Abstract
PolyA simple repeat sequence deletions are common in tumors with microsatellite instability (MSI+). Such deletions occur one base at a time in DNA mismatch repair (MMR)-deficient yeast suggesting larger deletions in human MSI+ tumors represent multiple sequential stepwise losses. Sum total deletions in four polyA repeats were variable (between −17 to −45 bp) in 20 sporadic MSI+ colorectal cancers. Progressive but less extensive total deletions (maximum of −12 bp) occurred in similar polyA sequences in MMR-deficient mice (mlh1−/−) up to 478 days old. PolyA repeat lengths were relatively stable but already shortened in the MMR-deficient cell line HCT116. A transgene with 26 A’s transfected into HCT116 shortened an average of 3.8 bases pairs after 469 days in culture, less than average deletions of BAT25 (−5.3) or BAT26 (−9.0) in MSI+ cancers. These findings further suggest that extensive polyA deletions common in MSI+ tumors likely reflect multiple stepwise smaller deletions that accumulate more than hundreds of divisions after loss of MMR.
Tumors with microsatellite instability (MSI) usually have defects in DNA mismatch repair (MMR). 1 However MMR-deficient tissues do not necessarily exhibit MSI. For example, mutations are relatively rare in MMR-deficient mice. 2-4 An explanation for varying mutation burdens are different intervals since loss of MMR. Longer intervals since loss of MMR should yield greater numbers of microsatellite (MS) mutations.
One approach counts MS mutations by measuring the proportion of loci with alleles different from germline lengths. This is the basis for tumor classification as stable (MSI-S), and with low (MSI-L) or high (MSI-H) instability. 5 MSI-H but not MSI-S or MSI-L tumors have MMR defects. However, proportions of mutated loci do not completely count total mutations because a single MS allele can mutate multiple times. 6-8 Therefore, MS alleles can record both loss of MMR (by changes from germline) and the interval since loss of MMR (by the extent of changes from germline).
One of the first alterations observed in tumors lacking MMR were deletions in polyA simple repeat sequences. 9,10 Counting polyA mutations may be simpler than stepwise mutations in dinucleotide repeat loci 7 because most mutations are deletions. PolyA sequences such as BAT25 and BAT26 are frequently and extensively deleted. 9-12 Yeast polyA deletions are predominately single base pair losses 13 suggesting larger deletions result from multiple smaller sequential or stepwise replication errors that accumulate throughout many divisions. Multiple successive single base pair polyA deletions likely also occur in mammalian MMR-deficient tissues. 8,9,12 To further characterize polyA deletion dynamics, MMR-deficient tissues were examined at multiple such loci. Similar to CA-repeat MS loci, 7 polyA deletions in MSI+ tumors likely reflect hundreds of divisions since loss of MMR.
Materials and Methods
Specimens
DNA extracted from formalin-fixed, paraffin-embedded colorectal cancer sections were screened for MSI at four CA-dinucleotide MS loci 14 and two polyA repeats (BAT25, BAT26). The 20 MSI+ colorectal cancers in this study would be classified as MSI-H. 5 None of the patients had histories consistent with hereditary nonpolyposis colorectal cancer.
DNA was also extracted from fixed normal small and large intestines of mlh1−/−-deficient mice 15 of different ages. Colorectal cancer cell lines HT29, SW480, and SW837 (MMR-proficient); HCT116 and SW48 (MMR (MLH1)-deficient 16 ); and HCT15 [MMR (MSH6)-deficient 17 ] were obtained from American Type Culture Collection (Manassas, VA). Their germline BAT allele lengths were estimated from the average germline lengths of the MSI+ cancers.
A 26-bp polyA sequence was inserted into the nontranscribed multiple cloning site of a pBluescript based plasmid vector and transfected into HCT116. Stable transfected G418-resistant clones were isolated by limiting dilution and propagated in mass culture for 469 days. Multiple copies (>10) of the A26 transgene were present per cell.
MS Analysis
DNA was amplified incorporating 33P-dCTP during 32 to 38 polymerase chain reaction (PCR) cycles. Deletions were estimated by comparing lengths between tumor and normal DNA after electrophoresis on 6% sequencing gels. The length of an allele was determined by its most intense band using a phosphoimager (Molecular Dynamics, Sunnyvale, CA). BAT25, BAT26, BAT40, 18 and BAT20 (Table 1) ▶ were analyzed for the human specimens. Similar sized mbat polyA murine sequences (Table 1) ▶ were used to analyze mlh1−/− mice. Germline murine sizes were determined by amplification of MMR-proficient mlh1+/+ littermates. Mononucleotide repeat lengths were confirmed by sequencing some of their PCR products. Lengths of the A26 transgene were determined by sequencing PCR products cloned into bacteria (TA cloning kit; Invitrogen, Carlsbad, CA). Although multiple copies of the A26 transgene were present per cell, each measured allele likely reflect mutations in different cells because PCR was performed with DNA extracted from mass cultures.
Table 1.
BAT Loci and Primers
| Name | Locus | GenBank accession | PolyA length | Primers 5′ to 3′ |
|---|---|---|---|---|
| BAT20 | STS SHGC-14162 | G16290 | 20 | ACTTCTCCCACCCATGTGAG |
| human | CAACCTGGGATTCTTTTCCA | |||
| mbat30 | Hif1A | Y13656 | 19 | CGGCGAGAACGAGAAGAAAA |
| murine | CCAAGATGGCGACGTGGA | |||
| mbat25 | S100 beta protein | L22144 | 25 | GGAGTTCATGGCCTTCGTC |
| murine | CCCTCATGTCTGTTGCAGAA | |||
| mbat26 | Pituitary tumor-transforming gene | AF060887 | 26 | TCACCATCCATTGCACAGTT |
| murine | CTGCGAGAAGGTACTCACCC | |||
| mbat37 | HYLTK, tyrosine kinase | X83972 | 37 | TCTGCCCAAACGTGCTTAAT |
| murine | CCTGCCTGGGCTAAAATAGA | |||
| mbat47 | Alpha-mannosidase | AF107018 | 47 | CCGTGGTCTGAGTGATGATG |
| murine | IIx gene | CCAGCTGTTCTATCCGGTTC |
Results
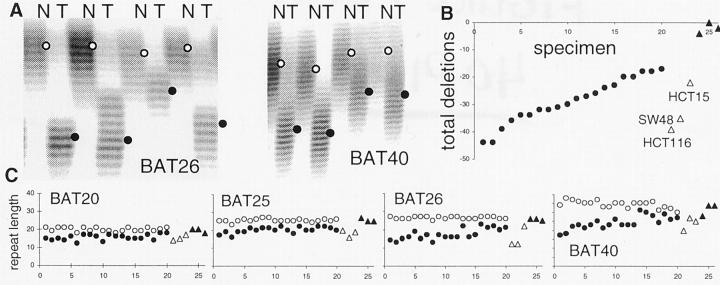
PolyA deletions were common and extensive in 20 MSI+ colorectal cancers (Figure 1) ▶ . Deletions, detected in all four examined polyA loci, were more extensive in some cancers. Total deletions throughout the four loci ranged from −17 to −45 bases with an average of −28.8 bases (Figure 1B) ▶ . For comparison, estimated deletions in MMR-proficient cell lines HT29, SW480, and SW837 were minimal to absent, and extensive in MMR (MLH1)-deficient HCT116 (−39 bases) and SW48 (−35 bases). Deletions in the MSH6-deficient cell line HCT15 (−22 bases) were at the minimum range observed in cancers.
Figure 1.
A: Examples of BAT26 and BAT40 deletions in MSI+ colorectal cancers. Lengths of the alleles are estimated from the densest band (open circle, normal; filled circle, tumor). B: Total deletions in BAT20, 25, 26, and 40 were variable for the 20 MSI+ colorectal cancers (specimens 1 to 20). For comparison, estimated total deletions for MMR-deficient (specimens 21 to 23; HCT116, SW48, HCT15) and MMR-proficient (specimens 24 to 26; HT29, SW480, SW837) colorectal cancer cell lines are illustrated. Deletions for the cell lines are estimates because matching normal tissues were unavailable and their starting germline sizes were assumed to be equal to the average lengths found in the 20 MSI+ tumor patients. C: Normal (open circles) and tumor (filled circles) allele lengths for MSI+ cancers and cell lines (triangles) at the four polyA loci.
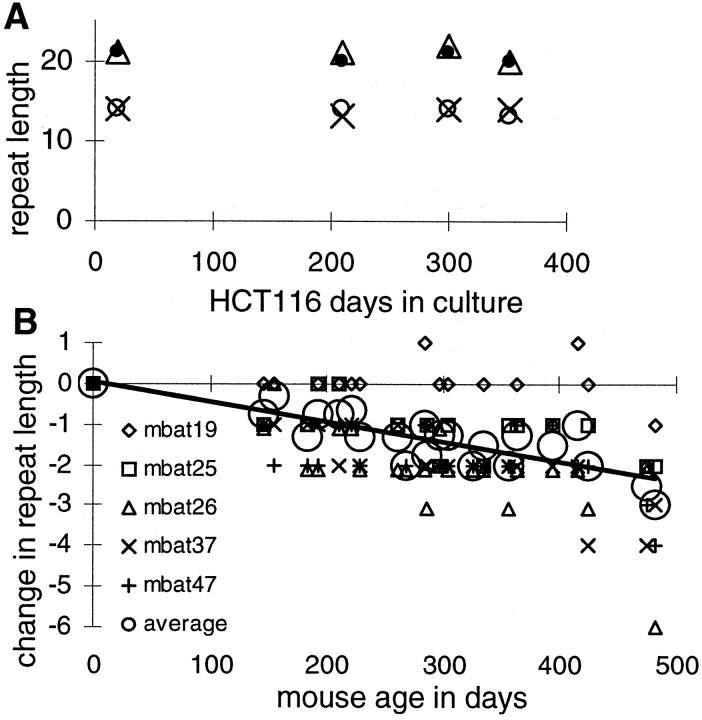
Serial observations allow sequence instability measurements. Already deleted polyA sequences were relatively stable in the MMR-deficient cell line HCT116 during 352 days in culture (Figure 2A) ▶ . In contrast, murine polyA (mbat) sequences of germline lengths similar to human BAT loci (Table 1) ▶ shortened in normal large and small intestines of MMR-deficient (mlh1−/−) mice (Figure 2B) ▶ . The mbat deletions were more frequent and extensive compared to HCT116, but less than the human MSI+ cancers. Losses increased with aging, with the largest deletion (−6 A’s) and greatest total deletion (−12 bases at four mbat loci similar to the BAT loci) in the oldest mouse (478 days old). Average deletions in the oldest mouse was −3.1 bp per locus.
Figure 2.
A: Serial measurements of BAT loci [BAT20 (X), 25 (filled circle), 26 (open circle), and 40 (triangle)] in a clonal isolate of HCT116. Further deletions in the already shortened BAT alleles were rare during 352 days in culture. B: Serial measurements of mbat loci (Table 1) ▶ in large and small intestines of mlh1−/− mice. In contrast to HCT116, progressive deletions occurred in these longer polyA repeats. Deletions in the oldest mouse (478 days old) were less than the MSI+ cancers (Figure 1) ▶ .
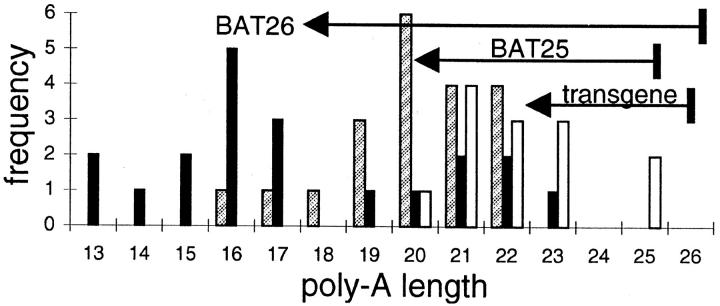
The relative lack of deletions in HCT116 during culture suggests deletion rates may be length-dependent. A lower length boundary was apparent for the MSI+ cancers (Figure 1C) ▶ as polyA lengths did not become shorter than 15 to 20 A’s. To investigate this potential lower length boundary, a transgene containing a stretch of 26 A’s was introduced into HCT116. After 469 days in culture (with a division rate of approximately once per day) the A26 transgene was deleted 1 to 6 bases (Figure 3) ▶ . The average A26 transgene deletion was less than BAT25 or BAT26 in cancers (−3.8 versus −5.3 or −9.0). Four other independently isolated A26-transfected HCT116 cell lines also demonstrated minimal changes during this culture period (data not shown).
Figure 3.
Distribution of polyA repeat lengths in MSI+ cancers (BAT25, shaded bars; BAT26, black bars) and the A26 transgene in HCT116 (open bars). Deletions in the A26 transgene were variable and less extensive than BAT loci in MSI+ cancers after 469 culture days. Arrows indicate average deletions.
Discussion
Genomic instability is a function of time and mutation rates. Here we demonstrate that extensive polyA deletions common in MSI+ cancers likely reflect several hundreds of divisions since loss of MMR. Deletions in MMR-deficient normal mouse intestines and an A26 transgene in a colon cancer cell line did not reach the extent observed in most MSI+ cancers even after ∼500 divisions. Their progressive and smaller deletions are consistent with a sequential stepwise deletion process.
These observations in mammalian cells are consistent with studies in yeast. Unrepaired slippage during DNA replication is the likely mechanism for MS mutations in MMR-deficient cells, 19,20 linking mutation with division. Yeast polyA mutations are predominantly single base deletions. 13 Therefore, multiple base pair deletions potentially count numbers of divisions needed to accumulate multiple individual smaller base pair deletion replication errors.
Counting mutations based on the extent of deletion may underestimate total mutations because less frequent insertions may also occur. Furthermore, deletions are nonlinear with respect to numbers of divisions because replication errors are a function of repeat length. For example, the mutation rate with an A14 repeat was estimated at 160 × 10−5 whereas it was 8.4 × 10−5 (∼19-fold lower) with an A12 repeat in yeast. 13 Similarly, by direct serial measurement deletions in already shortened polyA sequences in a MMR-deficient cell line were fewer compared to a longer repeat transfected into the same cell line, or longer repeats in normal murine intestines. Therefore, progressively more divisions are required for further deletion as a repeat shortens, which can account for the apparent lower length boundary (∼15 to 20 A’s) observed in MSI+ cancers and cell lines. Of note, CA-dinucleotide MS loci seem to be more suitable as molecular tumor clocks because additions are approximately equal to deletions that reduce a lower boundary effect. 7 Although counting dinucleotide MS mutations is more complex, mutations continue to be proportional to divisions throughout greater time intervals. 7
Further complicating division counting since loss of MMR is the stochastic nature of mutation. For example the extent of deletion varied for the A26 transgene within a single culture (Figure 3) ▶ . In addition, the underlying MMR deficiency may effect loss rates as compared to msh2-deficient cells, deletions with msh6-deficient yeast are ∼100-fold less frequent. 21 This observation is consistent with the fewer deletions estimated for the MSH6-deficient cell line HCT15 compared to MLH1-deficient cell lines. Therefore, variability of total deletions in MSI+ cancers (Figure 1B) ▶ may reflect factors other than numbers of divisions after loss of MMR. Nevertheless, it is apparent that deletions in MSI+ cancers are more extensive than in MMR-deficient normal and neoplastic cells observed throughout several hundred divisions.
Tumor analysis typically provides static genetic snapshots because progression is based on histological criteria. MS loci in MMR-deficient tumors provide an independent dimension of time because numbers of divisions since loss of MMR can be inferred by their amount of drift from germline regardless of tumor histology. The extensive polyA deletions observed in MSI+ cancers likely reflect hundreds of divisions since loss of MMR, consistent with a quantitative analysis of dinucleotide repeat loci that inferred ∼2000 divisions typically intervene between loss of MMR and tumor removal. 7 Some mutations recorded by MS loci may even precede visible progression or a gatekeeper mutation because loss of MMR and somatic mutations including deletions in BAT26 and BAT40 can accumulate in phenotypically normal cells. 22 The gradual accumulation of noncoding MS mutations during progression 9-12 may not be accompanied by recognizable changes in phenotype.
The ability to count mutations in noncoding MS loci may have important clinical implications because many selective mutations in MSI+ tumors seem to occur after loss of MMR. Frameshift mutations in short polynucleotide repeats of tumor suppressor genes are common in MSI+ tumors but rare in MSI− tumors. 18,23 Although progression to cancer can be limited by the infrequency of mutation, 24 mutations or time seem not to limit MSI+ cancers because most tumors present clinically within a few thousand divisions after loss of MMR. 7 Assuming one division per day, MSI+ colorectal tumors appear within a decade after loss of MMR compared to the decades estimated for progression of most colorectal cancers. 1 Limiting MSI+ cancers may be the initial loss of MMR, for example by methylation of MLH1 in sporadic colorectal cancers, 25 or host resistance factors such as immune surveillance. Interestingly, loss of HLA expression has been associated with MSI+ but not MSI− colorectal cancers. 26 Further studies may better define the analysis appropriate for reading tumor-specific histories from their mutations and deciphering the significance of these autobiographies.
Footnotes
Address reprint requests to Dr. Darryl Shibata, Department of Pathology, University of Southern California School of Medicine, 1200 N. State St. #736, Los Angeles, CA 90033. E-mail: dshibata@hsc.usc.edu.
Supported by National Institutes of Health Grants CN67010, CA58704 and CA70858.
References
- 1.Kinzler KW, Vogelstein B: Lessons from hereditary colorectal cancer. Cell 1996, 87:159-170 [DOI] [PubMed] [Google Scholar]
- 2.Tsao JL, Davis SD, Baker SM, Liskay RM, Shibata D: Intestinal stem cell divisions and genetic diversity: a computer and experimental analysis. Am J Pathol 1997, 151:573-579 [PMC free article] [PubMed] [Google Scholar]
- 3.Yao X, Buermeyer AB, Narayanan L, Tran D, Baker SM, Prolla TA, Glazer PM, Liskay RM, Arnheim N: Different mutator phenotypes in Mlh1- versus Pms2-deficient mice. Proc Natl Acad Sci USA 1999, 96:6850-6855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prolla TA, Baker SM, Harris AC, Tsao JL, Yao X, Bronner CE, Zheng B, Gordon M, Reneker J, Arnheim N, Shibata D, Bradley A, Liskay RM: Tumour susceptibility and spontaneous mutation in mice deficient in Mlh1, Pms1 and Pms2 DNA mismatch repair. Nat Genet 1998, 18:276-279 [DOI] [PubMed] [Google Scholar]
- 5.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S: A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 1998, 58:5248-5257 [PubMed] [Google Scholar]
- 6.Goldstein DB, Pollock DD: Launching microsatellites: a review of mutation processes and methods of inference. J Hered 1997, 88:335-342 [DOI] [PubMed] [Google Scholar]
- 7.Tsao JL, Yatabe Y, Salovaara R, Jarvinen HJ, Mecklin JP, Aaltonen LA, Tavaré S, Shibata D: Genetic reconstruction of individual colorectal tumor histories. Proc Natl Acad Sci USA 2000, 97:1236-1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shibata D, Peinado MA, Ionov Y, Malkhosyan S, Perucho M: Genomic instability in repeated sequences is an early somatic event in colorectal tumorigenesis that persists after transformation. Nat Genet 1994, 6:273-281 [DOI] [PubMed] [Google Scholar]
- 9.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M: Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature 1993, 363:558-561 [DOI] [PubMed] [Google Scholar]
- 10.Perucho M, Peinado MA, Ionov Y, Casares S, Malkhosyan S, Stanbridge E: Defects in replication fidelity of simple repeated sequences reveal a new mutator mechanism for oncogenesis. Cold Spring Harb Symp Quant Biol 1994, 59:339-348 [DOI] [PubMed] [Google Scholar]
- 11.Zhou XP, Hoang JM, Cottu P, Thomas G, Hamelin R: Allelic profiles of mononucleotide repeat microsatellites in control individuals and in colorectal tumors with and without replication errors. Oncogene 1997, 15:1713-1718 [DOI] [PubMed] [Google Scholar]
- 12.Percesepe A, Pedroni M, Sala E, Menigatti M, Borghi F, Losi L, Viel A, Genuardi M, Benatti P, Roncucci L, Peltomaki P, Ponz de Leon M: Genomic instability and target gene mutations in colon cancers with different degrees of allelic shifts. Genes Chromosom Cancer 2000, 27:424-429 [PubMed] [Google Scholar]
- 13.Tran HT, Keen JD, Kricker M, Resnick MA, Gordenin DA: Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol Cell Biol 1997, 17:2859-2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shibata D, Navidi W, Salovaara R, Li ZH, Aaltonen LA: Somatic microsatellite mutations as molecular tumor clocks. Nat Med 1996, 2:676-681 [DOI] [PubMed] [Google Scholar]
- 15.Baker SM, Plug AW, Prolla TA, Bronner CE, Harris AC, Yao X, Christie DM, Monell C, Arnheim N, Bradley A, Ashley T, Liskay RM: Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet 1996, 13:336-342 [DOI] [PubMed] [Google Scholar]
- 16.Boyer JC, Umar A, Risinger JI, Lipford JR, Kane M, Yin S, Barrett JC, Kolodner RD, Kunkel TA: Microsatellite instability, mismatch repair deficiency, and genetic defects in human cancer cell lines. Cancer Res 1995, 55:6063-6070 [PubMed] [Google Scholar]
- 17.Lettieri T, Marra G, Aquilina G, Bignami M, Crompton NE, Palombo F, Jiricny J: Effect of hMSH6 cDNA expression on the phenotype of mismatch repair-deficient colon cancer cell line HCT15. Carcinogenesis 1999, 20:373-382 [DOI] [PubMed] [Google Scholar]
- 18.Parsons R, Myeroff LL, Liu B, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B: Microsatellite instability and mutations of the transforming growth factor beta type II receptor gene in colorectal cancer. Cancer Res 1995, 55:5548-5550 [PubMed] [Google Scholar]
- 19.Strand M, Prolla TA, Liskay RM, Petes TD: Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature 1993, 365:274-276 [DOI] [PubMed] [Google Scholar]
- 20.Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye M: Frameshift mutations and the genetic code. Cold Spring Harb Symp Quant Biol 1966, 31:77-84 [DOI] [PubMed] [Google Scholar]
- 21.Sia EA, Kokoska RJ, Dominska M, Greenwell P, Petes TD: Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol Cell Biol 1997, 17:2851-2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parsons R, Li GM, Longley M, Modrich P, Liu B, Berk T, Hamilton SR, Kinzler KW, Vogelstein B: Mismatch repair deficiency in phenotypically normal human cells. Science 1995, 268:738-740 [DOI] [PubMed] [Google Scholar]
- 23.Schwartz S, Jr, Yamamoto H, Navarro M, Maestro M, Reventos J, Perucho M: Frameshift mutations at mononucleotide repeats in caspase-5 and other target genes in endometrial and gastrointestinal cancer of the microsatellite mutator phenotype. Cancer Res 1999, 59:2995-3002 [PubMed] [Google Scholar]
- 24.Loeb LA: Mutator phenotype may be required for multistage carcinogenesis. Cancer Res 1991, 51:3075-3079 [PubMed] [Google Scholar]
- 25.Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R: Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res 1997, 57:808-811 [PubMed] [Google Scholar]
- 26.Branch P, Bicknell DC, Rowan A, Bodmer WF, Karran P: Immune surveillance in colorectal carcinoma. Nat Genet 1995, 9:231-232 [DOI] [PubMed] [Google Scholar]