Abstract
Neuroblastoma, the most common solid extracranial neoplasm in children, shows an appreciable variability in clinical evolution. Amplification of the MYCN oncogene in this tumor is detected in 25 to 30% of cases and is associated with poor clinical outcome. In this study, quantitative polymerase chain reaction and fluorescence in situ hybridization were used to determine MYCN amplification status in 46 neuroblastoma tumors. MYCN amplification was detected in tumors from 11 patients. Fluorescence in situ hybridization revealed the presence of micronuclei containing amplified MYCN sequences in 8 of the 11 tumors. Micronuclei are indicative of spontaneous elimination or loss of amplified sequences by tumor cells. Because the elimination of amplified sequences can be enhanced in vitro by specific drugs such as hydroxyurea, our observations suggest a new therapeutic strategy specifically targeted to cells with amplified genes.
Neuroblastoma (NB), a sympathetic nervous system-derived tumor, is the second most frequent solid tumor of early childhood. This tumor shows a wide spectrum of clinical behaviors, from spontaneous regression of even widespread tumors to rapidly progressing, metastatic disease resistant to intensive chemo- and radiotherapy. 1,2 There have been several parameters proposed to refine diagnosis and to predict biological behavior. In addition to stage, patient age, and histology, important prognostic indicators include ploidy, MYCN amplification (MNA), 1p deletion, 17q overrepresentation and alteration in neurotrophin receptor gene expression. 3-7 In localized and stage IVS NB, amplification of the MYCN proto-oncogene is a highly significant marker for a poor prognosis independent of other biological or clinical parameters and the risk of treatment failure in patients whose tumors display MNA is still very high. 8,9 Experiments with animal models as well as clinical observations have confirmed that MNA clearly contributes to the malignant phenotype of this disease. 10,11
The MYCN oncogene is amplified in ∼30% of advanced stage tumors as well as in most cell lines, typically derived from advanced stage tumors. As shown previously in vitro, extrachromosomal amplified genes in acentric double-minutes chromosomes (dmin) can be expelled from the nucleus and lost from the cell. Loss of amplified sequences is correlated with a loss of malignant properties and cellular differentiation. 12-16 Expelled material is seen as micronuclei, small nuclear-like structures. Micronuclei can arise from the nucleus by budding during S phase 17 or after mitosis as the nuclear membrane reforms. They can contain not only amplified oncogenes but also acentric chromosome fragments or whole damaged chromosomes. 18,19
In this study we assayed 46 NB tumors for MNA, 1p, and 17q status. We found 11 tumors with MNA and in at least 8 of the 11 we observed micronuclei comprising amplified MYCN sequences. Our work is the second report of spontaneous elimination of amplified genes occurring in vivo from NB cells.
Materials and Methods
Tissue Handling
Forty-six samples of NB were analyzed in our institute between November, 1998 and March, 2000. Tumor tissue obtained by fine needle biopsy directly from the tumor and/or from metastatic bone marrow (BM) was smeared directly on slides (imprinted slides) at the time of diagnosis and/or placed in culture medium. Biopsy samples in culture medium were divided into two aliquots, one for fluorescence in situ hybridization (FISH) and the second for quantitative polymerase chain reaction (PCR) by TaqMan (PE Biosystems, Roche Molecular Systems, Inc., Branchburg, NJ). Histological analysis showed 70 to 90% tumor cells in samples NB1-8 (Table 1) ▶ and only 6 to 11% tumor cells in NB9-11 (data not shown).
Table 1.
Characterization of MNA Neuroblastoma Tumors Identified by FISH to Exhibit Micronuclei Containing Amplified MYCN Sequences
| NB number | Tissue sample | Age | Stage | Survival | Genetic aberrations | |||
|---|---|---|---|---|---|---|---|---|
| Del 1p | MYCN Copy number per haploid genome | 17q over- represent | Micronuclei per 100 nuclei | |||||
| NB1 | Bone marrow | 19 mo | IV | 12 months DOD | + | 110 | + | 3 |
| NB2 | Primary tumor | 5 years | IV | 20 months alive | + | 35 | + | 3 |
| NB3 | Primary tumor | 2 years | IV | 18 months DOD | + | 298 | + | 5 |
| NB4 | Primary tumor | 3 years | IV | 19 months alive | + | ND | − | 6 |
| NB5 | Primary tumor | 28 months | IIIb/IV | 5 months alive | + | ND | − | 2 |
| NB6 | Primary tumor | 8 months | IV | 3 months alive | + | 79 | + | 20 |
| NB7 | Primary tumor | 4 years | IV | 3 months alive | + | 50 | + | 1 |
| NB8 | Primary tumor | 2.5 years | IV | 1 month alive | + | 60 | + | 10 |
NB7–8 micronuclei were analysed only on imprinted slides, NB1–6 on cytogenetic slides.
ND, Not done; DOD, dead of disease.
Cytogenetic Preparation and FISH Analysis
Samples for FISH analysis were mechanically dissociated, washed, and cultured overnight in RPMI 1640 with 10% fetal calf serum and 10 ng/ml colchicine. Interphase nuclei and metaphase spreads were prepared by standard cytogenetic procedures. Material on imprinted slides was fixed in methanol:acetic acid (3:1) for 15 minutes and kept at room temperature overnight.
FISH was performed with commercial probes D1Z5 (centromeric region of chromosome 1), MYCN, D2Zcen, D17Zcen, and D17qter (Quantum Appligene, Illkirch, France; Vysis, Inc., Downers Grove, IL). Probe D1Z2 (for detection of 1p36 deletion) was obtained from the American Type Culture Collection (Rockville, MD) and labeled in our laboratory. We used one probe combination (centromere plus specific locus) per slide, which means that for each tumor we analyzed three slides (one for chromosome 1, one for chromosome 2, and one for chromosome 17). Whole chromosome-painting probes were obtained commercially (Quantum Appligene; Vysis). Conditions of hybridization have been described elsewhere. 20 For each primary tumor we surveyed 500 interphase nuclei on cytogenetic preparations (NB1-6) and 200 nuclei on imprinted slides (NB6-8) to establish the percentage of MYCN-containing micronuclei/100 nuclei. For detection of 1p deletion and 17q overrepresentation, we analyzed 200 interphasic nuclei and the rare metaphase cells present (NB3-6). In BM samples, 500 nuclei were analyzed.
Quantitative PCR Analysis
The second portion of biopsy material was frozen and cut into 6- to 8-μm slices for histological evaluation of tumor cell content and for DNA extraction. Nucleic acids were extracted using a RNA/DNA kit (Qiagen, Inc., Valencia, CA). Quality of genomic DNA and total RNA was assessed by gel electrophoresis. Quantification of MYCN copy number was performed by real-time quantitative PCR using the ABI PRISM 7700 sequence detection system (PE Biosystems). PCR first cycle conditions were: denaturation at 95°C for 10 minutes and reannealing/extension at 50°C for 2 minutes followed by 40 cycles at 95°C for 15 seconds and at 60°C for 1 minute. The PCR reaction mixture was prepared in accordance with the recommendations of the manufacturer and contained 5 mmol/L MgCl2. For MYCN copy number determination we used the following primes: forward primer 5′GGCGTTCCTCCTCCAACAC, reverse primer 5′CGTTTGAAGATCAGCTCGC, and the Taqman probe FAM5′ ACATTCACCATCACTGTGCGTCCCAAG 3′TAMRA. Briefly, during the real-time quantitative PCR process, 5′ to 3′ exonuclease activity of the Taq DNA polymerase (Gold Taq DNA pol, PE Applied Biosystems) cleaves the dual-labeled TaqMan probe annealed to a target sequence, thus releasing the reporter fluorescent dye (FAM) from the quencher dye (TAMRA). Excitation of released FAM dye by argon laser results in an increase in fluorescence that is analyzed through software (PE Applied Biosystems). Ct values (defined as the fractional cycle number at which the fluorescence generated by cleavage of the probe crosses a fixed threshold) were obtained at each cycle. A calibration curve of Ct versus the quantity of a reference DNA was generated and a gene copy number for the test sample is determined by extrapolation. Two controls with known MYCN gene copy number were used: normal healthy donor DNA with 1 copy/haploid genome and the cell line IGR-N-91 with 470 copies/haploid genome. Normalization of MYCN gene copy number for each sample is made by comparison to two internal control genes, GAPDH, and albumin.
Results
In tumor biopsy material from 46 NB patients, 11 (24%) showed MNA. Of the 11, three could be analyzed only for presence of MYCN amplification because of a lack of material and a poor quality of imprints. In these three tumors (NB 9-11) 200 well-separated nuclei analyzed by FISH were disomic/tetrasomic for chromosome 2. There were no apparent micronuclei in these three samples, but the very low percentage (6 to 11%) of malignant neuroblasts must be taken to consideration. Of the eight remaining tumors (Table 1) ▶ , seven were near-diploid and one (NB6) was near-tetraploid. All eight also had a 1p deletion and six of eight had a 17q overrepresentation.
In six MNA tumors the exact level of amplification was established by real-time quantitative PCR. The number of copies/haploid genome ranged from 35 to 298 (Table 1) ▶ .
In all these cases micronuclei containing numerous MYCN sequences were seen on either cytogenetic preparation after colcemid treatment for 12 hours (NB1-6) or direct preparations on imprinted slides (NB6-8) (Figure 1a) ▶ . In NB1 the micronuclei were detected in BM metastasis (the only tissue available), in NB2-8 they were seen in primary tumors. The size of micronuclei was very variable and was ∼5 to 30% of NB nuclei (Figure 1c) ▶ .
Figure 1.
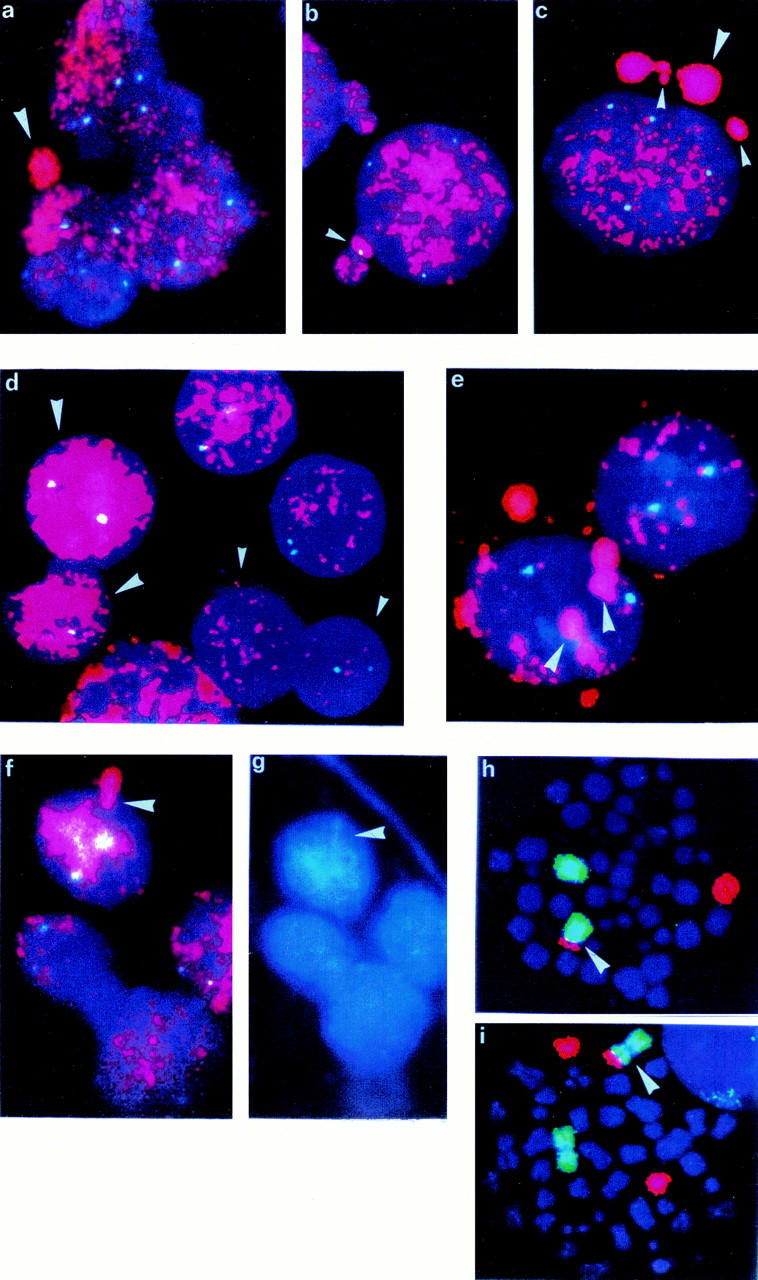
FISH with MYCN probe (red) and chromosome 2 centromeric probe (blue-green) a: Micronucleus seen directly in situ on touch imprint slide of NB6. b: Micronuclei in NB6 showing the centromeric signal of chromosome 2. c: Micronuclei of variable size seen in NB6. d: Amplification heterogeneity—dozens of copies of MYCN (small arrowhead) and hundreds of copies of MYCN (large arrowhead) in NB4. e: Accumulation of amplified material in well-defined area close to the nuclear membrane in NB2. f: Budding out of the nuclear membrane with amplified MYCN. g: The same picture in DAPI coloration showing drop-shape-like structure of extruded material in NB2 FISH with whole chromosome painting probes—chromosome 1 labeled by FITC, chromosome 17 by rhodamine. h: One der(1)t(1;17) (arrowhead) and one normal chromosome 1 and one normal chromosome 17 in NB4. i: One der(1)t(1;17) (arrowhead), one chromosome 1 normal, and two chromosomes 17 normal in NB3.
The frequency of micronuclei/100 nuclei varied from case to case (Table 1) ▶ and between samples of the tumor NB6 that was analyzed on both imprinted slides and cytogenetic preparations. Imprinted slides of NB6 showed a much lower frequency of micronuclei (2 mn/100 nuclei) compared to cytogenetic slides (20 mn/100 nuclei). Detailed analysis of 500 cells from the NB6 cytogenetic preparation showed the presence of 477 MYCN-amplified neuroblasts, 23 nonamplified cells (presumably infiltrating lymphocytes or surrounding normal tissue cells), and 98 micronuclei, which represents the highest frequency per 100 nuclei among these patients. In 16 of 98 micronuclei, together with MYCN-amplified sequences stained with rhodamine, there was a bright-green FITC signal of the chromosome 2 centromeric probe, indicating the co-expulsion of all or part of chromosome 2 (Figure 1b) ▶ . However, the size of micronuclei containing a centromere of chromosome 2 was not different from those that did not contain two centromeric sequences.
Apparent in our study is a marked heterogeneity in degree of amplification among cells from the same tumor (Figure 1d) ▶ . Some nuclei contain only a few (10 to 20) copies of the MYCN gene, whereas others show a very high level of amplification resulting in a uniform red staining. Also apparent are differences in the internal organization of amplified material in nuclei. Some nuclei are homogeneously stained, whereas in others we could easily distinguish one or several regions of accumulation of amplified material either in nuclei themselves (Figure 1e) ▶ or in buds on the surface of the nucleus (Figure 1, f and g) ▶ .
In addition to MNA, we also evaluated our eight tumor samples for presence of the 1p deletion (eight of eight) and the 17q over-representation (six of eight) (Table 1) ▶ . One of the two cases without 17q overrepresentation contained two apparently normal copies of chromosome 17 (NB5). The second tumor (NB4) had only one normal chromosome 17 and a structurally rearranged chromosome der(1)t(1;17), with simultaneous loss of genes on 17p and 1p (Figure 1h) ▶ . Moreover, a similar der(1)t(1;17) chromosome together with two (NB3, NB4, NB8) or four (NB6) normal chromosomes 17 were seen in rare metaphases (Figure 1i) ▶ .
Involved BM from patients NB2, NB3, NB7, and NB8 were also analyzed. These BM comprised only 0.3% (NB7), 0.5% (NB3), 2% (NB8), and 10% (NB2) neuroblasts unlike the BM from NB1 that comprised 90% neuroblasts. However, in matched BM we found the same genetic abnormalities as in primary tumors.
Discussion
Increase of gene dosage by DNA amplification is a common genetic mechanism for up-regulating gene expression and thus is important in tumorigenesis. Chromosomal abnormalities generally associated with DNA amplification are either dmin or homogeneously staining regions (hsr). 21 In metaphase spreads, dmin appear as small, spherical, usually paired chromosome-like structures that lack a centromere and do not segregate by the same mechanism used by chromosomes. 21,22 They can be lost during mitosis or extruded by a cell during S phase. 17 The elimination of acentric dmin from cancer cells has been demonstrated in vitro by many groups using different tumor cell lines, 12-16,23 including NB with amplified MYCN. 24
We now demonstrate that similar mechanisms may occur in the patient. In eight patients with MNA we observed the expulsion of MYCN sequences in micronuclei before therapy. Micronuclei size was variable even in the same tumor. One possible explanation is a variable DNA content. It was shown that the DNA content in radiation-induced micronuclei is influenced by several factors, eg, DNA synthesis in micronuclei or the presence of chromosome fragments or whole chromosomes. 25 In the NB6 case the presence of the some or all of chromosome 2 did not influence the size of observed micronuclei. The inclusion of chromosome 2 in micronuclei with dmin suggests a hypothesis that chromosome 2 is localized in interphase nuclei to the periphery of the nucleus, because a peripheral position is a prerequisite for DNA to be expelled. 17 This hypothesis is supported by the report of elevated micronucleus frequency in a healthy person associated with significant over-involvement of chromosome 2. 26 The specificity of involvement of chromosome 2 sequences in NB needs to be confirmed, however, because cells were treated for 12 hours with colchicine, a classic spindle poison that can induce chromosome loss in threshold concentrations. 27 The long colchicine incubation also could be a reason for the discrepancy in micronucleus frequency observed for NB6. The higher frequency of micronuclei in cytogenetic preparations could reflect the fact that colchicine can induce apoptosis. We cannot exclude the possibility, that at least some of the micronuclei containing MYCN-amplified sequences found in the colchicine-treated preparations could be apoptotic bodies. 28 Conversely, the lower frequency of micronuclei found on imprinted slides might be an underestimate, because nuclei were not as well separated and many micronuclei could have been hidden between compacted nuclei.
As seen in Table 1 ▶ and commonly found for NB, the number of amplified MYCN copies established by quantitative PCR is very variable. In our small series of patients we did not detect any correlation between number of micronuclei and the number of amplified MYCN gene copies. Seven of the eight patients were older than 1 year. All of them had MNA and clinically were stage IV NB (Table 1) ▶ . There are several factors and mechanisms that are involved in progression of a disease. One possible mechanism (which could be specifically induced) is expulsion of amplified genes by tumor cells, which has been observed to occur spontaneously. But why would a cell expel genes that it had previously amplified? There is a clear contradiction between gene amplification (which gives a cell a selective advantage) and spontaneous expulsion of amplified material (which can suppress the tumor phenotype). Spontaneous expulsion of amplified material could be considered as a self-defense mechanism to enable a cell to resume normal physiological functions (eg, replication, transcription) when the nucleus contains too many amplified sequences. There is a strict nuclear architecture and spatial distribution of different sequences into human chromosome territories in the nucleus 29-31 that must be respected. It is possible that, if amplification exceeds the available nuclear space, the cell has to eliminate the excess genes by expulsion.
Treatment in vitro with drugs that enhance micronuclear formation and thus expulsion of amplified genes have been shown to reduce the tumorigenicity and revert the phenotype of malignant cells. 12-16,32,33 In NB, MYCN amplification is associated with increased growth potential and tumorigenicity whereas down-regulation of MYCN expression correlates with decreased proliferation and differentiation induction. 34 Moreover, MNA late-stage tumors very often are refractory to chemotherapy, at least in part because the MYCN oncogene up-regulates expression of the MRP and MDR1 multidrug resistance genes in neuroblasts. 35-37 Enhancement of expulsion of amplified MYCN genes could be a way to reduce both tumorigenicity and the amplification-associated phenomenon of chemoresistance. Agents such as hydroxyurea, to which MDR1 over-expressing cells exhibit little or no cross resistance, have been shown to promote elimination of extrachromosomally amplified MYCN gene from tumor cells. 32,33 Inclusion of such drugs in conventional regimens constitutes an as yet unexplored therapeutic option specifically targeting aggressive cancer cells in the 30% of tumors with MYCN oncogene amplification.
Acknowledgments
We thank Dr. J. L. Biedler and Dr. R. A. Ross for helpful comments and their criticism of the manuscript; and B. Léon, B. Vasseur, and Y. Final for their technical assistance.
Footnotes
Address reprint requests to Dr. Alexander Valent, Laboratoire de Génomique Cellulaire des Cancers, Institut Gustave Roussy, 39, rue Camille Desmoulins, 94805 Villejuif, Cedex France. E-mail: avalent@igr.fr.
Supported by research grants from Ligue National Contre le Cancer, Association pour la Recherche sur le Cancer, and the Institute Gustave Roussy. A. Valent is a recipient of a fellowship from Ligue Contre le Cancer, Comité de l’Essonne, France.
References
- 1.Evans A, D’Angio G, Propert K, Anderson J, Hann HW: Prognostic factors in neuroblastoma. Cancer 1987, 59:1853-1859 [DOI] [PubMed] [Google Scholar]
- 2.Brodeur GM: Molecular basis for heterogeneity in human neuroblastomas. Eur J Cancer 1995, 31A:505-510 [DOI] [PubMed] [Google Scholar]
- 3.Brodeur GM, Fong CT, Morita M, Griffith R, Hayes FA, Seeger RC: Molecular analysis and clinical significance of N-myc amplification and chromosome 1 abnormalities in human neuroblastomas. Prog Clin Biol Res 1988, 271:3-15 [PubMed] [Google Scholar]
- 4.Schwab M, Alitalo K, Klempnauer KH, Varmus H, Bishop JM, Gilbert F, Brodeur G, Goldstein M, Trent J: Amplified DNA domain with limited homology to the myc cellular oncogene is shared by human neuroblastoma cell lines and a human neuroblastoma tumor. Nature 1883, 305:245-248 [DOI] [PubMed] [Google Scholar]
- 5.Kogner P, Barbany G, Dominici C, Castello MA, Raschella G, Persson H: Coexpression of messenger RNA for TRK protooncogene and low affinity nerve growth factor receptor in neuroblastoma with favorable prognosis. Cancer Res 1993, 53:2044-2050 [PubMed] [Google Scholar]
- 6.Meddeb M, Danglot G, Chudoba I, Vénuat AM, Bénard J, Avet-Loiseau H, Vasseur B, Le Paslier D, Hartmann O, Bernheim A: Additional copies of a 25 Mb chromosomal region originating from 17q23.1-17qter are present in 90% of high-grade neuroblastomas. Genes Chromosom Cancer 1996, 17:156-165 [DOI] [PubMed] [Google Scholar]
- 7.Bown N, Cotterill S, Lastowska M, O’Neil S, Pearson ADJ, Plantaz D, Meddeb M, Danglot G, Brinkschmidt C, Christiansen H, Laureys G, Speleman F: Gain of chromosome arm 17q and adverse outcome in patients with neuroblastoma. N Engl J Med 1999, 340:1954-1962 [DOI] [PubMed] [Google Scholar]
- 8.Rubie H, Hartmann O, Michon J, Frappaz D, Coze C, Chastagner P, Baranzelli MC, Plantaz D, Avet-Loiseau H, Benard J, Delattre O, Favrot M, Peyroulet MC, Thyss A, Perel Y, Bergeron C, Courbon-Collet B, Lemerle J, Sommelet D: N-myc gene amplification is a major prognostic factor in localized neuroblastoma: results of the French NLB90 study. J Clin Oncol 1997, 15:1171-1182 [DOI] [PubMed] [Google Scholar]
- 9.Katzenstein HM, Bowman LC, Brodeur GM, Thorner PS, Joshi VV, Smith EL, Look AT, Rowe ST, Nash MB, Holbrook T, Alvarado C, Rao PV, Castleberry RP, Cohn SL: Prognostic significance of age, MYCN oncogene amplification, tumor cell ploidy and histology in 110 infants with stage D(S) neuroblastoma: the Pediatric Oncology Group experience. J Clin Oncol 1998, 16:2007-2017 [DOI] [PubMed] [Google Scholar]
- 10.Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, Bishop JM: Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO J 1997, 16:2985-2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brodeur GM, Hogarty MD: Gene amplification in human cancers: biological and clinical significance. Vogelstein B Kintzler KW eds. The Genetic Basis of Human Cancer. 1998, :pp 161-172 McGraw-Hill, New York [Google Scholar]
- 12.Bonatti S, Miele M, Menichini P, Ottagio L, Abbondadolo A: Preferential loss of chromosomes containing amplified DNA regions in cultured cells. Prog Clin Biol Res 1989, 318:271-276 [PubMed] [Google Scholar]
- 13.Von Hoff DD, McGill JR, Forseth BJ, Davidson KK, Bradley TP, Van Devanter DR, Wahl GM: Elimination of extrachromosomally amplified myc genes from human tumor cells reduces their tumorigenicity. Proc Natl Acad Sci USA 1992, 89:8165-8169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ambros IM, Rumpler S, Luegmayr A, Hattinger CM, Strehl S, Kovar H, Gadner H, Ambros PF: Neuroblastoma cells can actively eliminate supernumerary MYCN gene copies by micronucleus formation—sign of tumor cell revertance? Eur J Cancer 1997, 33:2043-2049 [DOI] [PubMed] [Google Scholar]
- 15.Shimizu N, Nakamura H, Kadota T, Kitajima K, Oda T, Hirano T, Utiyama H: Loss of amplified c-myc genes in the spontaneously differentiated HL-60 cells. Cancer Res 1994, 54:3561-3567 [PubMed] [Google Scholar]
- 16.Eckhardt SG, Dai A, Davidson KK, Forseth BJ, Wahl GM, Von Hoff DD: Induction of differentiation in HL-60 cells by the reduction of extrachromosomally amplified c-myc. Proc Natl Acad Sci USA 1994, 91:6674-6678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimizu N, Itoh N, Utiyama H, Wahl GM: Selective entrapment of extrachromosomally amplified DNA by nuclear budding and micronuclei during S phase. J Cell Biol 1998, 140:1307-1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fenech M: The advantages and disadvantages of the cytokines-block micronucleus method. Mutat Res 1997, 392:11-18 [DOI] [PubMed] [Google Scholar]
- 19.Müller WU, Nüsse M, Miller BM, Slavotinek A, Viaggi S, Streffer C: Micronuclei: a biological indicator of radiation damage. Mutat Res 1996, 366:163-169 [DOI] [PubMed] [Google Scholar]
- 20.Valent A, Bénard J, Vénuat AM, Da Silva J, Duverger A, Duarte N, Hartmann O, Spengler BA, Bernheim A: Phenotypic and genotypic diversity of human neuroblastoma studied in 3 IGR cell lines models derived from bone marrow metastasis. Cancer Genet Cytogenet 1999, 112:124-129 [DOI] [PubMed] [Google Scholar]
- 21.Schwab M: Oncogene amplification in solid tumors. Cancer Biol 1999, 9:319-325 [DOI] [PubMed] [Google Scholar]
- 22.Carroll SM, Trotter J, Wahl GM: Replication timing control can be maintained in extrachromosomally amplified genes. Mol Cell Biol 1991, 11:4779-4785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimizu N, Shimura T, Tanaka T: Selective elimination of acentric double minutes from cancer cells through the extrusion of micronuclei. Mutat Res 2000, 448:81-90 [DOI] [PubMed] [Google Scholar]
- 24.Freeman-Edward J, O’Neill S, Lastowska M, Bown N: Expulsion of amplified MYCN from neuroblastoma tumor cells. Cancer Genet Cytogenet 2000, 116:87-88 [DOI] [PubMed] [Google Scholar]
- 25.Nüsse M, Miller BM, Viaggi S, Grawé J: Analysis of the DNA content distribution of micronuclei using flow sorting and fluorescent in situ hybridization with a centromeric DNA probe. Mutagenesis 1996, 11:405-413 [DOI] [PubMed] [Google Scholar]
- 26.Peace BE, Livingston G, Silberstein EB, Loper JC: A case of elevated spontaneous micronucleus frequency derived from chromosome 2. Mutat Res 1999, 430:109-119 [DOI] [PubMed] [Google Scholar]
- 27.Elhajouji A, Van Hummelen P, Kirch-Volders M: Indications for a threshold of chemically induced aneuploidy in vitro in human lymphocytes. Environ Mol Mutagen 1995, 26:292-304 [DOI] [PubMed] [Google Scholar]
- 28.Duncan AMV, Heddle JA: The frequency and distribution of apoptosis induced by three non-carcinogenic agents in mouse colonic crypts. Cancer Lett 1984, 23:307-311 [DOI] [PubMed] [Google Scholar]
- 29.Cremer C, Münkel C, Granzow M, Jauch A, Dietzel S, Eils R, Guan XY, Meltzer PS, Trent JM, Langowski J, Cremer T: Nuclear architecture and the induction of chromosomal aberrations. Mutat Res 1996, 366:97-116 [DOI] [PubMed] [Google Scholar]
- 30.Zink D, Bornfleth H, Visser A, Cremer C, Cremer T: Organization of early and late replicating DNA in human chromosome territories. Exp Cell Res 1999, 247:176-188 [DOI] [PubMed] [Google Scholar]
- 31.Tajbakhsh J, Luz H, Bornfleth H, Lampel S, Cremer C, Lichter P: Spatial distribution of GC- and AT-rich DNA sequences within human chromosome territories. Exp Cell Res 2000, 255:229-237 [DOI] [PubMed] [Google Scholar]
- 32.Canute GW, Longo SL, Longo JA, Winfield JA, Nevaldine BH, Hahn PJ: Hydroxyurea accelerates the loss of epidermal growth factor receptor genes amplified as double minute chromosomes in human glioblastoma multiforme. Neurosurgery 1996, 39:976-983 [DOI] [PubMed] [Google Scholar]
- 33.Van den Berg C, Von Hoff DD: Use of hydroxyurea to alter drug resistance of human tumor cells. Cancer Treat Res 1995, 78:95-114 [DOI] [PubMed] [Google Scholar]
- 34.Bordow S, Norris M, Haber P, Marschall G, Haber M: Prognostic significance of MYCN oncogene expression in childhood neuroblastoma. J Clin Oncol 1998, 16:3286-3294 [DOI] [PubMed] [Google Scholar]
- 35.Ferrandis E, Da Silva J, Riou G, Bénard J: Coactivation of the MDR1 and MYCN genes in human neuroblastoma cells during the metastatic process in the nude mouse. Cancer Res 1994, 54:2256-2261 [PubMed] [Google Scholar]
- 36.Cappellen D, Bénard J: Pleiotropic over-expression of multidrug-resistance-related gene is correlated to MYCN and max mRNA accumulation in the IGR-N91 human neuroblastoma model. Int J Cancer 1997, 70:430-436 [DOI] [PubMed] [Google Scholar]
- 37.Haber M, Bordow SB, Madafiglio M, Kavallaris M, Marshall GM, Mechetner EB, Fruehauft JP, Tee L, Cohn S, Salwen H, Schmid ML, Norris MD: Altered expression of MYCN oncogene, MRP gene expression and response to cytotoxic drugs in neuroblastoma cells. Oncogene 1999, 18:2777-2782 [DOI] [PubMed] [Google Scholar]


