Abstract
Angioimmunoblastic lymphadenopathy with dysproteinemia (AILD) is defined in the current lymphoma classifications as a T-cell non-Hodgkin’s lymphoma. However, in approximately one third of the cases of this lymphoproliferative disease rearrangements of T-cell receptor (TCR) genes indicating clonal expansion of T cells are not detectable. It is currently believed that these cases may represent early stages of a lymphoma with a minor oligoclonal T-cell population. In the present study, 18 lymph nodes with the characteristic histology of AILD were investigated for clonal T-cell receptor gene rearrangements by analysis of DNA extracted from whole tissue sections. Dominant T-cell clones were detected in 12 of these cases. Single CD4+ and CD8+ T cells and proliferating Ki67+ cells of seven cases were micromanipulated from frozen tissue sections. TCRβ gene rearrangements were amplified from these cells by polymerase chain reaction and sequenced. In all informative cases, the clonal gene rearrangements were only detected among CD4+, and not among CD8+ T cells, indicating that the tumor clones in AILD usually derive from CD4+ T cells. Minor clonal T-cell populations in those cases in which no clone was found by whole-tissue DNA analysis were not detectable even at single cell resolution. T-cell clones in 4 of 10 cases were found to express similar TCRβ chains, indicating a potential role of (super) antigen triggering in at least some cases of AILD.
Angioimmunoblastic lymphadenopathy with dysproteinemia (AILD) was initially described as a benign hyperimmune reaction 1 and also named immunoblastic lymphadenopathy or lymphogranulomatosis X. 2,3 These entities are now summarized as AILD, as they share clinical and histopathological features. Histologically, AILD may resemble Hodgkin’s disease showing a polymorphous infiltrate composed of lymphocytes, plasma cells, immunoblasts, eosinophils, and histiocytes. Typical Hodgkin or Reed/Sternberg cells, however, are usually not found in AILD tissue. 1,3 Characteristically arborising blood vessels with high endothelia, an amorphous eosinophilic intercellular material, and irregular networks of follicular dendritic cells are found in effaced lymph nodes. 2,4 AILD was identified as a neoplastic disorder and is listed in the current lymphoma classifications as a T-cell non-Hodgkin’s lymphoma. 5,6
The classification of AILD as a neoplastic lymphoproliferative disease was supported by the finding of clonal antigen-receptor gene rearrangements in DNA extracted from tumor tissue. 4,7-10 Southern blot hybridization experiments revealed clonal T-cell receptor (TCR)β gene rearrangements in ∼70% of the cases investigated and clonal immunoglobulin heavy chain (IgH) gene rearrangements in few cases. 9,11 Polymerase chain reaction (PCR) techniques for the amplification of TCR and IgH gene rearrangements yielded similar results. 12,13 In the remaining 30% of cases no dominant gene rearrangements were found. It remains unclear whether these cases lack clonal lymphocytic infiltrates, or whether such infiltrates escape detection because of limited sensitivity of the Southern blot and PCR techniques applied.
With the development of single target amplification of lymphocyte receptor gene rearrangements from single cells 14 analysis of even minor lymphocyte populations in lymph node tissue has become feasible. Recently, single target amplification of TCRβ gene rearrangements from individual T cells micromanipulated from tissue sections was established. 15 In the present study these techniques have been applied in seven cases of AILD to further characterize this tumor. This approach allows addressing the question of the phenotype of the tumor cells directly by analysis of single CD4+ and CD8+ T cells separately. Proliferating cells were investigated to detect minor lymphocyte proliferations in those cases with no obvious clone.
Materials and Methods
Tissue Samples
Lymph node specimens diagnostic of AILD were collected from the files of the Institutes of Pathology of the Universities of Kiel and Frankfurt, Germany. The diagnosis was based on the histological criteria as detailed above. In 12 cases frozen tissues and in six cases paraffin-embedded tissues were available.
Whole-Tissue DNA Analysis
Sections (10-μm thick) of lymph node tissue were cut and digested with proteinase K. Whole DNA was extracted using the Qiagen DNA extraction kit as recommended by the manufacturer (Qiagen, Hilden, Germany). For PCR analysis, 100 ng of DNA from frozen tissue and 500 ng of DNA from paraffin-embedded tissue were used for each reaction. TCRβ gene rearrangements were amplified and sequenced using the same conditions and primers as described below for second-round single-cell PCR. For DNA amplification from paraffin-embedded tissue an additional primer for the BJ2.4 segment was added to limit the length of PCR fragments to <450 bp (Jβ 2.4, Table 1 ▶ ).
Table 1.
Sequences of Primers Used in This Study
| VH con | 5′-CTGTCGACACGGCCGTGTATTACTG-3′ |
| JH con | 5′-AACTGCTGAGGAGACGGTGACC-3′ |
| Vγ 1–8 | 5′-ACCAGGAGGGGAAGGCCCCACAG-3′ |
| Vγ 9 | 5′-GGAAAGGAATCTGGCATTCCG-3′ |
| Vγ 10 | 5′-AATCCGCAGCTCGACGCAGCA-3′ |
| Vγ 11 | 5′-GCTCAAGATTGCTCAGGTGGG-3′ |
| Jγ 1/2 | 5′-ACCTGTGACACCAAGTGTTGTTC-3′ |
| Jγ P1/2 | 5′-AGTTACTATGAGCT(CT)TAGTCCC-3′ |
| Jγ P | 5′-TGTAATGATAAGCTTTGTTCC-3′ |
| Vβ 1/5 | 5′-ACAGCAAGTGAC〈TAG〉CTGAGATGCTC-3′ |
| Vβ 2 | 5′-GAGTGCCGTTCCCTGGACTTTCAG-3′ |
| Vβ 3 | 5′-GTAACCCAGAGCTCGAGATATCTA-3′ |
| Vβ 4 | 5′-TCCAGTGTCAAGTCGATAGCCAAGTC-3′ |
| Vβ 6.a | 5′-ATGTAACT〈CT〉TCAGGTGTGATCCAA-3′ |
| Vβ 6.b | 5′-GTGTGATCCAATTTCAGGTCATAC-3′ |
| Vβ 7 | 5′-TACGCAGACACCAA〈GA〉ACACCTGGTCA-3′ |
| Vβ 8 | 5′-GGTGACAGAGATGGGACAAGAAGT-3′ |
| Vβ 9 | 5′-CCCAGACTCCAAAATACCTGGTCA-3′ |
| Vβ 10 | 5′-AAGGTCACCCAGAGACCTAGACTT-3′ |
| Vβ 11 | 5′-GATCACTCTGGAATGTTCTCAAACC-3′ |
| Vβ 12 | 5′-CCAAGACACAAGGTCACAGAGACA-3′ |
| Vβ 13 | 5′-GTGTCACTCAGACCCCAAAATTCC-3′ |
| Vβ 14 | 5′-GTGACCCAGAACCCAAGATACCTC-3′ |
| Vβ 15 | 5′-GTTACCCAGACCCCAAGGAATAGG-3′ |
| Vβ 16 | 5′-ATAGAAGCTGGAGTTACTCAGTTC-3′ |
| Vβ 17 | 5′-CACTCAGTCCCCAAAGTACCTGTT-3′ |
| Vβ 18 | 5′-TGCAGAACCCAAGACACCTGGTCA-3′ |
| Vβ 19 | 5′-ACAAAGATGGATTGTACCCCCGAA-3′ |
| Vβ 20 | 5′-GTCAGATCTCAGACTATTCATCAATGG-3′ |
| Vβ 21 | 5′-CAGTCTCCCAGATATAAGATTA〈TC〉AGAG-3′ |
| Vβ 22 | 5′-GGTCACACAGATGGGACAGGAAGT-3′ |
| Vβ 23 | 5′-CTGATCAAAGAAAAGAGGGAAACAGCC-3′ |
| Vβ 24 | 5′-CAAGATACCAGGTTACCCAGTTTG-3′ |
| Vβ 25 | 5′-GACAGAAAGCAAAATTATATTGTGCC-3′ |
| 3′Jβ 1.2 | 5′-TACAACGGTTAACCTGGTCCCCGA-3′ |
| 5′Jβ 1.2 | 5′-TAACCTGGTCCCCGAACCGAAGG-3′ |
| 3′Jβ 1.3 | 5′-CACCTACAACAGTGAGCCAACTT-3′ |
| 5′Jβ 1.3 | 5′-GCCAACTTCCCTCTCCAAAATATATGG-3′ |
| 3′Jβ 1.5 | 5′-CCAACTTACCTAGGATGGAGAGTCGA-3′ |
| 5′Jβ 1.5 | 5′-GATGGAGAGTCGAGTCCCATCAC-3′ |
| 3′Jβ 1.6 | 5′-CCTGGTCCCATTCCCAAAGTGGA-3′ |
| 5′Jβ 1.6 | 5′-CCCATTCCCAAAGTGGAGGGGTG-3′ |
| 3′Jβ 2.2 | 5′-CCTTACCCAGTACGGTCAGCCTA-3′ |
| 5′Jβ 2.2 | 5′-AGTACGGTCAGCCTAGAGCCTTCT-3′ |
| 3′Jβ 2.6 | 5′-CAGCCGCCGCCTTCCACCTGAAT-3′ |
| 5′Jβ 2.6 | 5′-CGGCCCCGAAAGTCAGGACGTT-3′ |
| 3′Jβ 2.7 | 5′-TCCATCGTTCACCTTCTCTCTAAACA-3′ |
| 5′Jβ 2.7 | 5′-GCCCGAATCTCACCTGTGACCGTG-3′ |
| Jβ 2.4 | 5′-CCAGCTTACCCAGCACTGAGAGC-3′ |
〈 〉 denotes a mixture of nucleotides at that position.
Vβ primers 7, 9, and 18 expectedly cross-react with members of all of these three Vβ families and are, therefore, used in a mix at a concentration of 14 nmol/L and 50 nmol/L each for first round and second round amplifications, respectively.
Analysis of IgH and TCRγ chain complementarity-determining region III (CDR3) length distributions was performed as described elsewhere. 16,17 Briefly, IgH gene rearrangements were amplified by 40 cycles of PCR at an annealing temperature of 61°C in a standard reaction mixture containing 125 nmol/L of each VH and JH consensus primers (Table 1) ▶ , 2 mmol/L MgCl2 and 0.7 U Taq polymerase (Boehringer Mannheim, Mannheim, Germany). TCRγ gene rearrangements were amplified in two separate reactions for each DNA sample containing standard reagents, 400 nmol/L of each J and V primer, 1.5 mmol/L MgCl2, and 0.7 U Taq polymerase (Boehringer Mannheim). A mixture of three J primers was used with either the Vγ 1-8 primer or a mixture of Vγ 9, Vγ 10, and Vγ 11 primers (Table 1) ▶ at an annealing temperature of 60°C for 40 cycles of amplification. VH consensus primer for IgH analysis and Jγ primers for TCRγ detection were fluorescence labeled with FAM6 (MWG, Ebersberg, Germany). PCR products were analyzed on an automated sequencer using the GeneScan software as recommended by the manufacturer (ABI377; Applied Biosystems, Weiterstadt, Germany).
Major histocompatibility complex class II (MHC II) gene polymorphism (DRB1, DQB1, DPB1) was typed by PCR amplification of genomic DNA extracted from paraffin-embedded tissue. HLA-DRB1 and HLA-DQB1 alleles were defined using sequence-specific oligonucleotides as recommended by the manufacturer (ELPHA HLA-DRB/HLA-DQB; Biotest, Dreieich, Germany). HLA-DPB1 typing was performed by reverse dot-blot hybridization (INNO-LiPA DPB; Innogenetics, Zwinjndrecht, Belgium).
Immunostaining and Micromanipulation of Cells from Frozen Tissue Sections
Seven frozen lymph-node specimens showing the typical morphology of AILD were chosen for micromanipulation. Immunostaining of frozen tissue sections was performed as described, 14 using monoclonal antibodies against CD4 (MT310), CD8 (DK25), and the proliferation marker Ki67 (all antibodies by DAKO, Glostrup, Denmark). Alkaline phosphatase was developed using Fast Red TR (DAKO). CD4+, CD8+, and Ki67+ cells were isolated from adjacent sections by micromanipulation and transferred into PCR tubes containing 20 μl of PCR buffer as described. 14 Samples of the buffer covering the sections during the micromanipulation procedure and tubes containing PCR buffer but no cell served as negative controls.
Amplification of TCR Vβ Gene Rearrangements from Single Cells
Amplification of rearranged TCR Vβ genes was performed according to a recently established protocol. 15 Briefly, micromanipulated cells were incubated with proteinase K. A first round of PCR was performed in the same tube using a mix of 25 Vβ family- and 7 Jβ-specific primers (Table 1) ▶ in a 50-μl volume containing standard reagents, 42 nmol/L of each primer, 2 mmol/L MgCl2, and 2.5 U of Expand HF polymerase mix (Boehringer Mannheim) for 35 cycles of amplification at an annealing temperature of 61°C. A second round of amplification was performed in 96-well plates, adding 1 μl of the first round reaction to eight separate reaction mixtures, each containing a mixture of internal Jβ primers (Table 1) ▶ and 2 to 5 of the 25 Vβ primers in the following combinations: Vβ 2, 3, 22; Vβ 4, 6a, 14; Vβ 6b, 8, 21; Vβ 1/5, 11, 12; Vβ 13, 15; Vβ 7/9/18, 17, 20; Vβ 23, 24, 25; and Vβ 10, 16, 19. Second round amplification was performed in a 50-μl volume containing standard reagents, 150 nmol/L of each primer, 2 mmol/L MgCl2, and 0.7 U Taq polymerase (Boehringer Mannheim) for 44 cycles at an annealing temperature of 61°C.
PCR products were gel-purified and directly sequenced using the Ready Reaction dRhodamine cycle sequencing kit (Perkin Elmer, Foster City, CA) and an automatic sequencer (ABI377) as recommended by the manufacturer. Sequences were deposited in the European Molecular Biology Laboratory database under accession numbers AJ301370 to AJ301551.
Results
Whole-Tissue DNA Analysis
Whole tissue DNA of 18 lymph nodes displaying the characteristic histology of AILD was extracted. TCRβ gene rearrangements were amplified in eight separate PCR reactions using Vβ family-specific primers. Ten cases harboring a dominant TCRβ gene rearrangement were identified (Table 2) ▶ . Clonality was confirmed by direct sequencing of the PCR product. For case 8 only a nonfunctional gene rearrangement was obtained. In case 11, two in-frame gene rearrangements were amplified indicating the presence of either two clones in the lymph node tissue or one clone with in-frame rearrangements on both alleles. The eight other cases showed smeared bands for each of the eight PCR reactions, indicating a polyclonal T-cell population.
Table 2.
Results of Whole-Tissue DNA Analysis and MHC Class II Typing in AILD
| Case | TCRβ PCR whole tissue | TCRγ† whole tissue | IgH† whole tissue | MHC II | ||
|---|---|---|---|---|---|---|
| DRB1* | DQB1* | DPB1* | ||||
| 1 | Clonal | Clonal | Polyclonal | 03, 07 | 02, 0302 | 0101, 0401 |
| 2 | Clonal | Clonal | Polyclonal | 03, 13 | 02, 06 | 0301, 0401 |
| 3 | Clonal | Clonal | Polyclonal | 11, 16 | 0301, 05 | 0201, 0401 |
| 4 | Polyclonal | Polyclonal | Polyclonal | 01, 07 | 05, 02 | nd |
| 5 | Polyclonal | Clonal | Polyclonal | 10, 13 | 05, 06 | 0201, 0402 |
| 6 | Polyclonal | Polyclonal | Polyclonal | 13, 13 | 06, 06 | nd |
| 7 | Polyclonal | Polyclonal | Polyclonal | 15, − | 06,− | nd |
| 8 | Clonal | nd | nd | nd | nd | nd |
| 9 | Polyclonal | nd | nd | nd | nd | nd |
| 10 | Clonal | nd | nd | nd | nd | nd |
| 11 | Clonal | nd | nd | nd | nd | nd |
| 12 | Polyclonal | nd | nd | nd | nd | nd |
| 13 | Polyclonal | Clonal | Polyclonal | 03, 11 | 02, 0301 | nd |
| 14 | Clonal | Clonal | Polyclonal | 14, − | 15,− | nd |
| 15 | Clonal | Clonal | Clonal | 11, 13 | 06, 0301 | nd |
| 16 | Clonal | Clonal | Polyclonal | 11, 15 | 06, 0301 | 0401, 0402 |
| 17 | Polyclonal | Polyclonal | Polyclonal | 11, − | 02, 0301 | nd |
| 18 | Clonal | Clonal | Polyclonal | 01, 13 | 05, 06 | 0401, 0402 |
†CDR3 length distribution analysis of IgH and TCRγ gene rearrangements.
The dominant T-cell clones identified by TCRβ PCR were also detected by analysis of CDR3 length distributions of TCRγ gene rearrangements. This technique revealed the presence of a dominant clone in two further cases that had not been detected by PCR analysis of the TCRβ gene locus (Table 2) ▶ . Analysis of CDR3 length distributions of IgH gene rearrangements revealed a clonal B-cell population in addition to a T-cell clone in one of the cases (case 15, Table 2 ▶ ).
Micromanipulation and Single-Cell PCR
Two hundred thirteen specific PCR products were obtained from 1,004 cells micromanipulated from frozen tissue sections of seven cases of AILD (Table 3) ▶ . This PCR efficiency of 21% fits with our previous results. 15,18 No specific PCR product was obtained from any of the 334 negative control samples (Table 3) ▶ . In four of these cases a dominant clone had been found by whole-tissue DNA analysis. In these cases the clonal gene rearrangement was repeatedly obtained from CD4+ cells and the respective CD4+ T-cell clone seemed to dominate the population of CD4+ T cells. The clonal rearrangements were also detected in single Ki67+ cells micromanipulated from adjacent sections. Thus, a proliferating CD4+ T-cell clone was present in each of these cases. Only two clonally related T cells were identified among the CD8+ T cells of one case (case 3), which were unrelated to the clone identified by whole-tissue DNA analysis and only unrelated rearrangements were obtained from CD4+ and CD8+ T cells of the remaining three cases that had been negative for clonal expansions in the initial analysis of whole-tissue DNA. All sequences of rearranged TCR Vβ gene segments obtained in this study were completely identical to the respective germline sequences, supporting the view that TCR-V genes are generally not subject to somatic hypermutation. 19
Table 3.
Single-Cell PCR Analysis in AILD
| Case | Single CD4+ cells | Single CD8+ cells | Single Ki67+ cells | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Number of specific PCR products/ number of cells | Functional/ nonfunctional sequences | Clonal*/unique sequences | Number of specific PCR products/ number of cells | Functional/ nonfunctional sequences | Clonal*/unique sequences | Number of specific PCR products/ number of cells | Functional/ nonfunctional sequences | Clonal*/unique sequences | |
| 1 | 9 /32 | 9 /0 | 7 /2 | 12 /32 | 10 /2 | 0 /10 | 3 /32 | 3 /0 | 3 /0 |
| 2 | 2 /32 | 2 /0 | 2 /0 | 9 /52 | 6 /3 | 0 /7 | 3 /48 | 3 /0 | 3 /0 |
| 3 | 11 /32 | 9 /2 | 6 /3 | 19 /84 | 13 /6 | 2 /11 | 6 /32 | 6 /0 | 5 /1 |
| 4 | 16 /48 | 15 /1 | 0 /15 | 12 /32 | 8 /4 | 0 /8 | 19 /48 | 17 /2 | 0 /17 |
| 5 | 8 /48 | 8 /0 | 5 /3 | 7 /48 | 5 /1† | 0 /5 | 11 /44 | 11 /0 | 7 /4 |
| 6 | 23 /72 | 20 /3 | 0 /20 | 10 /72 | 8 /1† | 0 /8 | 4 /72 | 4 /0 | 0 /4 |
| 7 | 11 /54 | 11 /0 | 0 /11 | 13 /36 | 12 /† | 0 /12 | 5 /54 | 3 /† | 0 /3 |
| Controls‡ | 0 /106 | 0 /118 | 0 /110 | ||||||
*In each of the four cases harboring clones, the same clonal rearrangements were detected in the CD4+ and Ki67+ cells. Only potentially functional sequences were considered.
†The reading frame of a few sequences could not be unequivocally determined.
‡Controls include aliquots of the buffer covering the sections during micromanipulation and PCR buffer.
TCRβ Gene Segment Usage
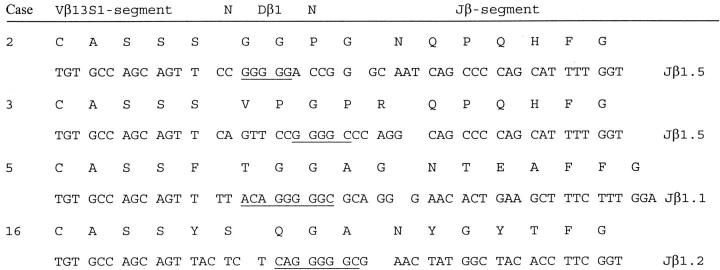
Table 4 ▶ displays the gene segments used in functional rearrangements of dominant T-cell expansions. In four of 10 cases in which a functional TCRβ gene rearrangement of an expanded T-cell clone was detected, this rearrangement used Vβ13S1. The overrepresentation of this gene segment raised the question whether the CD4+ T-cell clones of different cases of AILD possibly shared common antigen specificity. Comparison of deduced CDR3 amino acid sequences supported this hypothesis to some degree as the CD4+ clones identified in cases 2 and 3 not only shared the Vβ13S1 gene segment but also the length of the CDR3 and the Jβ element (Jβ1.5, Figure 1 ▶ ). However, MHC II-typing did not reveal a MHC II allele common to all of the cases that used the Vβ13S1 gene segment (Table 2) ▶ .
Table 4.
Gene Segment Usage of Functional TCRβ Rearrangements in T-Cell Clones of AILD
| Case | Vβ segment | Jβ segment |
|---|---|---|
| 1 | 2S1 | 1.6 |
| 2 | 13S1 | 1.5 |
| 3 | 13S1 | 1.5 |
| 5 | 13S1 | 1.1 |
| 10 | 15S1 | 2.7 |
| 11* | 17S1/18S1 | 2.6/1.1 |
| 14 | 9S1 | 2.2 |
| 15 | 5S1 | 1.1 |
| 16 | 13S1 | 1.2 |
| 18 | 2S1 | 1.6 |
*Two in-frame gene rearrangements were amplified in case 11 (Vβ17S1Jβ2.6/Vβ18S1Jβ1.1).
Figure 1.
CDR3 comparison of clonal TCR Vβ13S1 gene rearrangements in four cases of AILD. Segments are aligned according to the conserved motifs CAS of the Vβ and FG of the Jβ segment. The end of the Vβ and the beginning of the Jβ segment are marked by an empty space. Nucleotides encoded by the Dβ1 segment are underlined.
Discussion
The neoplastic nature of AILD has been a matter of debate since this disease was first described. 1-3 Still some authors distinguish between AILD as a nonneoplastic hyperimmune reaction and AILD-like T-cell lymphoma because some patients experience regression even without treatment. 20 Most patients affected by AILD, however, have a poor outcome, even with polychemotherapy, and frequently die of fatal infections in a state of immunodeficiency or progress to immunoblastic lymphoma. 21 The malignant clinical behavior and the detection of clonal TCR gene rearrangements in ∼70% of the cases support the concept that AILD is a T-cell lymphoma. No histological, immunohistochemical, or molecular biological parameter was found that could distinguish between patients with a favorable outcome and those with a severe prognosis. 12 Nature, origin, and biological relevance of the clonal lymphocyte populations are until now only poorly understood.
In the present study, in 12 of 18 cases of AILD a dominant T-cell clone was detected in DNA extracted from whole tissue specimens. We investigated CD4+, CD8+, and proliferating (Ki67+) cells isolated from histological sections of seven of these cases. In four of these seven cases a dominant T-cell clone had been found. In one of these cases (case 5), the clone was only detected by single-cell PCR and analysis of TCRγ CDR3 length distribution. In this case many unique T cells were found besides those of the clone (Table 3) ▶ . Hence, the clone was not detected by TCRβ PCR in whole tissue DNA because it was most likely below the detection limit of this technique. This might also apply for case 13 of which only paraffin-embedded tissue was investigated (Table 2) ▶ . In the four cases in which a dominant clone had been found, the clonal TCRβ gene rearrangements could be assigned to CD4+ and Ki67+ cells by single cell amplification. These clones of proliferating CD4+ cells seemed to account for the majority of T-helper cells in the tumor tissue. Only two clonally related cells were identified among the CD8+ T cells of one case. Whereas clonal expansions of CD8+ T cells have been described even in healthy individuals, 22,23 the presence of a dominant CD4+ T-cell clone is uncommon in normal lymphoid tissue. Therefore, it seems likely that the dominant clones of proliferating CD4+ T cells found in the four cases of AILD represent the tumor cell population. Previous investigations that tried to identify the lineage of the neoplastic T cells by double-immunohistochemical stainings with proliferation markers and T-cell markers failed to unequivocally determine the tumor cell phenotype. Some identified CD4+ cells, others reported CD8+ cells as the major proliferating T-cell population. 4,24-26 Furthermore, by double-immunohistochemical stainings alone it is not possible to make a statement regarding the clonality of cells stained. Our data support the view that the neoplastic T-cell clones in AILD are of CD4+ phenotype.
The peculiar histology of AILD, which displays a polymorphous cellular infiltrate of reactive cells as outlined above, is thought to be caused by an abnormal production of cytokines. Enhanced expression of tumor necrosis factor-α, lymphotoxin, interleukin-6, and interleukin-1β transcripts has been reported in AILD tissue. 27-29 These findings fit well with a CD4+ T-cell derivation of the neoplastic cells in AILD. Presuming that the reactive cells in the lymph node tissue are attracted by cytokines produced by the neoplastic cells this expression pattern would resemble that of inflammatory CD4+ TH1 cells. 30 The etiological factor that causes the abnormal proliferation of these cells, however, has yet to be established.
In three cases of the single cell analysis no dominant T-cell clone could be detected. By analysis of proliferating cells and T-cell subsets only sequences of unrelated TCR gene rearrangements were obtained. One might speculate that the neoplastic T cells do not necessarily dominate the CD4+ T-cell population at all stages of the disease and hence, the clone was not detected because of the limited number of cells investigated. By aberrant cytokine expression even a minor tumor cell infiltrate could account for the peculiar histology of AILD, as it is also seen in Hodgkin’s disease. It also has to be considered that the oligonucleotide primers applied for amplification of TCRβ gene rearrangements cover all but two Vβ gene segments. 15 T-cell clones using one of those two Vβ-gene segments in their respective gene rearrangements are thus not detected and, therefore, would have been missed in the single cell investigation. Furthermore, in a complex mixture of oligonucleotides as applied for amplification of TCRβ gene rearrangements in this study, PCR failure because of degradation of single primers cannot be totally ruled out.
At least in two cases of the present study, however, a different explanation seems more likely. In the single cell analysis, the fraction of micromanipulated proliferating cells that yielded a PCR product was very low in comparison to the CD4+ and CD8+ cells, especially in cases 6 and 7, suggesting that most of these cells were not T cells. Immunoblastic B-cell lymphomas developing in patients suffering from AILD have been reported frequently. 21,31-34 In these cases the AILD tumor clone may have been of B-cell origin. Indeed, in cases 4 and 6 of the present study dominant clonal B-cell proliferations were detected by single cell amplification of Ig gene rearrangements (manuscript in preparation, Tilmann Spieker, Andreas Bräuninger, personal communication).
Tumor clones in 4 of 10 cases used the Vβ13S1 gene segment. Given the limited number of cases studied, this overrepresentation might merely be coincidental, but in comparison to a control population of T cells from the peripheral blood where the Vβ13S1 segment is only found in 3.7% of the gene rearrangements 18 the usage of this segment is surprisingly high. The overexpression of a particular V gene segment raises the question whether the neoplastic CD4+ T-cell clones of different patients shared a common antigen specificity. CDR3 similarities between two of the rearrangements using Vβ13S1 supported this hypothesis to some degree. Alternatively, this overrepresentation could have been caused by stimulation of these clones by a superantigen. There was, however, no MHC II allele common to all of the cases with Vβ13S1-expressing tumor clones. Restricted usage of certain Vβ gene segments in clones of AILD has also been reported by Smith and colleagues. 13 In their panel of cases the Vβ2S1 gene segment, which was also found in 2 of 10 cases of the present study, was repeatedly found. Clones using the Vβ13S1 gene segment in their rearrangements were not reported. However, the Vβ13S1 gene segment may not have been efficiently amplified by Smith and colleagues 13 given that their consensus primer carried several mismatches to the Vβ13S1 gene segment. Both Vβ2 and Vβ13 gene segments have been found to be predominantly expressed by CD4+ T cells in lesions of Sjögren’s syndrome, 35,36 an autoimmune disorder associated with AILD. 37-39 One might speculate on basis of this data that an unknown type of antigen or superantigen triggering of the tumor cells or their precursors could be involved in the pathogenesis of at least some cases of AILD.
Acknowledgments
We thank Andreas Bräuninger and Klaus Rajewsky for critical reading of the manuscript and helpful discussion, and Christiane Gerhardt and Tanja Schaffer for technical support.
Footnotes
Address reprint requests to Klaus Willenbrock, Senckenberg Institute of Pathology, Theodor Stern Kai 7, 60590 Frankfurt am Main, Germany. E-mail: willenbrock@em.uni-frankfurt.de.
Supported by a grant from the Deutsche Krebshilfe, Dr. Mildred Scheel Stiftung, and by the Deutsche Forschungsgemeinschaft through SFB502. Ralf Küppers is supported by the Heisenberg program of the Deutsche Forschungsgemeinschaft.
References
- 1.Frizzera G, Moran EM, Rappaport H: Angio-immunoblastic lymphadenopathy with dysproteinaemia. Lancet 1974, 1:1070-1073 [DOI] [PubMed] [Google Scholar]
- 2.Lukes RJ, Tindle BH: Immunoblastic lymphadenopathy. A hyperimmune entity resembling Hodgkin’s disease. N Engl J Med 1975, 292:1-8 [DOI] [PubMed] [Google Scholar]
- 3.Radaszkiewicz T, Lennert K: Lymphogranulomatosis X (immunoblastic adenopathy): clinical features, treatment and prognosis. Dtsch Med Wschr 1975, 100:1157-1163 [DOI] [PubMed] [Google Scholar]
- 4.Feller AC, Griesser H, Schilling CV, Wacker HH, Dallenbach F, Bartels H, Kuse R, Mak TW, Lennert K: Clonal gene rearrangement patterns correlate with immunophenotype and clinical parameters in patients with angioimmunoblastic lymphadenopathy. Am J Pathol 1988, 133:549-556 [PMC free article] [PubMed] [Google Scholar]
- 5.Harris NL, Jaffe ES, Stein H, Banks PM, Chan JK, Cleary ML, Delsol G, De Wolf-Peeters C, Falini B, Gatter KC: A revised European-American classification of lymphoid neoplasms: a proposal from the International Lymphoma Study Group. Blood 1994, 84:1361-1392 [PubMed] [Google Scholar]
- 6.Harris NL, Jaffe ES, Diebold J, Flandrin G, Müller-Hermelink HK, Vardiman J, Lister TA, Bloomfield CD: The World Health Organization classification of neoplastic diseases of the hematopoietic and lymphoid tissues. Report of the Clinical Advisory Committee meeting, Airlie House, Virginia, November, 1997. Ann Oncol 1999, 10:1419-1432 [DOI] [PubMed] [Google Scholar]
- 7.O’Connor NT, Crick JA, Wainscoat JS, Gatter KC, Stein H, Falini B, Mason DY: Evidence for monoclonal T lymphocyte proliferation in angioimmunoblastic lymphadenopathy. J Clin Pathol 1986, 39:1229-1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiss LM, Strickler JG, Dorfman RF, Horning SJ, Warnke RA, Sklar J: Clonal T-cell populations in angioimmunoblastic lymphadenopathy and angioimmunoblastic lymphadenopathy-like lymphoma. Am J Pathol 1986, 122:392-397 [PMC free article] [PubMed] [Google Scholar]
- 9.Lipford EH, Smith HR, Pittaluga S, Jaffe ES, Steinberg AD, Cossman J: Clonality of angioimmunoblastic lymphadenopathy and implications for its evolution to malignant lymphoma. J Clin Invest 1987, 79:637-642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tobinai K, Minato K, Ohtsu T, Mukai K, Kagami Y, Miwa M, Watanabe S, Shimoyama M: Immunoblastic lymphadenopathy (IBL)-like T-cell lymphoma: clinico-pathologic, immunophenotypic and immunogenotypic analyses. Nippon Ketsueki Gakkai Zasshi - Acta Haematologica Japonica 1987, 50:1668-1676 [PubMed] [Google Scholar]
- 11.Griesser H, Feller A, Lennert K, Minden M, Mak TW: Rearrangement of the beta chain of the T cell antigen receptor and immunoglobulin genes in lymphoproliferative disorders. J Clin Invest 1986, 78:1179-1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lorenzen J, Li G, Zhao-Höhn M, Wintzer C, Fischer R, Hansmann ML: Angioimmunoblastic lymphadenopathy type of T-cell lymphoma and angioimmunoblastic lymphadenopathy: a clinicopathological and molecular biological study of 13 Chinese patients using polymerase chain reaction and paraffin-embedded tissues. Virchows Arch 1994, 424:593-600 [DOI] [PubMed] [Google Scholar]
- 13.Smith JL, Hodges E, Quin CT, McCarthy KP, Wright DH: Frequent T and B cell oligoclones in histologically and immunophenotypically characterized angioimmunoblastic lymphadenopathy. Am J Pathol 2000, 156:661-669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Küppers R, Zhao M, Hansmann ML, Rajewsky K: Tracing B cell development in human germinal centres by molecular analysis of single cells picked from histological sections. EMBO J 1993, 12:4955-4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roers A, Montesinos-Rongen M, Hansmann ML, Rajewsky K, Küppers R: Amplification of TCRbeta gene rearrangements from micromanipulated single cells: T cells rosetting around Hodgkin and Reed-Sternberg cells in Hodgkin’s disease are polyclonal. Eur J Immunol 1998, 28:2424-2431 [DOI] [PubMed] [Google Scholar]
- 16.Kneba M, Bolz I, Linke B, Bertram J, Rothaupt D, Hiddemann W: Characterization of clone-specific rearrangement T-cell receptor gamma-chain genes in lymphomas and leukemias by the polymerase chain reaction and DNA sequencing. Blood 1994, 84:574-581 [PubMed] [Google Scholar]
- 17.Linke B, Bolz I, Fayyazi A, von Hofen M, Pott C, Bertram J, Hiddemann W, Kneba M: Automated high resolution PCR fragment analysis for identification of clonally rearranged immunoglobulin heavy chain genes. Leukemia 1997, 11:1055-1062 [DOI] [PubMed] [Google Scholar]
- 18.Willenbrock K, Roers A, Blöhbaum B, Rajewsky K, Hansmann ML: CD8(+) T cells in Hodgkin’s disease tumor tissue are a polyclonal population with limited clonal expansion but little evidence of selection by antigen. Am J Pathol 2000, 157:171-175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roers A, Hansmann ML, Rajewsky K, Küppers R: Single-cell PCR analysis of T helper cells in human lymph node germinal centers. Am J Pathol 2000, 156:1067-1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frizzera G, Kaneko Y, Sakurai M: Angioimmunoblastic lymphadenopathy and related disorders: a retrospective look in search of definitions. Leukemia 1989, 3:1-5 [PubMed] [Google Scholar]
- 21.Knecht H, Schwarze EW, Lennert K: Histological, immunohistological and autopsy findings in lymphogranulomatosis X (including angio-immunoblastic lymphadenopathy). Virchows Arch 1985, 406:105-124 [DOI] [PubMed] [Google Scholar]
- 22.Posnett DN, Sinha R, Kabak S, Russo C: Clonal populations of T cells in normal elderly humans: the T cell equivalent to “benign monoclonal gammapathy” [published erratum appears in J Exp Med 1994, 179:1077]. J Exp Med 1994, 179:609-618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen DF, Doukhan L, Kalams S, Delwart E: High-resolution analysis of T-cell receptor beta-chain repertoires using DNA heteroduplex tracking: generally stable, clonal CD8+ expansions in all healthy young adults. J Immunol Methods 1998, 215:113-121 [DOI] [PubMed] [Google Scholar]
- 24.Watanabe S, Sato Y, Shimoyama M, Minato K, Shimosato Y: Immunoblastic lymphadenopathy, angioimmunoblastic lymphadenopathy, and IBL-like T-cell lymphoma. A spectrum of T-cell neoplasia. Cancer 1986, 58:2224-2232 [DOI] [PubMed] [Google Scholar]
- 25.Namikawa R, Suchi T, Ueda R, Itoh G, Koike K, Ota K, Takahashi T: Phenotyping of proliferating lymphocytes in angioimmunoblastic lymphadenopathy and related lesions by the double immunoenzymatic staining technique. Am J Pathol 1987, 127:279-287 [PMC free article] [PubMed] [Google Scholar]
- 26.Nakamura S, Suchi T: A clinicopathologic study of node-based, low-grade, peripheral T-cell lymphoma. Angioimmunoblastic lymphoma, T-zone lymphoma, and lymphoepithelioid lymphoma. Cancer 1991, 67:2566-2578 [DOI] [PubMed] [Google Scholar]
- 27.Foss HD, Anagnostopoulos I, Herbst H, Grebe M, Ziemann K, Hummel M, Stein H: Patterns of cytokine gene expression in peripheral T-cell lymphoma of angioimmunoblastic lymphadenopathy type. Blood 1995, 85:2862-2869 [PubMed] [Google Scholar]
- 28.Yamaguchi S, Kitagawa M, Inoue M, Tomizawa N, Kamiyama R, Hirokawa K: Cell dynamics and expression of tumor necrosis factor (TNF)-alpha, interleukin-6, and TNF receptors in angioimmunoblastic lymphadenopathy-type T cell lymphoma. Exp Mol Pathol 2000, 68:85-94 [DOI] [PubMed] [Google Scholar]
- 29.Ohshima KS, Suzumiya J, Kawasaki C, Kanda M, Kikuchi M: Cytoplasmic cytokines in lymphoproliferative disorders: multiple cytokine production in angioimmunoblastic lymphadenopathy with dysproteinemia. Leuk Lymphoma 2000, 38:541-545 [DOI] [PubMed] [Google Scholar]
- 30.Arai KI, Lee F, Miyajima A, Miyatake S, Arai N, Yokota T: Cytokines: coordinators of immune and inflammatory responses. Annu Rev Biochem 1990, 59:783-836 [DOI] [PubMed] [Google Scholar]
- 31.Boros L, Bhaskar AG, D’Souza JP: Monoclonal evolution of angioimmunoblastic lymphadenopathy. Am J Clin Pathol 1981, 71:856-860 [DOI] [PubMed] [Google Scholar]
- 32.Bauer TW, Mendelsohn G, Humphrey RL, Mann RB: Angioimmunoblastic lymphadenopathy progressing to immunoblastic lymphoma with prominent gastric involvement. Cancer 1982, 50:2089-2098 [DOI] [PubMed] [Google Scholar]
- 33.Pirker R, Schwarzmeier JD, Radaszkiewicz T, Lenzhofer R, Konrad K, Bettelheim P, Bauer K, Prischl F: B-immunoblastic lymphoma arising in angioimmunoblastic lymphadenopathy. Acta Haematol 1986, 75:105-109 [DOI] [PubMed] [Google Scholar]
- 34.Abruzzo LV, Schmidt K, Weiss LM, Jaffe ES, Medeiros LJ, Sander CA, Raffeld M: B-cell lymphoma after angioimmunoblastic lymphadenopathy: a case with oligoclonal gene rearrangements associated with Epstein-Barr virus. Blood 1993, 82:241-246 [PubMed] [Google Scholar]
- 35.Yonaha F, Sumida T, Maeda T, Tomioka H, Koike T, Yoshida S: Restricted junctional usage of T cell receptor V beta 2 and V beta 13 genes, which are overrepresented on infiltrating T cells in the lips of patients with Sjögren’s syndrome. Arthritis Rheum 1992, 35:1362-1367 [DOI] [PubMed] [Google Scholar]
- 36.Sumida T, Yonaha F, Maeda T, Tanabe E, Koike T, Tomioka H, Yoshida S: T cell receptor repertoire of infiltrating T cells in lips of Sjögren’s syndrome patients. J Clin Invest 1992, 89:681-685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diebold J, Zittoun R, Tulliez M, Reynes M, Tricot G, Bernadou A, Audouin J: Pseudolymphoma and lymphoproliferative syndromes in Gougerot-Sjögren syndrome. Sem Hop 1978, 54:1033-1040 [PubMed] [Google Scholar]
- 38.Bisson M, Massias M, Segond P, Jacquot JM, Brunaud MD: Association of angioimmunoblastic lymphadenopathy and Gougerot-Sjögren syndrome. Nouv Presse Med 1978, 7:2393. [PubMed] [Google Scholar]
- 39.Bignon YJ, Janin-Mercier A, Dubost JJ, Ristori JM, Fonck Y, Alphonse JC, Sauvezie BJ: Angioimmunoblastic lymphadenopathy with dysproteinaemia (AILD) and sicca syndrome. Ann Rheum Dis 1986, 45:519-522 [DOI] [PMC free article] [PubMed] [Google Scholar]