Abstract
We studied the relative RNA expression of clock genes throughout one 24-hour period in biopsies obtained from the oral mucosa and skin from eight healthy diurnally active male study participants. We found that the human clock genes hClock, hTim, hPer1, hCry1, and hBmal1 are expressed in oral mucosa and skin, with a circadian profile consistent with that found in the suprachiasmatic nuclei and the peripheral tissues of rodents. hPer1, hCry1, and hBmal1 have a rhythmic expression, peaking early in the morning, in late afternoon, and at night, respectively, whereas hClock and hTim are not rhythmic. This is the first human study to show a circadian profile of expression for all five clock genes as documented in rodents, suggesting their functional importance in man. In concurrent oral mucosa biopsies, thymidylate synthase enzyme activity, a marker for DNA synthesis, had a circadian variation with peak activity in early afternoon, coinciding with the timing of S phase in our previous study on cell-cycle timing in human oral mucosa. The major peak in hPer1 expression occurs at the same time of day as the peak in G1 phase in oral mucosa, suggesting a possible link between the circadian clock and the mammalian cell cycle.
In mammals, most physiological, biochemical, and behavioral processes vary in a regular and predictable periodic manner with respect to time of day (endogenous circadian rhythm). 1 The suprachiasmatic nuclei (SCN) of the anterior hypothalamus are the site of the circadian pacemaker in mammals. 2 Such evolutionary distant taxa as Synechococcus, Neurospora, Drosophila, and mammals share a common mechanism and in some cases homologous genes for circadian clock control. 3 Five mammalian clock genes, Clock, 4 Per, 5,6 Bmal1, 7 Tim, 8 and Cry 9 have been cloned and their interactions in an intracellular transcriptional/translational feedback loop studied. 3,10 Clock genes have a characteristic circadian expression in the SCN of nocturnally active rodents. The three mouse mPer genes (‘m ’ prefix for mouse) 11 peak at different times, with mPer1 peaking soon after subjective dawn (early activity period) and mPer3 and mPer2 peaking 2 hours and 4 hours later, respectively. mBmal1 RNA levels have a circadian rhythm with a peak after subjective dusk (late activity period). 12 mCry1 oscillates in the SCN with a rhythm similar to the phase of mPer2. 10 mClock 5,6 and mTim RNA levels do not oscillate in the SCN. 8 Clock genes are also expressed in rodent extra-neural tissues with a phase delay of 4 to 6 hours with respect to the SCN rhythm. 3,10,11,13,14 The human circadian pacemaker has been shown to be as stable and precise in measuring time as that of other mammals. 15 This suggests that the molecular mechanisms regulating the circadian rhythm in rodents should also be detectable in humans.
We previously reported that the nuclear expression of cell-cycle protein markers varies in an orderly and sequential manner throughout the day in the oral mucosa of diurnally active human study participants. 16 Specifically, a marker of late G1 phase (p53), is maximal at 11:00 hours, whereas markers of G1 to S phase (cyclin E), G2 phase (cyclin A), and M phase (cyclin B) are maximal at 15:00 hours, 16:00 hours, and 21:00 hours, respectively. The normal physiological progression throughout time for the peak expression of these proteins is consistent with previous rodent and human data showing a circadian variation in cell proliferation in gastrointestinal tract mucosa. 17-20
Thymidylate synthase (TS) catalyzes the reductive methylation of deoxyuridine-5′-monophosphate to deoxythymidine-5′-monophosphate, the only pathway for de novo synthesis of deoxythymidine-5′-monophosphate and subsequent DNA synthesis. 21 TS activity is highest during S phase, and can therefore be used to mark this event in the cell cycle. 22-24
We hypothesized that the human (‘h ’ prefix for human) clock genes hPer1, hCry1, hBmal1, hClock, and hTim, would be expressed in human oral mucosa and skin with a circadian profile analogous to that reported for rodent peripheral tissues (relative to the activity/rest cycle). Based on the known circadian coordination of proliferation in oral mucosa, we hypothesized that TS enzyme activity in oral mucosa would have a circadian rhythm with peak activity in early afternoon. We hypothesized also that there might be a predictable association between clock gene expression and cell-cycle phases.
Materials and Methods
Study Participants, Tissue Biopsies, and Serum Samples
The Sunnybrook Health Science Center Ethics Review Board approved the study protocol, and all study participants signed an informed consent. All study participants were healthy males, mean age 27.3 years (range, 22 to 33 years). The average time of sleep onset was 23:30 hours (range, 22:30 to 01:00 hours) and that for awakening was 07:15 hours (range, 06:30 to 09:00 hours). No attempt was made to synchronize the sleeping habits of the study participants before the study. The study participants slept during the night of the procedure except for the 30 minutes required to perform the biopsies at midnight and at 04:00 hours. All study participants had lunch at 12:30 hours and dinner at 20:30 hours. Snacks and cold drinks were allowed at the study participant’s discretion at other times. The six biopsies, from each of the eight study participants, were obtained during a single 24-hour period beginning at 08:00 hours on December 5, 1998.
Buccal mucosa and skin biopsies for RNA studies were obtained under local anesthesia using a 3-mm dermatological punch at 4-hour intervals beginning at 08:00 hours. Skin biopsies were obtained from the right hip, in an area not exposed to sun. All tissues were immediately snap-frozen in liquid nitrogen and stored at −70°C. A second biopsy was taken from the oral mucosa from all study participants at the same times for a separate study of TS enzyme activity. Serum was separated from blood and stored at −70°C until assayed for cortisol (microparticle enzyme immunoassay, Abbot Diagnostics) and melatonin. 25
RNA Isolation
Tissues were thawed and then minced using a homogenizer. Total RNA was extracted using Trizol (Gibco BRL) according to the manufacturer’s specifications. cDNA was generated by reverse transcription (Gibco BRL) of 2 μg of total RNA in a volume of 20 μl of buffer containing 2.5 mmol/L DTT, 20 U RNase inhibitor, 50 pmol random primer, 100 nmol dNTP, and 200 U of reverse transcriptase. The mix was incubated for 1 hour at 37°C, followed by three times dilution, and inactivation at 65°C for 10 minutes before storage at −20°C.
Polymerase Chain Reaction (PCR)
Primers were designed based on published data on human homologues of the clock genes in GenBank as follows: hClock: forward (f) AAGTTAGGGCTGAAAGACGACG, reverse (r) GAACTCCGAGAAGAGGCAGAAG, product size 171 bp. hPer1: f-CTGAGGAGGCCGAGAGGAAAGAA, r-AGGAGGAGGAGGCACATTTACGC, 132 bp; hBmal1: f-AAGGATGGCTGTTCAGCACATGA, r-CAAAAATCCATCTGCTGCCCTG, 132 bp; hTim: f-GGAGAAAGCTCAGCAGCATGATGA, r-TGCTCAATGAAGTGGAAGGTACGG, 138 bp; hCry1: f-ATCTAGCCAGGCATGCAGTT, r-CTCCAATCTGCATCAAGCAA, 132 bp; β-actin: f-GGGGCTGTGCTGTGGAAGCTAA, r-GTGCCAGGGCAGTGATCTCCTT, 208 bp; TS: f-CCAAAGCTCAGGATTCTTCG, r-GCACCCTAAACAGCCATTTC, 119 bp.
Duplex PCR was performed in a 25-μl reaction mixture containing 1 μl of cDNA, 0.2 μmol/L of each primer, 200 μmol/L dNTP, 2 U of Taq polymerase (Gibco BRL), and buffer (10 mmol/L Tris-HCl, pH 8.3, 50 mmol/L potassium chloride, 1.5 mmol/L magnesium chloride, and 1% Triton X-100). Each PCR reaction consisted of one cycle at 94°C for 5 minutes, followed by 30 cycles at 94°C for 30 seconds, 60°C for 1 minute, and 72°C for 90 seconds, with an additional 7-minute cycle at 72°C at the end of the reaction. First, primers for hClock were co-amplified with β-actin. After showing that there was no variation in the expression of hClock throughout a 24-hour period, primers for hBmal1, hTim, hPer1, hCry1, and TS were amplified together with hClock in a duplex PCR. Products were analyzed by the electrophoresis of 10 μl of PCR product through 2% agarose gel. The relative intensity of each band was compared using computer densitometry, and the results expressed as a ratio of the density for the gene of interest to that of the control gene in each sample.
TS Enzyme Activity
TS enzyme activity was studied only in oral mucosa to allow us to correlate the clock gene data with our previous data on cell-cycle progression in oral mucosa. Frozen tissues were thawed and homogenized in fixed volumes of ice-cold 200 mmol/L Tris-HCl, pH 7.4, with 100 mmol/L NaF, 20 mmol/L β-mercaptoethanol, and a protease inhibitor cocktail (Boehringer Mannheim, Mannheim, Germany) by mechanical homogenization using 40 complete strokes with a glass Dounce homogenizer. The homogenates were centrifuged at 4°C at 12,000 × g for 10 to 20 minutes, the supernatant fluid recovered, and protein content determined by the dye-binding method of Bradford. 26
TS catalytic activity was determined using a tritium release assay by the method of Armstrong and Diasio 27 with modifications. TS activity was determined on 35-μl freshly prepared 12,000 × g tissue supernatants in a final volume of 50 μl in 200 mmol/L Tris-HCl, pH 7.4, 100 mmol/L NaF, 20 mmol/L β-mercaptoethanol, protease inhibitors, 5 μl of cofactor solution (6.5 mmol/L tetrahydrofolic acid, 65 mmol/L NaHCO3, 65 mmol/L formalin, 0.25 mol/L β-mercaptoethanol, and 40 mmol/L sodium ascorbate, pH 7.0) and 10 μl of 50 μmol/L [5-3H]-2′-deoxyuridine 5′-monophosphate (20 Ci/mmol; Moravek Biochemical, Brea, CA) and 25 mmol/L CMP. Linearity was established for the working range of protein concentrations and incubation times at 37°C. Samples were incubated in triplicate at 37°C. At one time point, the radioactivity in the acid soluble fraction, after acid charcoal treatment, was determined. Background counts were consistently <5% of total input counts and standard deviations of triplicate determinations were <15%.
Data Analysis
The single cosinor method 28 was used to analyze for circadian rhythm individually and as a group, using both original data and data normalized from 0 to 1 (the six original data points for each variable throughout 24 hours divided by the largest value). This inferential method involves fitting a curve of a predefined period(s) by the method of least squares. The rhythm characteristics and their dispersions standard error (SE) and 95% confidence interval (CI)) estimated by this method include the mesor (middle value of the fitted cosine representing a rhythm-adjusted mean), the amplitude (half the difference between the minimum and maximum of the fitted cosine function), and the high point or acrophase (time of peak value in the fitted cosine function expressed as the lag in hours and minutes from midnight). The waveform of a time series may sometimes be more accurately approximated by the least squares fit of a multiple-component cosine model involving a concomitant fit of two or more components (ie, 24 hours plus 12 or 8 or 3 hours, etc). Each time series was tested for a circadian rhythm by the fit of a 24-hour single cosine and a 24- and 12-hour composite cosine model. The latter model was informative (ie, the 12-hour component significantly decreased residual error estimates beyond that of the 24-hour component alone) only for the analysis of hPer1. Detection of rhythm was achieved by rejection of the zero amplitude hypothesis with 95% certainty as reflected by the P value resulting from a comparison of residuals before and after the cosine curve fit. Rhythm characteristics (mesor, amplitude, acrophase) for each variable were compared between sites (skin versus oral mucosa) by a parameter test. 29 The six sampling time-normalized means were also analyzed for time-effect by one-way analysis of variance (F and P values).
Results
Circadian Coordination of Study Participants
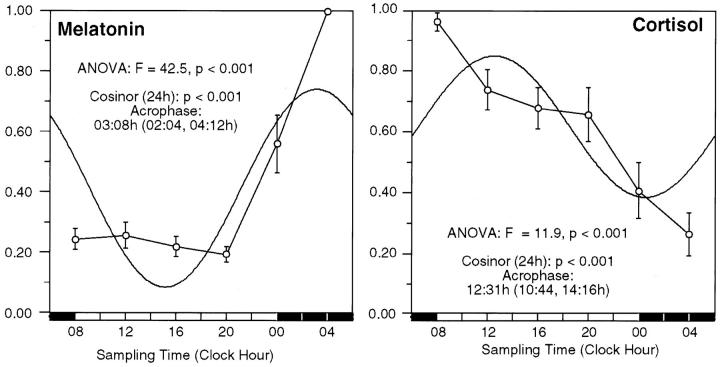
Cortisol and melatonin rhythms were studied in each individual during the biopsy period to further objectively document normal circadian time keeping and complement the daily patterns of reported sleep and activity. During the 24-hour biopsy period, all study participants showed the expected circadian variation in serum cortisol and melatonin (Figure 1) ▶ with peak values early in the day for cortisol (12:31 hours, cosinor, P < 0.001; analysis of variance: F = 11.9, P < 0.001) and at night for melatonin (03:08 hours, cosinor, P < 0.001; analysis of variance: F = 42.5, P < 0.001).
Figure 1.
Melatonin and cortisol rhythms during the 24-hour biopsy period. Normalized data for the serum melatonin and cortisol in eight study participants during the 24-hour biopsy period are shown. Time point means (normalized value) and SE are depicted along the 24-hour time scale. The best fitting 24-hour single component cosine curve is shown. The time for the acrophase (95% confidence limits), P value from cosinor analysis, and time-effect by analysis of variance are shown.
The peak time for cortisol by the cosinor curve fit is later than expected because of 12-hour components that are particularly prominent in two individuals (study participants 7 and 8). The raw data for cortisol show a peak at 08:00 hours for study participants 1 to 6. Study participant 7 has a primary peak at 0800 hours and a secondary peak at 2000 hours. Study participant 8 has a primary peak at 1200 hours and a secondary peak at 0800. When a 12-hour component is added to the model for cortisol, the peak is at 0924 hours. The raw data for melatonin show a peak at 0400 hours for all study participants.
Circadian Pattern of Clock Gene Expression
Figure 2, A through F ▶ , shows representative panels for the duplex PCR with hClock co-amplified with β-actin, and hPer1, hCry1, hBmal1, hTim, and TS. There was no significant 24-hour variation in hClock expression using β-actin as control (analysis of variance: F = 0.7, P = 0.593; cosinor: P = 0.162). Therefore, hClock was used in subsequent experiments as control for hBmal1, hPer1, hTim, hCry1, and TS. The data are summarized in Tables 1, 2, and 3 ▶ ▶ ▶ .
Figure 2.
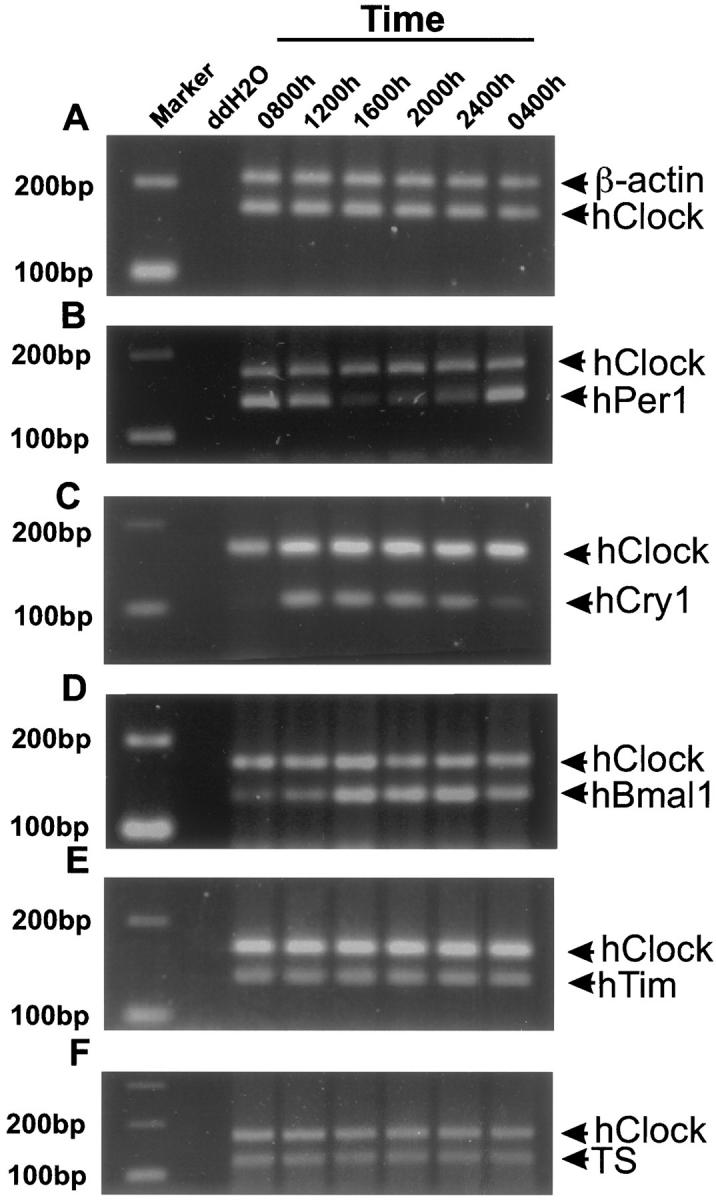
Examples of the results for duplex PCR for gene expression in human oral mucosa. Primers for hClock were co-amplified with β-actin (A). After showing that there was no significant variation throughout 24 hours in the expression of hClock versus β-actin, hPer1 (B), hCry1 (C), hBmal1 (D), hTim (E), and TS (F) were co-amplified with hClock. A to F show the relative expression of all four clock genes throughout 24 hours, a control lane (ddH2O), and a marker lane.
Table 1.
Normalized Data for the Expression of Clock Genes, TS mRNA, and TS Activity in Human Oral Mucosa
| Variable | Mesor, ± SE | Amplitude, ± SE | Amplitude, % | Acrophase and 95% CI for 24 hour fit (hour:min) | Cosinor P value (24 hour) | Analysis of variance P value |
|---|---|---|---|---|---|---|
| hPer1 | 0.55 ± 0.03 | 0.29 ± 0.04 | 52.7 | 08:29 (07:16 to 09:40) | <0.001 | <0.001 |
| hCry1 | 0.66 ± 0.03 | 0.22 ± 0.04 | 33.3 | 17:04 (15:36 to 18:36) | <0.001 | <0.001 |
| hBmal1 | 0.65 ± 0.02 | 0.35 ± 0.03 | 53.8 | 21:40 (21:00 to 22:20) | <0.001 | <0.001 |
| hClock | 0.77 ± 0.03 | 0.07 ± 0.04 | — | — | 0.162 | 0.593 |
| hTim | 0.78 ± 0.03 | 0.06 ± 0.04 | — | — | 0.265 | 0.638 |
| TS mRNA | 0.83 ± 0.02 | 0.04 ± 0.03 | — | — | 0.497 | 0.218 |
| TS Activity | 0.67 ± 0.03 | 0.16 ± 0.05 | 23.9 | 13:27 (11:00 to 16:00) | 0.008 | 0.076 |
SE, standard error; CI, confidence interval for the best fitting 24-hour single component cosine curve.
Table 2.
Normalized Data for the Expression of Clock Genes in Human Skin
| Variable | Mesor ± SE | Amplitude ± SE | Amplitude, % | Acrophase and 95% CI for 24 hour fit (hour:min) | Cosinor P value (24 hour) | Analysis of variance P value |
|---|---|---|---|---|---|---|
| hPer1 | 0.74 ± 0.03 | 0.13 ± 0.05 | 17.6 | 14:44 (11:44 to 17:44) | 0.024 | 0.005 |
| hBmal1 | 0.75 ± 0.02 | 0.16 ± 0.03 | 21.3 | 22:14 (20:48 to 23:40) | <0.001 | <0.001 |
| hClock | 0.78 ± 0.03 | 0.06 ± 0.04 | — | — | 0.293 | 0.722 |
| hTim | 0.84 ± 0.02 | 0.05 ± 0.03 | — | — | 0.294 | 0.562 |
SE, standard error; CI, confidence interval for the best fitting 24-hour single component cosine curve.
Table 3.
Normalized Data for the Expression of hPer1 in Human Oral Mucosa and Skin (12/24 Composite Cosinor Model)
| Variable | Mesor ± SE | Amplitude ± SE | Amplitude, % | Acrophase and 95% CI for 24/12 hour fit (hour:min) | Cosinor P value (24/12 hour) | Analysis of variance P value |
|---|---|---|---|---|---|---|
| Mucosa hPer1/pp | 0.55 ± 0.03 | 0.36 | 65.5 | 07:58 (06:28 to 09:28) | <0.001 | <0.001 |
| Skin hPer1/pp | 0.74 ± 0.03 | 0.23 | 31.1 | 10:52 (09:40 to 12:04) | 0.013 | 0.005 |
SE, standard error; CI, confidence interval for the best fitting 24-hour single component cosine curve; pp, primary peak.
hClock and hTim
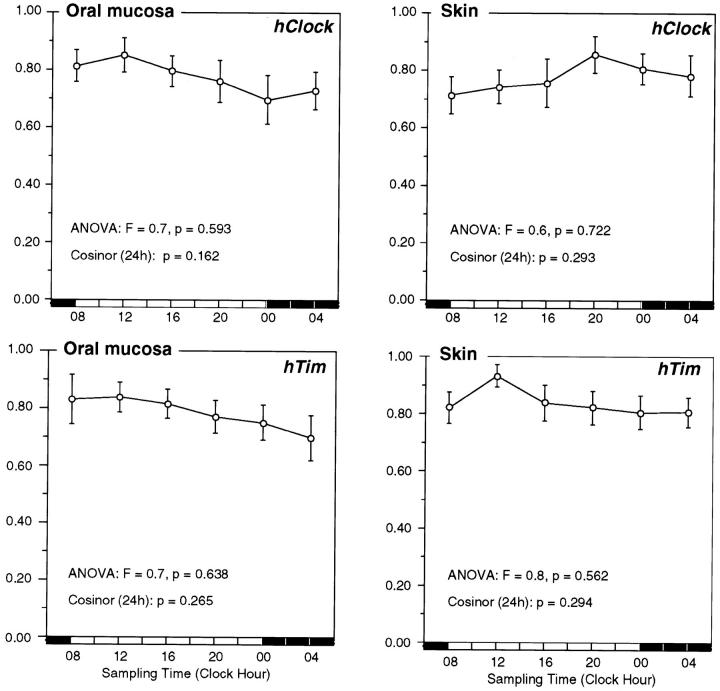
There was no significant 24-hour variation in hClock expression (Figure 3) ▶ in oral mucosa (analysis of variance: F = 0.7, P = 0.593; cosinor, P = 0.162) or skin (analysis of variance: F = 0.6, P = 0.722; cosinor, P = 0.293). The expression of hTim (Figure 3) ▶ throughout 24 hours was also nonrhythmic in both oral mucosa (analysis of variance: F = 0.7, P = 0.638; cosinor, P = 0.265) and skin (analysis of variance: F = 0.8, P = 0.638; cosinor, P = 0.294).
Figure 3.
The timing of hClock and hTim expression in oral mucosa and skin. Normalized data for the circadian expression of hClock, and hTim in human oral mucosa and skin of eight study participants. Time point means (normalized values) and SE are depicted along the 24-hour time scale.
hPer1
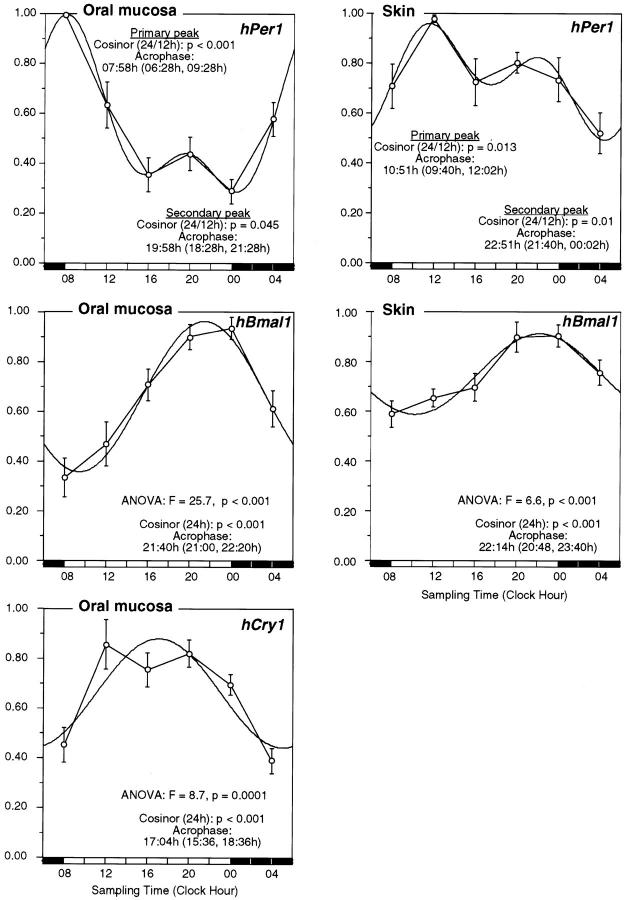
Although there is a significant 24-hour variation in the expression of hPer1 in both oral mucosa and skin (Tables 1 and 2) ▶ ▶ , a 12/24-hour composite cosinor model, explains a greater amount of the variance in the data and predicts better the times of day for peak values than the 24-hour single cosinor model (Table 3 ▶ and Figure 4 ▶ ). With the 12/24-hour composite cosine model, two significant daily peaks in hPer1 expression were detected in both tissues. The first and dominant peak was at 0758 hours (95% CI, 0628 to 0928 hours; cosinor, P < 0.001) and 1051 hours (95% CI, 09:40 to 1202 hours; cosinor, P = 0.013) in oral mucosa and skin, respectively. The predicted peak times for the major peaks in oral mucosa and skin are now much closer in time than with the 24-hour fit, but the timing of the peaks is still significantly different (P < 0.001 from parameter test). The second and smaller peak in hPer1 expression was at 1958 hours (95% CI, 1828 to 2128 hours; cosinor, P = 0.045) and 2251 hours (95% CI, 21:40 to 00:02 hours, cosinor, P = 0.01) in oral mucosa and skin, respectively. The timing of these peaks was also significantly different (P = 0.036 from parameter test). The percent amplitude of the primary peak for hPer1 was significantly lower in skin than in oral mucosa (65.5% versus 31.1%; P = 0.0182). In the oral mucosa, all eight study participants had peak measured (raw data) hPer1 expression at 0800 hours. In the skin, five of eight study participants had peak measured hPer1 expression at 1200 hours, with the remaining three peaking between 0000 and 0800 hours.
Figure 4.
The timing of hPer1, hBmal1, and hCry1 expression in oral mucosa and skin. Normalized data for the circadian expression of hPer1, hCry1, and hBmal1 in human oral mucosa and skin of eight study participants. Time point means (normalized values) and SE are depicted along the 24-hour time scale. For hBmal1 and hCry1, a 24-hour single-component cosine curve is shown along with the predicted acrophase (95% confidence limits) and P values from cosinor analysis and time-effect by analysis of variance. For hPer1, a 12/24 hours composite cosinor curve is shown along with the predicted acrophases for each peak (95% confidence limits) and P values for each rhythm component from cosinor analysis.
hBmal1 and hCry1
There was a significant 24-hour variation (Figure 4) ▶ in the expression of hBmal1, in both oral mucosa (analysis of variance: F = 25.7, P < 0.001) and skin (analysis of variance: F = 6.6, P < 0.001). A 24-hour cosinor fit to these data demonstrates a significant circadian rhythm with peak expression after dusk in both oral mucosa (2140 hours; 95% CI, 2100 to 2220 hours; cosinor, P < 0.001) and skin (2214 hours; 95% CI, 2048 to 2340 hours; cosinor, P < 0.001). The timing of the peak for hBmal1 in oral mucosa was identical to that in skin (P = 0.462 for difference). The percent amplitude of the variation in hBmal1 was significantly lower in skin than in oral mucosa (21.3% versus 53.8%; P < 0.0001). In all eight study participants, peak measured hBmal1 expression occurred between 2000 hours and 0000 hours both in the oral mucosa and skin.
There was a significant 24-hour variation (Figure 4) ▶ in the expression of hCry1 in oral mucosa with peak expression at 17:04 hours (95% CI, 1536 to 1836 hours; cosinor, P < 0.001; analysis of variance: F = 8.7, P < 0.001). In seven of eight study participants peak measured hCry1 expression occurred between 1200 hours and 1600 hours, with one study participant peaking at 2000.
Circadian Pattern of TS Activity and TS mRNA Levels in Oral Mucosa
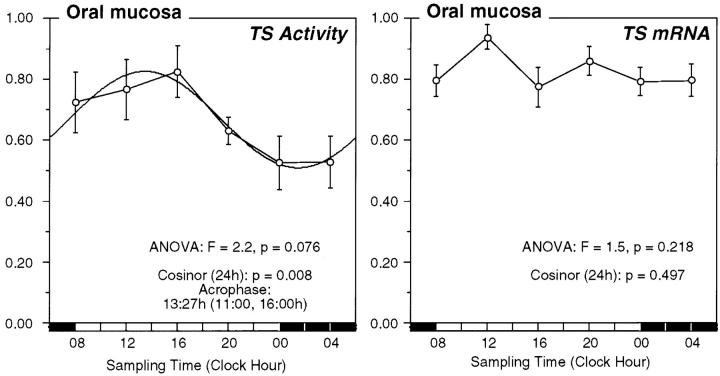
There was a significant circadian variation in TS activity (Figure 5) ▶ in oral mucosa with peak values in early afternoon at 1327 hours (95% CI, 1100 to 1600 hours, cosinor, P = 0.008; analysis of variance: F = 2.2, P = 0.076). The mean value for oral mucosal TS activity in the eight study participants was 8.5 ± 0.73 pmol/min/mg with a range of individual’s mean values of 6.7 to 16.6 pmol/minute/mg. Within individuals, oral mucosa TS activity varied 1.4- to 5.1-fold (mean, 3.2-fold) throughout the day, with values varying from 1.5 to 27.8 pmol/minute/mg. In all eight study participants, peak measured TS activity occurred between 0800 hours and 1600 hours, with six study participants having peak TS activity between the 4-hour time span from 1200 hours to 1600 hours. Protein yields in the tissue supernatant preparations failed to vary with time of day suggesting that these time of day differences in TS activity are not the result of differential dilution by variable amounts of total protein throughout the day.
Figure 5.
Circadian TS activity and TS mRNA expression in oral mucosa. Normalized data for the circadian variation in TS enzyme activity and TS mRNA expression in human oral mucous in eight study participants. Time point means (normalized value) and SE are depicted along the 24-hour time scale. The best fitting 24-hour single-component cosine curve is shown. The time for the acrophase (95% confidence limits), P value from cosinor analysis, and time-effect by analysis of variance are shown.
There was no circadian variation in TS mRNA expression in oral mucosa (cosinor, P = 0.497; analysis of variance: F = 1.5, P = 0.218).
Discussion
Clock Gene Expression in Human Tissues
This is the first study to show a rhythmic expression of clock genes in peripheral human tissue throughout 24 hours. Study participants had a normal circadian activity profile as documented by their reported sleep onset/awakening times and their rhythm in cortisol and melatonin. We have shown that hClock, hTim, hPer1, hCry1, and hBmal1 are expressed in human oral mucosa and skin, with a circadian profile that is consistent with that found in the SCN and peripheral tissues of mice and rats. 3 hPer1, hCry1, and hBmal1 show a rhythmic expression, peaking early in the morning (early activity), in late afternoon and at night (late activity), respectively, whereas hClock and hTim are not rhythmic. In the mouse SCN, mPer1 peaks soon after subjective dawn (early activity), mCry1 peaks later in the day, and mBmal1 peaks after subjective dusk (late activity), whereas mClock and mTim expression is not rhythmic. In rodent peripheral tissues the clock gene expression follows the same pattern, but Per1, Cry1, and Bmal1 peak 4 to 6 hours later than in the SCN. 10,11 These data suggest that the human clock genes may be functionally important for the molecular control of the human circadian pacemaker, thus extending the homology that exists between the clock genes in Drosophila and rodents to humans. The functional importance of the clock genes in man is further suggested by the recent finding of hClock expression in the human SCN 30 and that a hClock polymorphism is associated with an inherited human diurnal preference (eveningness). 31
When the data for hPer1 expression in oral mucosa and skin are analyzed using a 24/12 composite cosine model, two significant peaks (a major and a minor one) are detected in both tissues. In rodents, Per1, Per2, and Per3 expression peaks several hours apart during a 4- to 6-hour period within a given tissue. 11 Previous reports of per gene expression in peripheral tissues of rodents have documented one major 24-hour peak for each per gene. Some of these studies, however, used a limited number of sampling times and analysis for multiple rhythm components were not performed. Although the two daily peaks for hPer1, seen here, could represent hybridization of the hPer1 primers with more than one variant of hPer with different times of peak expression, a truly biphasic expression of hPer1 is possible and warrants further study.
Sampling every 4 hours is used commonly in human studies and rodent studies in chronobiology. Although more frequent sampling might allow for a better detection of rhythms with a shorter period than 24 hours, it could also interfere with and distress the study participants to the extent of masking the circadian rhythm. In discussions with our ethics committee, every 4-hour sampling was seen as the most we could expect our study participants to endure. This sampling frequency proved adequate to detect the expected circadian variation in clock gene expression.
TS in Oral Mucosa
This is the first study to look at the enzyme activity of TS in human tissue throughout 24 hours. The finding of a significant circadian rhythm in this S-phase marker further supports our previous finding of a circadian coordination of cell-cycle events in oral mucosa. 16 The timing of the peak of TS activity correlates well with the timing of S phase in the previous study (Figure 6, A ▶ and B3). A circadian variation has been demonstrated in rodent tissue TS enzyme activity 32 and rodent and in vitro studies have shown that peak TS activity coincides with S phase. 22-24
Figure 6.
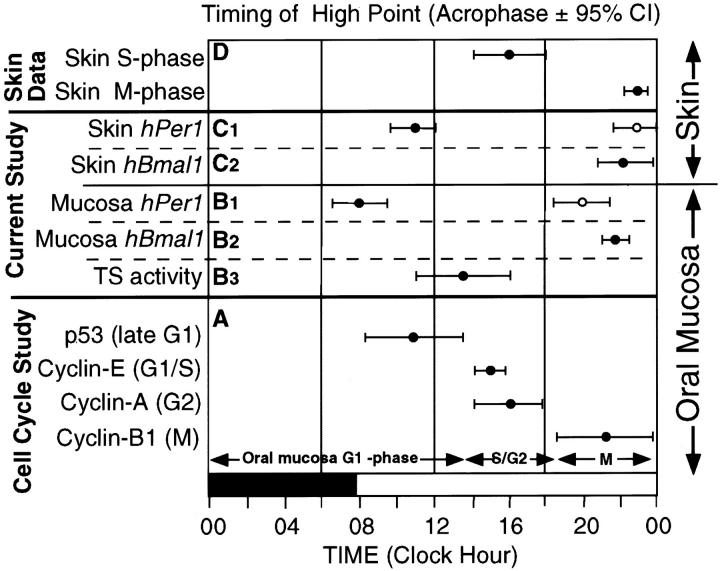
Association of circadian clock gene expression with the timing of cell-cycle phases. A: The high points for the best-fitting cosine for p53, cyclin-E, cyclin-A, and cyclin-B1 are depicted along the 24-hour time scale based on our previous study on human oral mucosa. The 95% confidence limits for each variable are shown. The cell-cycle phase, for which each protein is a marker, is shown in parentheses on the y axis and on the graph. B: The acrophases for the major (filled circle) and minor (open circle) peaks of expression for hPer1 (B1) and for the single peak in expression of hBmal1 (B2) in oral mucosa are depicted along the 24-hour time scale. In B3, the acrophase for the peak activity of TS (S phase) in oral mucosa is depicted along the 24-hour time scale. The 95% confidence limits for each variable are shown. C: The acrophases for the major (filled circle) and minor (open circle) peaks of expression for hPer1 (C1) and for the single peak in expression of hBmal1 (C2) in skin are depicted along the 24-hour time scale. The 95% confidence limits for each variable are shown. D: The acrophases for S phase and M phase in human skin based on pooled data from 12 studies looking at M phase and 14 studies looking at S phase throughout 24 hours are depicted along a 24-hour time scale. The 95% confidence limits for each variable are shown.
There was no rhythm in the expression of TS mRNA in oral mucosa, implying that the control of the rhythmic TS enzyme activity is at the posttranscriptional level. This is consistent with several studies showing that the acute induction of TS as cells enter S phase is controlled primarily at the translational level. 23,24,33,34
TS is the target for chemotherapy drugs such as 5-FU, FdUrd, and methotrexate, and also for several new folate-based TS inhibitors and multi-targeted antifolates. Studies of the circadian timing of FdUrd and 5-FU in rodents and humans show large differences in gastrointestinal toxicity and antitumor response depending on the time of day of therapy. 35-37 In a prospective randomized clinical trial, optimal timing (for reduced toxicity) of 5-FU-based chemotherapy reduced fivefold (14% versus 76%, P < 0.0001) the rate of severe oral mucosal toxicity. 37 Our findings of a circadian variation in both cell-cycle progression 16 and TS enzyme activity in human oral mucosa may partly explain this significant time-dependent incidence of 5-FU-induced oral mucosal toxicity.
Human Cell-Cycle Progression May Be Gated by the Circadian Clock
Gating of the cell division cycle by the circadian clock has been demonstrated in several unicellular organisms including Euglena, Cyanobacteria, Chlamydomonas, Paramecium, Tetrahymena, and Gonyaulax polyedra. 38-43 The data on the TS enzyme activity in oral mucosa allow timing of the peak in S phase in relation to the peak expression of the clock genes (Figure 6 ▶ , B3). The timing of the TS peak in early afternoon agrees well with the time of peak S-phase markers in our previous study (Figure 6A) ▶ of cell-cycle progression in human oral mucosa. 16 In oral mucosa, the major peak in hPer1 expression coincides with the peak in a G1 phase marker (p53) but precedes the peak in markers of S phase (cyclin E and TS). The peak for hBmal1 coincides with an M-phase marker (cyclin B1).
The information on human skin proliferation (Figure 6D) ▶ is based on our cosinor analysis of pooled data from 14 studies looking at the timing of S phase (peak, 16:06 hours; 95% CI, 14:04 to 18:08 hours), and 12 studies looking at the timing of M phase (peak, 22:54 hours; 95% CI, 22:12 to 23:36 hours). 44 Although we have not measured proliferation markers in the skin samples in this study, the coincidence of peaks in specific cell-cycle stages in skin compared to the oral mucosa is striking. With these limitations in mind, the major peak in hPer1 expression in skin may also coincide with the peak in G1 phase, whereas the peak in hBmal1 may coincide with the peak in M phase (Figure 6, C and D) ▶ .
Our correlation of the timing of clock gene expression in oral mucosa with the timing of S phase (TS activity), suggests that the circadian clock may, in part, control the timing of cell-cycle events in tissues undergoing continuous circadian rhythms in proliferation (gastrointestinal mucosa, skin, and bone marrow). That the cell cycle may be influenced by the circadian rhythm, is supported by the observation that phase-shifting of mice leads to a corresponding shift (throughout 3 to 4 weeks) in the timing of cell-cycle events both in gut and bone marrow. 45,46 Such phase-shifting has recently been shown to be associated with a corresponding rapid shift (1 day) in the circadian expression of Per1 in rat SCN and a delayed shift (≥6 days) in the Per1 rhythm in peripheral tissues. 47 Per1 peak expression has been found to coincide with the peak expression of clock-controlled genes that control downstream circadian processes. 48,49 In our study, the hPer1 peak coincides with the peak in oral mucosa G1 phase, where the main restriction point controlling progression through the cell cycle resides. 50 Although no cause and effect relationships between clock gene expression and cell proliferation can be claimed by our results, these data set the stage for a testable hypothesis to relate these two fundamental processes.
Many mouse and rat organs that are not actively proliferating (muscle, kidney, etc) have been shown to have a circadian expression of Per, Bmal1, and Cry. 10,11,13,14 Active tissue proliferation is therefore not a requisite for synchronous clock gene expression. In these tissues the clock genes may provide clock control of nonproliferative metabolic and physiological processes via the expression of clock-controlled genes. 51
Footnotes
Address reprint requests to Georg A. Bjarnason, M.D., Division of Medical Oncology, Toronto-Sunnybrook Regional Cancer Centre, 2075 Bayview Ave., Toronto, Ontario, M4N 3M5 Canada.
Supported by the Pharmaceutical Manufacturers Association of Canada Health Research Foundation/Medical Research Council of Canada (to G. A. B.), The Medical Research Council of Canada (to R. C. K. J. and Y. B.-D.), the National Cancer Institute of Canada (to G. A. B., R. C. K. J., and Y. B.-D.), the Office of Research and Development, Medical Research Service, Department of Veterans Affairs (to P. A. W. and W. J. M. H.), the Robison Family Foundation (to W. J. M. H.), and the National Institutes of Health (grants RO1 CA31635 to W. J. M. H. and RO1 CA50749 to W. J. M. H.).
References
- 1.Pittendrigh CS: Temporal organization: reflections of a Darwinian clock-watcher. Annu Rev Physiol 1993, 55:16-54 [DOI] [PubMed] [Google Scholar]
- 2.Weaver DR: The suprachiasmatic nucleus: a 25-year retrospective. J Biol Rhythms 1998, 13:100-112 [DOI] [PubMed] [Google Scholar]
- 3.Dunlap JC: Molecular bases for circadian clocks. Cell 1999, 96:271-290 [DOI] [PubMed] [Google Scholar]
- 4.Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald D, Dove WF, Pinto LH, Turek FW, Takahashi JS: Mutagenesis and mapping of a mouse gene, clock, essential for circadian behavior. Science 1994, 264:719-725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tei H, Okamura H, Shigeyoshi Y, Fukuhara C, Ozawa R, Hirose M, Sakaki Y: Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature 1997, 389:512-516 [DOI] [PubMed] [Google Scholar]
- 6.Sun ZS, Albrecht U, Zhuchenko O, Bailey J, Eichele G, Lee CC: RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell 1997, 90:1003-1011 [DOI] [PubMed] [Google Scholar]
- 7.Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ: Role of the CLOCK protein in the mammalian circadian mechanism. Science 1998, 280:1564-1569 [DOI] [PubMed] [Google Scholar]
- 8.Sangoram AM, Saez L, Antoch MP, Gekakis N, Staknis D, Whiteley A, Fruechte EM, Vitaterna MH, Shimomura K, King DP, Young MW, Weitz CJ, Takahashi JS: Mammalian circadian autoregulatory loop: a timeless ortholog and mPer1 interact and negatively regulate CLOCK-BMAL1-induced transcription. Neuron 1998, 21:1101-1113 [DOI] [PubMed] [Google Scholar]
- 9.Todo T, Ryo H, Yamamoto K, Toh H, Inui T, Ayaki H, Nomura T, Ikenaga M: Similarity among the Drosophila (6-4) photolyase, a human photolyase homolog, and the DNA photolyase-blue-light photoreceptor family. Science 1996, 272:109-112 [DOI] [PubMed] [Google Scholar]
- 10.Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM: mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 1999, 98:193-205 [DOI] [PubMed] [Google Scholar]
- 11.Zylka MJ, Shearman LP, Weaver DR, Reppert SM: Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron 1998, 20:1103-1110 [DOI] [PubMed] [Google Scholar]
- 12.Honma S, Ikeda M, Abe H, Tanahashi Y, Namihira M, Honma K, Nomura M: Circadian oscillation of BMAL1, a partner of a mammalian clock gene clock, in rat suprachiasmatic nucleus. Biochem Biophys Res Commun 1998, 250:83-87 [DOI] [PubMed] [Google Scholar]
- 13.Sakamoto K, Nagase T, Fukui H, Horikawa K, Okada T, Tanaka H, Sato K, Miyake Y, Ohara O, Kako K, Ishida N: Multitissue circadian expression of rat period homolog (rPer2) mRNA is governed by the mammalian circadian clock, the suprachiasmatic nucleus in the brain. J Biol Chem 1998, 273:27039-27042 [DOI] [PubMed] [Google Scholar]
- 14.Oishi K, Sakamoto K, Okada T, Nagase T, Ishida N: Antiphase circadian expression between BMAL1 and period homologue mRNA in the suprachiasmatic nucleus and peripheral tissues of rats. Biochem Biophys Res Commun 1998, 253:199-203 [DOI] [PubMed] [Google Scholar]
- 15.Czeisler CA, Duffy JF, Shanahan TL, Brown EN, Mitchell JF, Rimmer DW, Ronda JM, Silva EJ, Allan JS, Emens JS, Dijk DJ, Kronauer RE: Stability, precision, and near-24-hour period of the human circadian pacemaker. Science 1999, 284:2177-2181 [DOI] [PubMed] [Google Scholar]
- 16.Bjarnason GA, Jordan R, Sothern RB: Circadian variation in the expression of cell-cycle proteins in human oral epithelium. Am J Pathol 1999, 154:613-622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheving LE, Burns ER, Pauly JE, Tsai TH: Circadian variation in cell division of the mouse alimentary tract, bone marrow and corneal epithelium. Anat Rec 1978, 191:479-486 [DOI] [PubMed] [Google Scholar]
- 18.Buchi KN, Moore JG, Hrushesky WJM, Sothern RB, Rubin NH: Circadian rhythm of cellular proliferation in the human rectal mucosa. Gastroenterology 1991, 101:410-415 [DOI] [PubMed] [Google Scholar]
- 19.Marra G, Anti M, Percesepe A, Armelao F, Ficarelli R, Coco C, Rinelli A, Vecchio FM, D’arcangelo E: Circadian variation of epithelial cell proliferation in human rectal crypts. Gastroenterology 1994, 106:982-987 [DOI] [PubMed] [Google Scholar]
- 20.Warnakulasuriya KAAS, MacDonald DG: Diurnal variation in labelling index in human buccal epithelium. Arch Oral Biol 1993, 38:1107-1111 [DOI] [PubMed] [Google Scholar]
- 21.Radparvar S, Houghton PJ, Houghton JA: Characteristics of thymidylate synthase purified from a human colon adenocarcinoma. Arch Biochem Biophys 1988, 260:342-350 [DOI] [PubMed] [Google Scholar]
- 22.Mirjolet JF, Barberi-Heyob M, Merlin JL, Marchal S, Etienne MC, Milano G, Bey P: Thymidylate synthase expression and activity: relation to S-phase parameters and 5-fluorouracil sensitivity. Br J Cancer 1998, 78:62-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Navalgund LG, Rossana C, Muench AJ, Johnson LF: Cell cycle regulation of thymidylate synthetase gene expression in cultured mouse fibroblasts. J Biol Chem 1980, 255:7386-7390 [PubMed] [Google Scholar]
- 24.Ayusawa D, Shimizu K, Koyama H, Kaneda S, Takeishi K, Seno T: Cell-cycle-directed regulation of thymidylate synthase messenger RNA in human diploid fibroblasts stimulated to proliferate. J Mol Biol 1986, 190:559-567 [DOI] [PubMed] [Google Scholar]
- 25.Grota LJ, Snieckus V, De Silva SO, Tsui HW, Holloway WR, Lewy AJ, Brown GM: Radioimmunoassay of melatonin in rat serum. Prog Neuropsychopharmacol 1981, 5:523-526 [DOI] [PubMed] [Google Scholar]
- 26.Bradford MM: A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976, 72:248-254 [DOI] [PubMed] [Google Scholar]
- 27.Armstrong RD, Diasio RB: Improved measurement of thymidylate synthetase activity by a modified tritium-release assay. J Biochem Biophys Methods 1982, 6:141-147 [DOI] [PubMed] [Google Scholar]
- 28.Mojón A, Fernandez JR, Hermida RC: Chronolab: an interactive software package for chronobiologic time series analysis written for the Macintosh computer. Chronobiol Int 1992, 9:403-412 [DOI] [PubMed] [Google Scholar]
- 29.Bingham C, Arbogast B, Cornelissen-Guillaume G, Lee JK, Halberg F: Inferential statistical methods for estimating and comparing cosinor parameters. Chronobiologia 1982, 9:392-439 [PubMed] [Google Scholar]
- 30.Steeves TD, King DP, Zhao Y, Sangoram AM, Du F, Bowcock AM, Moore RY, Takahashi JS: Molecular cloning and characterization of the human CLOCK gene: expression in the suprachiasmatic nuclei. Genomics 1999, 57:189-200 [DOI] [PubMed] [Google Scholar]
- 31.Katzenberg D, Young T, Finn L, Lin L, King DP, Takahashi JS, Mignot E: A CLOCK polymorphism associated with human diurnal preference. Sleep 1998, 21:569-576 [DOI] [PubMed] [Google Scholar]
- 32.Lincoln DW, Hrushesky WJM, Wood PA: Circadian organization of thymidylate synthase activity in normal tissues: a possible basis for 5-fluorouracil chronotherapeutic advantage. Int J Cancer 2000, 88:479-485 [PubMed] [Google Scholar]
- 33.Jenh CH, Rao LG, Johnson LF: Regulation of thymidylate synthase enzyme synthesis in 5-fluorodeoxyuridine-resistant mouse fibroblasts during the transition from the resting to growing state. J Cell Physiol 1985, 122:149-154 [DOI] [PubMed] [Google Scholar]
- 34.Chu E, Koeller DM, Casey JL, Drake JC, Chabner BA, Elwood PC, Zinn S, Allegra CJ: Autoregulation of human thymidylate synthase messenger RNA translation by thymidylate synthase. Proc Natl Acad Sci USA 1991, 88:8977-8981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peters GJ, Van Dijk J, Nadal JC, Van Groeningen CJ, Lankelman J, Pinedo HM: Diurnal variation in the therapeutic efficacy of 5-fluorouracil against murine colon cancer. In Vivo 1987, 1:113-118 [PubMed] [Google Scholar]
- 36.von Roemeling R, Hrushesky WJM: Determination of the therapeutic index of floxuridine by its circadian infusion pattern. J Natl Cancer Inst 1990, 82:386-393 [DOI] [PubMed] [Google Scholar]
- 37.Lévi F, Zidani R, Misset JL: Randomized multicentre trial of chronotherapy with oxaliplatin, fluorouracil and folinic acid in metastatic colorectal cancer. Lancet 1997, 350:681-686 [DOI] [PubMed] [Google Scholar]
- 38.Carré IA, Edmunds LN: Oscillator control of cell divisions in Euglena: cyclic AMP oscillations mediate the phasing of the cell division cycle by the circadian clock. J Cell Sci 1993, 104:1163-1173 [DOI] [PubMed] [Google Scholar]
- 39.Mori T, Binder B, Johnson CH: Circadian gating of cell division in cyanobacteria growing with average doubling times of less than 24 hours. Proc Natl Acad Sci USA 1996, 93:10183-10188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goto K, Johnson CH: Is the cell division cycle gated by a circadian clock? The case of Chlamydomonas reinhardtii. J Cell Biol 1995, 129:1061-1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barnett A: Cell division: a second circadian clock system in Paramecium multimicronucleatum. Science 1969, 164:1417-1419 [DOI] [PubMed] [Google Scholar]
- 42.Wille JJ, Jr, Ehret CF: Light synchronization of an endogenous circadian rhythm of cell division in Tetrahymena. J Protozool 1968, 15:785-789 [DOI] [PubMed] [Google Scholar]
- 43.Sweeney BM, Hastings JW: Rhythmic cell division in populations of gonyaulax polyedra. J Protozool 1958, 5:217-224 [Google Scholar]
- 44.Brown WR: A review and mathematical analysis of circadian rhythms in cell proliferation in mouse, rat, and human epidermis. J Invest Dermatol 1991, 97:273-280 [DOI] [PubMed] [Google Scholar]
- 45.Scheving LE, Tsai TH, Scheving LA: Chronobiology of the intestinal tract of the mouse. Am J Anat 1983, 168:433-465 [DOI] [PubMed] [Google Scholar]
- 46.Tampellini M, Filipski E, Liu XH, Lemaigre G, Li XM, Vrignaud P, Francois E, Bissery MC, Levi F: Docetaxel chronopharmacology in mice. Cancer Res 1998, 58:3896-3904 [PubMed] [Google Scholar]
- 47.Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H: Resetting central and peripheral circadian oscillators in transgenic rats. Science 2000, 288:682-685 [DOI] [PubMed] [Google Scholar]
- 48.Jin X, Shearman LP, Weaver DR, Zylka MJ, De Vries GJ, Reppert SM: A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell 1999, 96:57-68 [DOI] [PubMed] [Google Scholar]
- 49.Ripperger JA, Shearman LP, Reppert SM, Schibler U: CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev 2000, 14:679-689 [PMC free article] [PubMed] [Google Scholar]
- 50.Murray AW: Creative blocks: cell cycle checkpoints and feedback controls. Nature 1992, 359:599-604 [DOI] [PubMed] [Google Scholar]
- 51.Claret CQ, Queiroz O: Multiple levels in the control of rhythms in enzyme synthesis and activity by circadian clocks: recent trends. Chronobiol Int 1990, 7:25-33 [DOI] [PubMed] [Google Scholar]