Abstract
Primary amyloidosis is a fatal disorder characterized by low numbers of clonal plasma cells in the bone marrow and the systemic deposition of light chain fragments in the form of amyloid. The molecular pathobiology of amyloidosis is primarily unknown. Recently, a novel karyotypically undetectable t(4;14)(p16.3;q32) translocation has been identified in ∼20% of multiple myeloma patients. The translocation leads to the apparent deregulation of two genes located on 4p16.3, the fibroblast growth-factor receptor 3 (FGFR3), and the putative transcription factor multiple myeloma SET domain (MMSET), and to the generation of IGH/MMSET hybrid transcripts. In this study, we investigated the presence of the t(4;14) translocation in 42 AL patients using a reverse transcriptase-polymerase chain reaction assay for the detection of IGH/MMSET transcripts. Chimeric transcripts were found in six patients (14%) and were consistent with a 4p16.3 breakpoint involving intron 3 and juxtaposing IGH regions to exon 4. In three of these cases, hybrid transcripts juxtaposing IGH regions to exon 5 were also observed and were probably the result of an alternative splicing skipping exon 4. Because all of the fusion transcripts (six of six) excluded exon 3, the first translated MMSET exon, only putative 5′ truncated MMSET proteins could be generated. In conclusion, our results demonstrate that the t(4;14)(p16.3;q32) translocation is a recurrent genetic lesion in primary amyloidosis.
Primary systemic amyloidosis (AL) is a plasma cell (PC) dyscrasia characterized by a deposition of monoclonal light chains in the form of amyloid fibrils that leads to progressive organ dysfunction and eventual death. 1 Knowledge of the biology of the underlying PC clone is needed to design and optimize therapeutic strategies and identify prognostic factors. Structural chromosomal alterations or genetic lesions affecting proto-oncogenes or tumor suppressor genes are important features that still have to be addressed in AL.
Over the last few years, we and others have demonstrated that chromosomal translocations involving the immunoglobulin heavy chain (IGH) switch regions at 14q32 are very frequent in PC dyscrasias involving a variety of chromosome loci, 2 mainly 11q13, 4p16.3, 16q23, and 6p25 where the putative target genes cyclin D1, FGFR3 and multiple myeloma SET domain (MMSET ), c-MAF and MUM1/IRF4 are respectively located. 3-8 The t(4;14)(p16.3;q32) translocation is of particular interest because it seems to be specifically associated with multiple myeloma (MM) (20% of cases), 9 and leads to the apparent deregulation of two potential proto-oncogenes, FGFR-3 (fibroblast growth factor receptor 3) 4,5 and MMSET. 6 It also leads to the formation of fusion IGH/MMSET hybrid transcripts, which are specific molecular markers that can be detected by reverse transcriptase-polymerase chain reaction (RT-PCR). 6,9
In this study, we investigated the presence of the t(4;14) translocation in primary AL by using a recently described sensitive RT-PCR assay 9 to look for related IGH/MMSET transcripts in bone marrow taken from 42 patients.
Materials and Methods
Patients
The patient population consisted of 42 randomly chosen patients with primary AL who underwent bone marrow aspiration at the coordinating center of the Italian Amyloid Program (Pavia, Italy). Amyloid was identified by means of Congo-red staining on tissue biopsies and/or abdominal fat aspirates taken after the patients had given their informed consent. Marrow PC clonality was assessed by means of double-staining immunofluorescence on Ficoll-separated mononuclear cells using fluorochrome-conjugated anti-light-chain isotype antisera (DAKO, Glostrup, Denmark); clonality is indicated by a κ/λ isotype ratio <1.1 (λ PC clone) or >2.6 (κ PC clone). 10 The patients showed a monoclonal component at serum or urine immunofixation using anti-isotype-specific rabbit-antisera on high-resolution agarose gel electrophoresis (DAKO). 10 Any association with clinically overt MM (percentage of PC >15% and renal failure or hypercalcemia or osteolytic bone lesions) was excluded by clinical and laboratory findings. There was no family history suggestive of hereditary AL in any patient.
Bone marrow from 11 normal donors, 9 patients with secondary thrombocytosis, 11 patients with primary thrombocythemia, and 3 patients with polycythemia vera were investigated as negative controls for the presence of the IGH/MMSET transcripts.
RT-PCR Analysis of IGH/MMSET Hybrid Transcripts and MMSET Gene Expression
Total RNA from the Ficoll-separated bone-marrow mononuclear cells of the AL patients, PC lines (KMS-11, NCI-H929, and OPM-2, used as positive controls for the three known translocation breakpoints), and 34 negative controls was extracted using Trizol (Life Technologies, Inc., Grand Island, NY). RT-PCR analysis was performed as described previously. 9 Briefly, 1 μg of total RNA was transcribed using Superscript RT (Gibco-BRL) and random hexamers (Pharmacia Biotech, Uppsala, Sweden). The first PCR round was performed using a portion of the first-strand cDNA as template, JH (5′-CCCTGGTCACCGTCTCCTCA-3′) or Iμ1 (5-AGCCCTTGTTAATGGACTTG-3′) as 5′ primers, and the 3′ primer mmset exon 6 reward (ms6r) (5′-CCTCAATTTCCCTGAAATTGGTT-3′) (see Figure 1 ▶ for primer positions). For nested-PCR, 1 μl of the first PCR round was re-amplified using Iμ2 (5-CTTTGCAAGGCTCGCAGTGAC-3′) and ms5r (5-AAGAACTGTACGTGATACT-3′) as internal primers (Figure 1) ▶ . The PCR products were electrophoresed on a 1.8% agarose gel in Tris borate-ethylenediaminetetraacetic acid and visualized by staining with ethidium bromide.
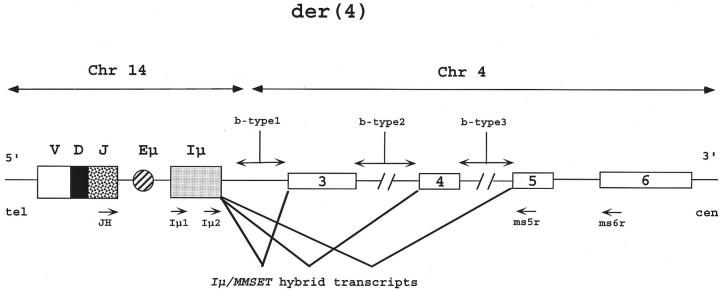
Figure 1.
Schematic representation of der(4) chromosome generated by the t(4;14)(p16;q32) translocation. As a consequence of the translocation, the VDJ unit, the immunoglobulin enhancer (Eμ) and the intron μ (Iμ) are telomeric to the MMSET gene and in the same transcriptional orientation. In this example, the translocation breakpoint is in MMSET intron 2 (breakpoint type 1, b-type 1); other characterized breakpoints involve intron 3 (b-type 2) or intron 4 (b-type 3). 6,9 IGH/MMSET fusion transcripts are generated from both the VDJ and the Iμ promoter, but only Iμ/MMSET transcripts are illustrated here for the sake of simplicity: VDJ or Iμ are spliced to the MMSET exon 3 (b-type 1), exon 4 (b-type 2), or exon 5 (b-type 3), depending on the translocation breakpoint. The approximate position of the primers (→) used for the first (JH or Iμ1 plus ms6r) and second (Iμ2 plus ms5r) RT-PCR rounds are shown below the map. The PCR fragment length localizes the 4p16.3 breakpoint. The figure is not to scale.
Dilution experiments with each of the control cell lines showed that the sensitivity of the nested RT-PCR for IGH/MMSET transcripts is such as to be able to detect one positive cell in a background of 10 5 normal cells. 9
DNA Sequencing
Direct DNA sequencing was performed on PCR-amplified fragments using the appropriate primers described above. The DNA fragments were purified by agarose gel extraction (QIA quick kit; Qiagen, Valencia, CA) and sequenced in both directions using an automated DNA sequencer (Applied Biosystems, Foster City, CA) and the Taq Dye Deoxy Terminator cycle sequencing kit (Perkin-Elmer, Norwalk, CT).
Results
Patient Population
Table 1 ▶ summarizes the clinical findings and laboratory values of the 42 AL patients included in our study. Median PC infiltration was 7%. Expansions of monoclonal PCs were detected by means of bone marrow light-chain κλ isotype ratio analysis in 37 cases (88%) and a monoclonal component was identified at serum and urine immunofixation on high-resolution agarose gel electrophoresis in 93% of the patients. These results are compatible with the sensitivities of the methods used, 10 and a combination of the two analyses 10 confirmed a monoclonal disorder in all cases. Twenty-four of the 42 patients were alive at the time of our study, the others having died from AL-related causes. Median survival was 32 months.
Table 1.
Clinical Findings and Laboratory Values for the 42 AL Patients Studied
| Median age (range) | 56 years (45–80) |
| Men/women | 26/16 |
| Median survival from diagnosis (range) | 32 months (1–80+) |
| Bone marrow aspirate | |
| Clonal plasma cell isotype ratio | 88% of patients |
| Plasma cell infiltration | 7% (median, range 1–15) |
| Monoclonal component | 93% of patients |
| λ light chain isotype | 69% |
| κ light chain isotype | 31% |
| Major organs involved (hierarchical) | no. of patients (%) |
| Heart | 18 (43) |
| Kidney | 12 (29) |
| Liver | 5 (12) |
| Peripheral nervous system | 2 (5) |
| Others | 5 (12) |
| No. of organs involved | no. of patients (%) |
| 1 | 15 (36) |
| 2 | 22 (52) |
| 3 | 3 (7) |
| 4 | 2 (5) |
IGH/MMSET Hybrid Transcripts in AL Patients
The t(4;14)(p16.3;q32) chromosomal translocation leads to the juxtaposition of the IGH locus to the MMSET gene in the same transcriptional orientation on both der(4) and der(14) chromosomes. 6 4p16 breakpoints occur within the 5′ introns of the MMSET gene or upstream of its coding sequence (probably in its regulatory regions); three different types of breakpoints have so far been characterized in MMs (Figure 1) ▶ . 6,9 The translocation leads to hybrid JH-MMSET and Iμ-MMSET transcripts from der(4) chromosome, probably initiating from the VDJ or Iμ (intron μ) promoters (Figure 1) ▶ . Hybrid MMSET-IGH transcripts from the reciprocal der(14) have been less frequently identified. 6
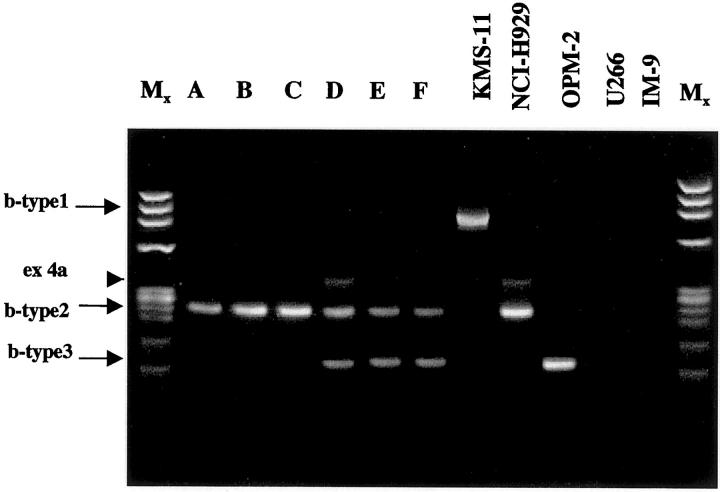
The presence of IGH-MMSET fusion transcripts was investigated in the panel of 42 patients using the previously described RT-PCR assay. No hybrid transcripts were detected in the first PCR round using both sets of IGH-MMSET primers (JH-msr6 or Iμ-msr6), a finding that is consistent with the scarce PC infiltration typical of primary AL. Nested-PCR (sensitivity, 10−5) was then performed using the Iμ2 and ms5r internal primers on the fragments obtained using the Iμ1 and ms6r primers (Figure 1) ▶ . Figure 2 ▶ shows the results from the six positive cases for IGH-MMSET rearrangements. A breakpoint-type 2 transcript was detected in three patients (Figure 2 ▶ , lanes A to C), whereas transcripts corresponding to b-type 2 and b-type 3 were observed in the other three cases (Figure 2 ▶ , lanes D to F). Finally, no hybrid IGH/MMSET transcripts were detected by single-round and nested RT-PCR in RNAs from the bone marrow of 34 negative controls.
Figure 2.
Translocation t(4;14)(p16;q32) in AL. Nested RT-PCR products from the six positive patients (lanes A–F); the KMS-11, NCI-H929, and OPM-2 cell lines are representative of the various types of fusion transcripts. U266 and IM-9 are negative controls. The arrowhead indicates a fusion transcript containing the alternative-splicing MMSET exon 4a. 9
Direct DNA sequencing of the amplified fragments confirmed that the types of the IGH-MMSET fusion transcripts were the same as those previously described (data not shown). 6,9
Clinicopathological Features of the Patients with a t(4;14) Translocation
Table 2 ▶ shows the clinicopathological features of the patients harboring the t(4;14) translocation. Two patients (Table 2 ▶ , PG and AR) had long survival times (36 and 63 months) and no subsequent MM was observed. In two cases (Table 2 ▶ , MG and AR), the PC clone was very small and the PC light chain isotype ratio was normal, a situation that is observed in ∼15% of AL patients. 10,11 Therapy consisted of melphalan and prednisone in all but nine cases treated with high-dose dexamethasone.
Table 2.
Clinicopathological Features of the Six AL Patients Positive for the t(4;14) Translocation (Breakpoint Types Are Reported)
| Patient | Age (yr) | Main organ involved | b-type | MC* | PC% | BMPC k/λ ratio | Survival (months) |
|---|---|---|---|---|---|---|---|
| MC | 45 | Heart | 2 | BJλ | 10 | 0.13 | 7 |
| OG | 62 | Kidney | 2 | IgGk | 7 | 7.3 | 4 |
| MG | 62 | Kidney | 2 | IgGk | 3 | 2.0 | 25 |
| PG | 80 | Heart | 2+ 3 | BJλ | 3 | 0.14 | 36 |
| AR | 62 | Heart | 2+ 3 | BJλ | 6 | 1.2 | 63 |
| OS | 65 | Heart | 2+ 3 | BJλ | 10 | 0.76 | 15+ |
Abbreviations: b-type, breakpoint type; MC, monoclonal component; PC, plasma cells; BMPC, bone marrow plasma cell.
*Detectable only by immunofixation.
Discussion
Chromosomal translocations affecting the IGH locus on 14q32 are a major mechanism of proto-oncogene activation in B-cell neoplasia. 12 Throughout recent years, it has been demonstrated that 14q32 translocations are very frequent in MMs, involving the IGH switch regions and a variety of proto-oncogenes including cyclin D1 on 11q13, FGFR3 and MMSET on 4p16.3, c-maf on 16q23, and MUM1/IRF4 on 6p25. 3-8 Fluorescence in situ hybridization studies have also demonstrated that 14q32 translocations involve virtually all clonal cells, thus suggesting that they occur very early in myelomagenesis. 13 These findings prompted us to study the presence of IGH translocations in primary AL, a PC-related disorder with a primarily unknown molecular pathobiology. In particular, we investigated the novel karyotypically undetectable t(4;14)(p16.3;q32) translocation, which has been shown by us and others to occur in ∼20% of MMs and leads to the apparent deregulation of the FGFR3 and MMSET genes located on 4p16.3. 4-6,9 Interestingly, it also leads to the formation of hybrid IGH/MMSET transcripts initiating from (V)DJ and Iμ IGH promoters relocated on chromosome der(4); 6,9 reciprocal MMSET/IGH transcripts from der(14) have also been found but significantly less frequently. It has been predicted that most of the chimeric IGH/MMSET transcripts do not encode putative fusion proteins, 6 but full-length MMSET proteins if the breakpoint is upstream of exon 3 (b-type 1), or 5′ truncated MMSET proteins respectively lacking the terminal 238 or 323 amino acids if the breakpoints are upstream of exon 4 (b-type 2) or 5 (b-type 3) (Figure 1) ▶ ; in-frame AUG codons are present in exons 3, 4, and 6. 6
The recent availability of a specific and reliable (fully fluorescence in situ hybridization-concordant) RT-PCR assay (single-round or nested PCR) for detecting IGH/MMSET hybrid transcripts from all of the of 4p16.3 breakpoints so far identified in MM 9 prompted us to search for t(4;14) in a panel of samples from 42 patients with primary AL. Given the scarce PC infiltration in AL, this assay should be considered a method of choice for detecting the possible presence of t(4;14). The patient series was fairly representative of a typical AL population in terms of PC infiltration, the distribution of organ involvement and survival. 14 IGH-MMSET hybrid transcripts were detected only by means of nested RT-PCR in 6 of the 42 patients (14%): the fusion transcripts involved MMSET exon 4 (b-type 2) in three cases and exon 4 and exon 5 (b-type 2 and 3) in the remaining three cases (Table 2 ▶ and Figure 2 ▶ ). None of the fusion transcripts included exon 3 (b-type 1). In a previous series of myeloma patients investigated using the same RT-PCR assay, 9 6 of 11 hybrid transcripts (54%) were type 1, whereas the only positive MGUS case had a b-type 2 transcript like our amyloid patients. Thus, if the various full-length and 5′ truncated MMSET proteins were translated from chimeric transcripts, these amyloid cell clones would generate 5′ truncated MMSET proteins from translocated alleles.
Concerning the three patients showing both the b-type 2 and 3 transcripts (Table 2 ▶ and Figure 2 ▶ , lanes D to F), these data were confirmed by direct sequencing (data not shown) and represent a novel finding because none of the 11 previously described MM patients with a t(4;14) translocation showed this pattern. 9 One possible explanation is that these transcripts represent different mRNA spliced forms of the MMMSET gene from a b-type 2 translocation.
Although IGH/MMSET transcripts were only detected by means of nested RT-PCR (which is consistent with the low level of PC infiltration in our patients), this finding raised the question as to whether this highly sensitive method (10−5) could detect hybrid transcripts from t(4;14) that may occur in the hematopoietic cells of normal individuals, as has been demonstrated for the t(14;18) chromosomal translocation. 15 Although we did not address this question specifically, our analysis showing no IGH/MMSET transcripts in bone marrows from 34 negative controls supports the validity of the nested RT-PCR approach in AL.
The analysis of various clinicopathological features in the six positive patients did not reveal any apparent correlation with the presence of fusion transcripts. However, because the number of positive cases was small, any clinicobiological relevance of this alteration remains to be elucidated in larger studies. This could be relevant because, although survival in AL is related to organ dysfunction, 14 it is also influenced by clone dimensions 11 and therapeutic response; biological variables are therefore needed to identify categories of patients whose worse prognosis warrant more aggressive therapies.
In conclusion, this study shows that the t(4;14) translocation is a recurrent lesion in AL patients (6 of 42, 14%). Its incidence seems to be higher than in MGUS (1 of 16 positive, 7%) and slightly lower than in MM (11 of 53 positive, 20%). 9 Taken together, these findings further support the concept that IGH translocations are common and relevant genetic events in PC dyscrasias.
Footnotes
Address reprint requests to Giampaolo Merlini, M.D., Biotechnology Research Laboratories, University Hospital-IRCCS Policlinico S. Matteo, P.le Golgi, 2, 27100 Pavia, Italy. E-mail: gmerlini@smatteo.pv.it.
Supported by grants from Associazione Italiana per lu Ricerca sul Cancro (to A. N. and G. M.), European Biomed 2 (program no. BMH4-CT 98-3689 to G. M.), Progetto di Ateneo, Ministero della Università e della Ricerca Scientifica e Tecnologica 1999 (nos. 9906038391-010 and 9906038391-007 to A. N. and G. M., respectively), Fondazione Ferrata-Storti, and Instituto di Ricovero e Cura a Carattere Scientifico Policlinico S. Matteo.
References
- 1.Perfetti V, Colli Vignarelli M, Casarini S, Ascari E, Merlini G: Biological features of the clone involved in primary amyloidosis (AL). Leukemia 2001, 15:195-202 [DOI] [PubMed] [Google Scholar]
- 2.Hallek M, Bergsagel PL, Anderson KC: Multiple myeloma: increasing evidence for a multistep transformation process. Blood 1998, 91:3-21 [PMC free article] [PubMed] [Google Scholar]
- 3.Ronchetti D, Finelli P, Richelda R, Baldini L, Rocchi M, Viggiano L, Cuneo A, Bogni S, Fabris S, Lombardi L, Maiolo AT, Neri A: Molecular analysis of 11q13 breakpoints in multiple myeloma. Blood 1999, 93:1330-1337 [PubMed] [Google Scholar]
- 4.Chesi M, Nardini E, Brents LA, Schroch E, Ried T, Kuehl WM, Bergsagel PL: Frequent translocation t(4;14)(p16.3;q32.3) in multiple myeloma is associated with increased expression and activating mutations of fibroblast growth factor receptor 3. Nat Genet 1997, 16:260-264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richelda R, Ronchetti D, Baldini L, Cro L, Viggiano L, Marzella R, Rocchi M, Otsuki T, Lombardi L, Maiolo T, Neri A: A novel chromosomal translocation t(4;14)(p16.3;q32) in multiple myeloma involves the fibroblast growth-factor receptor 3 gene. Blood 1997, 90:4062-4070 [PubMed] [Google Scholar]
- 6.Chesi M, Nardini E, Lim RSC, Smith KD, Kuehl MW, Bergsagel PL, Ried T: The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood 1998, 92:3025-3034 [PubMed] [Google Scholar]
- 7.Chesi M, Bergsagel PL, Shonukan OO, Martelli ML, Brents LA, Chen T, Schröck E, Ried T, Kuehl WM: Frequent dysregulation of the c-maf protooncogene at 16q23 by translocation to an immunoglobulin locus in multiple myeloma. Blood 1998, 91:4457-4463 [PubMed] [Google Scholar]
- 8.Iida S, Rao PH, Butler M, Corradini P, Boccadoro M, Klein B, Chaganti RS, Dalla-Favera R: Deregulation of MUM1/IRF4 by chromosomal translocation in multiple myeloma. Nat Genet 1997, 17:226-230 [DOI] [PubMed] [Google Scholar]
- 9.Malgeri U, Baldini L, Perfetti V, Colli Vignarelli M, Colombo G, Lotti V, Compasso S, Bogni S, Lombardi L, Maiolo AT, Neri A: Detection of t(4;14)(p16.3;q32) chromosomal translocation in multiple myeloma by reverse transcription-polymerase chain reaction analysis of IGH-MMSET fusion transcripts. Cancer Res 2000, 60:4058-4061 [PubMed] [Google Scholar]
- 10.Perfetti V, Garini P, Colli Vignarelli M, Marinone MG, Zorzoli I, Merlini G: Diagnostic approach to and follow-up of difficult cases of AL amyloidosis. Haematologica 1995, 80:409-415 [PubMed] [Google Scholar]
- 11.Perfetti V, Colli Vignarelli M, Anesi E, Garini P, Quaglini S, Ascari E, Merlini G: The degrees of plasma cell clonality and marrow infiltration adversely influence the prognosis of AL amyloidosis patients. Haematologica 1999, 84:218-221 [PubMed] [Google Scholar]
- 12.Gaidano G, Dalla Favera R: Molecular biology of lymphomas. De Vita VT Hellman S Rosenberg SA eds. Cancer: Principles and Practice of Oncology. 1997, :pp 2131-2145 Lippincott-Raven, Philadelphia [Google Scholar]
- 13.Finelli P, Fabris S, Zagano S, Baldini L, Intini D, Nobili L, Lombardi L, Maiolo AT, Neri A: Detection of t(4;14) (p16.3;q32) translocation in multiple myeloma by double-color fluorescent in situ hybridization. Blood 1999, 94:724-732 [PubMed] [Google Scholar]
- 14.Merlini G: AL amyloidosis. Kyle RA Gertz MA eds. Amyloid and Amyloidosis 1998. 1999, :pp 88-95 The Parthenon Publishing Group, New York [Google Scholar]
- 15.Aster JC, Kobayashi Y, Shiota M, Mori S, Sklar J: Detection of the t(14;18) at similar frequencies in hyperplastic lymphoid tissues from American and Japanese patients. Am J Pathol 1992, 141:291-299 [PMC free article] [PubMed] [Google Scholar]