe D, Kream B: Construction of DNA sequences complementary to rat alpha 1 and alpha 2 collagen mRNA and their use in studying the regulation of type I collagen synthesis by 1,25-dihydroxyvitamin D. Biochemistry 1984, 23:6210-6216 [DOI] [PubMed] [Google Scholar]
- 25.Herbst H, Heinrichs O, Schuppan D, Milani S, Stein H: Temporal and spatial patterns of transin/stromelysin RNA expression following toxic injury in rat liver. Virchows Arch B Cell Pathol Incl Mol Pathol 1991, 60:295-300 [DOI] [PubMed] [Google Scholar]
- 26.Herbst H, Wege T, Milani S, Pellegrini G, Orzechowski HD, Bechstein WO, Neuhaus P, Gressner AM, Schuppan D: Tissue inhibitor of metalloproteinase-1 and -2 RNA expression in rat and human liver fibrosis. Am J Pathol 1997, 150:1647-1659 [PMC free article] [PubMed] [Google Scholar]
- 27.Milani S, Herbst H, Schuppan D, Grappone C, Pellegrini G, Pinzani M, Casini A, Calabro A, Ciancio G, Stefanini F, Burroughs A, Surrenti C: Differential expression of matrix-metalloproteinase-1 and -2 genes in normal and fibrotic human liver. Am J Pathol 1994, 144:528-537 [PMC free article] [PubMed] [Google Scholar]
- 28.Chomczynski P, Sacchi N: Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 1987, 162:156-159 [DOI] [PubMed] [Google Scholar]
- 29.Tso JY, Sun XH, Kao TH, Reece KS, Wu R: Isolation and characterization of rat and human glyceraldehyde-3-phosphate dehydrogenase cDNAs: genomic complexity and molecular evolution of the gene. Nucleic Acids Res 1985, 13:2485-2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong HH, Uzel MI, Duan C, Sheff MC, Trackman PC: Regulation of lysyl oxidase, collagen, and connective tissue growth factor by TGF-beta1 and detection in human gingiva. Lab Invest 1999, 79:1655-1667 [PubMed] [Google Scholar]
- 31.Di Mola FF, Friess H, Martignoni FE, Di Sebastiano P, Zimmermann A, Innocenti P, Graber H, Gold LI, Korc M, Buchler MW: Connective tissue growth factor is a regulator for fibrosis in human chronic pancreatitis. Ann Surg 1999, 230:63-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakanishi T, Kimura Y, Tamura T, Ichikawa H, Yamaai Y, Sugimoto T, Takigawa M: Cloning of a mRNA preferentially expressed in chondrocytes by differential display-PCR from a human chondrocytic cell line that is identical with connective tissue growth factor (CTGF) mRNA. Biochem Biophys Res Commun 1997, 234:206-210 [DOI] [PubMed] [Google Scholar]
- 33.Ito Y, Aten J, Bende RJ, Oemar BS, Rabelink TJ, Weening JJ, Goldschmeding R: Expression of connective tissue growth factor in human renal fibrosis. Kidney Int 1998, 53:853-861 [DOI] [PubMed] [Google Scholar]
- 34.Milani S, Herbst H, Schuppan D, Riecken EO, Stein H: Procollagen expression by nonparenchymal rat liver cells in experimental biliary fibrosis. Gastroenterology 1990, 98:175-184 [DOI] [PubMed] [Google Scholar]
- 35.Milani S, Herbst H, Schuppan D, Stein H, Surrenti C: Transforming growth factors β1 and β2 are differentially expressed in fibrotic liver disease. Am J Pathol 1991, 139:1221-1229 [PMC free article] [PubMed] [Google Scholar]
- 36.Ruehl M, Somasundaram R, Schoenfelder I, Schmid M, Huet S, Riecken EO, Schuppan D: Preferential binding of connective tissue growth factor to liver collagens type I, III and VI. Hepatology 1999, 30:493A [Google Scholar]
- 37.Bedossa P, Paradis V: Transforming growth factor-beta (TGF-beta): a key-role in liver fibrogenesis. J Hepatol 1995, 22:37-42 [PubMed] [Google Scholar]
- 38.Roberts AB: Molecular and cell biology of TGF-beta. Miner Electrolyte Metab 1998, 24:111-119 [DOI] [PubMed] [Google Scholar]
- 39.Letterio JJ, Roberts AB: Regulation of immune responses by TGF-beta. Annu Rev Immunol 1998, 16:137-161 [DOI] [PubMed] [Google Scholar]
- 40.Markowitz SD, Roberts AB: Tumor suppressor activity of the TGF-beta pathway in human cancers. Cytokine Growth Factor Rev 1996, 7:93-102 [DOI] [PubMed] [Google Scholar]
- 41.Schuppan D, Cho JJ, Jia JD, Hahn EG: Interplay of matrix and myofibroblasts during hepatic fibrogenesis. Curr Top Pathol 1999, 93:205-218 [DOI] [PubMed] [Google Scholar]