Abstract
Splenic marginal zone lymphoma (SMZL) has recently been recognized in the World Health Organization classification of hematological diseases as distinct type of non-Hodgkin’s lymphoma. In contrast to the well-established chromosomal changes associated with other B-cell non-Hodgkin’s lymphoma, few genetic alterations have been found associated with SMZL. The aim of our study was to analyze by comparative genomic hybridization (CGH) the chromosomal imbalances in 29 patients with SMZL and to correlate these findings with clinical and biological characteristics and patient outcome. In 21 cases, cytogenetic studies were simultaneously performed. Most of the patients (83%) displayed genomic imbalances. A total of 111 DNA copy number changes were detected with a median of four abnormalities per case (range, 1 to 12). Gains (n = 92) were more frequent than losses (n = 16), while three high-level amplifications (3q26-q29, 5p11-p15, and 17q22-q25) were observed. The most frequent gains involved 3q (31%), 5q (28%), 12q and 20q (24% each), 9q (21%), and 4q (17%). Losses were observed in 7q (14%) and 17p (10%). SMZL patients with genetic losses had a shorter survival than the remaining SMZL patients (P < 0.05). In summary, chromosomal imbalances in regions 3q, 4q, 5q, 7q, 9q, 12q, and 20q have been detected by CGH in SMZL. Patients with SMZL displaying genetic losses by CGH had a short survival.
Splenic marginal zone lymphoma (SMZL) has been considered a subtype of B-cell non-Hodgkin’s lymphoma (NHL) in the recent classification of hematological diseases. 1 SMZL has been recognized as a separate entity on the basis of its morphological, phenotypic, and clinical characteristics. 2 Several studies have been performed to assess the genetic abnormalities in marginal lymphomas. Recently, two translocations, t(1;14)(p22;q32) involving BCL10/IgH 3 and t(11;18)(q21;q21) -API2/MLT- 4 have been described, associated with high-grade and low-grade MALT lymphomas, respectively. However, genetic information of SMZL is scanty. Cytogenetic analyses have shown involvement of chromosomes 1, 3, 7, and 8. 5,6 Fluorescence in situ hybridization studies on interphase nuclei have demonstrated the presence of trisomy 3 in a proportion of cases ranging from 18 to 47% of SMZL. 6-9 In previous studies we have reported that in some SMZL cases, the only cytogenetic abnormality displayed was del(7q) that could suggest that del(7q) is associated with SMZL. 6,10 Recently, loss of heterozygosity studies demonstrated that the frequency of allelic loss in SMZL (40%) is higher than that observed in other B-cell lymphoproliferative syndromes. The most frequently deleted microsatellite was D7S487. 11 These results are in accordance with our fluorescence in situ hybridization studies that mapped the commonly deleted region in chronic B-cell lymphoproliferative disorders at 7q31.3. 12
Comparative genomic hybridization (CGH) is a double-color hybridization procedure that provides, in a single experiment, a general view of genomic imbalances, including partial or complete trisomies, monosomies, or amplifications within the tumor genome. 13 This technique can be used to identify previously unexpected genetic abnormalities. Several groups have studied chromosomal changes by CGH in different subtypes of B-cell NHL. 14-16 Thus, in marginal zone lymphomas, gains on chromosomes 3 and 18 are frequent. 17 In extranodal lymphomas such as gastrointestinal lymphomas, total or partial gains of chromosomes 11, 12, and 1q are frequently observed. 18 However, little is known about the genomic imbalances in SMZL.
The present study was designed to screen DNA copy changes by CGH in a series of 29 SMZL and to correlate the results obtained from CGH with the most relevant clinical, biological, and cytogenetic characteristics, including disease outcome.
Materials and Methods
Patients
Tumor specimens from 29 patients with SMZL were included in the study. All cases were classified according to the criteria proposed by Isaacson and colleagues 19 and Mollejo and colleagues. 2 All samples were studied at diagnosis and were reviewed and classified by three of the authors (MM, TF, and MAP).
Cytogenetics
Cytogenetic studies were available in 21 patients. Most chromosome analyses were performed on the spleen (11 cases), whereas in six cases studies were performed on peripheral blood, in three cases on the lymph nodes, and in the remaining case, on the bone marrow. Cytogenetic analysis of the splenic tissue was performed after 24 and 48 hours of culture without stimulating agent, and 48 hours culture stimulated with phorbol 12-myristate 13-acetate. Peripheral blood and bone marrow cells were cultured for 3 days in presence of phorbol 12-myristate 13-acetate. Metaphases were G-banded and karyotypes were described according to the International System of Human Cytogenetic Nomenclature. 20 At least 20 mitoses were analyzed in each patient.
CGH
Tumor DNA was isolated from the spleen (25 cases), peripheral blood (2 cases), or bone marrow (2 cases). Reference DNA was obtained from peripheral blood lymphocytes of healthy donors (same sex as patients). The phenol-chloroform method was used for DNA extraction according to standard procedure. 21 CGH analysis was performed according to the method described by Lichter and Ried 22 Briefly, tumor DNA (test DNA) was labeled with biotin-16-dUTP (Boehringer Mannheim, Mannheim, Germany) and normal DNA (reference DNA) was labeled with digoxigenin-11-dUTP (Boehringer Mannheim) by a standard nick translation reaction. The size of the nick-translated fragments ranged from 300 to 1,000 bp. Equal amounts (1 μg) of labeled tumor and normal DNAs, and 70 μg of unlabeled human Cot-1 DNA (Life Technologies, Inc., Gaithersburg, MD) were co-hybridized to slides with human metaphase chromosome spreads prepared from phytohemagglutinin-stimulated lymphocytes from normal individuals. After hybridization for 1 to 2 days in a moist chamber at 37°C, posthybridization washes were performed to a stringency of 0.1× standard saline citrate at 42°C. Tumor and normal DNA were detected by avidin-fluorescein isothiocyanate and rhodamine-conjugated anti-digoxigenin, respectively. The slides were counterstained with 4,6-diamidino-2-phenylindole and mounted with an antifade solution. Image acquisition was performed with an epifluorescence microscope (Olympus BX60) equipped with a cooled charge-coupled device camera. Calculation of the tumor DNA to normal DNA fluorescent ratios along the length of each chromosome was performed by means of an automated CGH software package (Cytovision; Applied Imaging, Sunderland, UK). Ratio values obtained from at least 10 metaphase cells for each case were averaged. Ratio values >1.25 and <0.75 were considered to represent chromosomal gain and loss, respectively. Overrepresentation was defined as high-level amplification when the profile exceeded the cut-off value of 1.5. Chromosomal gains exceeding 1.5 involving the whole chromosome or large areas of a chromosomal arm were not considered as high-level DNA amplification. Negative control experiments were performed using differentially labeled male versus male and female versus female DNA. Additional control experiments included the interchange of the digoxigenin-dUTP and biotin-dUTP labels between normal and tumor DNA. According to previous recommendations, we measured the average number of copy alterations (ANCA) index. 23 This index represents the average number of copy alterations in a tumor type and is calculated by dividing the total number of copy alterations by the number of tumors analyzed.
Results
Patients
The most relevant clinical data from the 29 patients is shown in Table 1 ▶ . No sex predominance was observed (male to female,14 to 15) and the median age was 64 years (range, 39 to 79 years). According to the Ann-Arbor classification, all patients presented with advanced stage because of bone marrow infiltration. However, only 20% of patients displayed high levels of serum lactic dehydrogenase (normal value, 460 IU/L). In 43% of cases, a lymph node involvement was present. Lymphocytosis was present in 55% of patients. All but two patients (cases 26 and 27) received a splenectomy and in 10 cases, chemotherapy was administrated. Overall survival of the whole series was 72 months (95% CI, 44 to 100 months). At the time of the study, 18 patients remained alive (Table 1) ▶ .
Table 1.
Clinical Characteristics and Genomic Imbalances in 29 Patients with Splenic Marginal Zone Lymphoma (SMZL)
| Case | Reference | Age/sex | LDH (IU/L) | Organ involvement | Treatment | Follow-up (months) | Chromosomal changes | ||
|---|---|---|---|---|---|---|---|---|---|
| Losses | Gains | Amplifications | |||||||
| 1 | P1 | 67 /F | 499 | Spleen, BM, PB | Splenectomy | 53+ | 1 | 10 | 1 |
| 2 | P2 | 62 /M | 935 | Spleen, BM | Splenectomy | 59+ | 0 | 0 | 0 |
| 3 | P5 | 72 /F | 503 | Spleen, LN, BM | Splenectomy, CT | 72 | 0 | 5 | 0 |
| 4 | P10 | 56 /M | 350 | Spleen, BM, PB | Splenectomy, CT | 77+ | 0 | 0 | 0 |
| 5 | P12 | 65 /F | 350 | Spleen, BM, PB | Splenectomy, CT | 53+ | 0 | 5 | 0 |
| 6 | P13 | 58 /M | 350 | Spleen, BM, PB | Splenectomy, CT | 17 | 2 | 0 | 0 |
| 7 | P33 | 61 /M | 688 | Spleen, LN, BM, PB | Splenectomy | 42+ | 0 | 5 | 0 |
| 8 | P74 | 63 /M | 350 | Spleen, LN, BM, PB | Splenectomy, CT | 57 | 2 | 0 | 0 |
| 9 | P149 | 66 /F | 703 | Spleen, BM, PB | Splenectomy, CT | 29 | 0 | 0 | 0 |
| 10 | P162 | 53 /M | 368 | Spleen, BM, Liver | Splenectomy, CT | 38+ | 0 | 4 | 0 |
| 11 | P202 | 72 /F | 398 | Spleen, BM, PB | Splenectomy | 36+ | 2 | 5 | 0 |
| 12 | P206 | 69 /F | 401 | Spleen, BM, PB | Splenectomy | 13+ | 0 | 7 | 0 |
| 13 | P220 | 74 /M | 350 | Spleen, LN, BM, PB | Splenectomy | 3 | 2 | 0 | 0 |
| 14 | 1034 | 66 /F | 635 | Spleen, LN, BM | Splenectomy | 22 | 1 | 0 | 0 |
| 15 | P222 | 72 /M | NA | Spleen, BM | Splenectomy | 36 | 0 | 7 | 0 |
| 16 | 1443 | 67 /M | 312 | Spleen, LN, BM, PB | Splenectomy | 2 | 1 | 8 | 0 |
| 17 | P254 | 51 /M | 287 | Spleen, BM, PB | Splenectomy, CT | 6+ | 1 | 0 | 1 |
| 18 | P256 | 69 /F | 350 | Spleen, BM | Splenectomy | 4 | 0 | 2 | 0 |
| 19 | 1020 | 70 /F | 444 | Spleen, BM, PB | Splenectomy | 34+ | 0 | 0 | 0 |
| 20 | 2184 | 68 /M | 388 | Spleen, LN, BM, PB | Splenectomy | 16+ | 0 | 7 | 0 |
| 21 | 2299 | 72 /M | 250 | Spleen, LN, BM, PB | Splenectomy | 15+ | 3 | 9 | 0 |
| 22 | 2760 | 60 /F | 397 | Spleen, LN, BM, PB | Splenectomy | 8+ | 0 | 2 | 0 |
| 23 | 4066 | 39 /M | 432 | Spleen, BM | Splenectomy | 28+ | 0 | 1 | 0 |
| 25 | 4722 | 45 /F | 415 | Spleen | Splenectomy, CT | 136 | 0 | 4 | 0 |
| 25 | 4340 | 51 /F | 450 | Spleen, LN, BM | Splenectomy | 24+ | 1 | 2 | 0 |
| 26 | 4941 | 55 /M | 428 | Spleen, LN, BM | Not treatment | 15+ | 0 | 3 | 1 |
| 27 | 5787 | 79 /F | 397 | Spleen, BM | No treatment | 26+ | 0 | 5 | 0 |
| 28 | 5388 | 48 /F | 317 | Spleen, LN, BM | Splenectomy | 21 | 0 | 2 | 0 |
| 29 | 1742 | 52 /F | 310 | Spleen, LN, BM | Splenectomy, CT | 113+ | 0 | 0 | 0 |
F, female; M, male; BM, bone marrow; PB, peripheral blood; LN, lymph node; CT, chemotherapy; NA, not available; +, patient alive at time of analysis.
Cytogenetics
In 16 of the 21 patients (76%) in which cytogenetic studies were available, an abnormal karyotype was present. Most cases were pseudodiploid. In two cases (cases 6 and 25) the modal number of chromosomes was more than 49. Only one case (case 8) had a hypodiploid karyotype. Seven out of the 16 patients (44%) had a complex karyotype. The most frequent cytogenetic abnormality was deletion of 7q (six cases) whereas a trisomy of chromosome 3 was observed in only three cases. Trisomies of chromosomes 5 and 12 were detected in two cases, respectively. Several chromosomal breakpoints were found to be repetitively involved in SMZL patients. Thus, abnormalities involving 7q22-q33 were detected in seven cases; whereas rearrangements in 1p22, 1q21, and 5q31, were found in two patients each.
CGH
Twenty-four of the 29 patients with SMZL (83%) showed DNA sequence copy number changes and five cases displayed normal profiles. A total of 111 DNA copy number changes were detected with a median of four abnormalities per case (range, 1 to 12): 92 gains, 16 losses, and three high-level amplifications (Figure 1) ▶ . All abnormal cases except cases 14, 22, and 23 showed more than one chromosomal imbalance (Table 2) ▶ . In 19 patients, overepresentations of chromosomal material were detected. Gains of chromosomal material involved 3q (31%), 5q (28%), 12q and 20q (24% each), 9q (21%), 4q (17%), and 11q and 12p (14% each). The consensus regions of gains were located at 3q23-q25, 4q25-q28, 5q13-q15, 9q31, 12q15-q21, and 20q. In 10 cases (35%), a genetic loss was present. In four of the 10 cases, the losses of chromosomal material were the only changes detected. Losses were located in 7q (14%) with a commonly deleted region at 7q31-q32, 17p (10%), and 8p, 13q, and 15q (7% each) (Figure 1) ▶ . Only three cases showed high-level amplifications located at 3q26-q29, 5p11-p15, and 17q22-q25. These amplifications were located in chromosomal regions in which known fragile sites have been identified (3q26-29 and FRA3C, 5p11-p15 and FRA-5A, and 17q22-25 and FRA17B). As indicated in Material and Methods, ANCA index represents the average number of copy alterations in a tumor type. In the present study, the ANCA index was 3.8.
Figure 1.
Summary of the genomic imbalances in 29 patients with SMZL. Left: Lines indicate loss of chromosomal material. Right: Lines indicate gain of chromosomal material. High-level DNA amplifications are represented as solid squares. Each line represents a gained or lost region in a single tumor. The numbers on top of each line refer to the patient analyzed.
Table 2.
Results of CGH and G-Banding in Patients with SMZL
| Case | Sample | CGH | Cytogenetics | ||||
|---|---|---|---|---|---|---|---|
| Gains | Losses | Amplifications | Sample | Karyotype | |||
| 1 | Spleen | 1p34–p36,2q12–q32, 3q12–q26,4q12–q31,7q11–q21, 9q21–q22,10q21–q22,11q12–q21,12q13–q15, 13q12–q14,20q11–q13 | 7q31–q36 | 3q26–q29 | LN | 46,XX,t(1;5)(p11;q11),del(7) (q22q33)del(8)(q12),del(14) (q21),del(18)(q13) | [14] |
| 2 | Spleen | – | – | – | PB | 46,XY | [23] |
| 3 | Spleen | 5q31–q35,9q22–q34,15q24–q26, 16q21–q24,20q11–q13 | – | – | – | ND | |
| 4 | Spleen | – | – | – | – | ND | |
| 5 | Spleen | 1q21–q32,3q12–q29,4q12q26, 7,18q11–q21 | – | – | – | ND | |
| 6 | Spleen | – | 15q24–q26,17p11p13 | – | LN | 85–86,XXY,del(1)(q21q33),t(1;2) (p31;q22),del(3)(p12p24), add(4)(q31),del(5)(p12p15), del(6)(q21q25),del(9)(p13p23), dup(10)(q21q25),add(14)(q32), add(17)(q23),der(20)(q13) | [12] |
| 7 | Spleen | 2q22–q31,3q24–q26,18p11, 19q13,20q11–q13 | – | – | ND | ||
| 8 | Spleen | – | 8p12–p23,17p11–p13 | – | Spleen | 44,XY,t(1;3)(q21;q26),del(8)(q22), del(7)(q32),−7,+der(7)t(1;7) (p12;p12),−20,−21 | [16] |
| 9 | Spleen | – | – | – | Spleen | 46,XX,del(6)(q21q24)46,XX | [4] [16] |
| 10 | Spleen | 5q11–q14,12q12–q24,20,Xq27–q28 | – | – | ND | ||
| 11 | Spleen | 1q31–q32,4q22–q28,12p11–p13, 12q21–q24,Xq13–q25 | 8q22q24,15q24–q26 | – | Spleen | 46,XX,t(2;17)(p13;q21)46,XX | [5] [10] |
| 12 | Spleen | 1p21–p31,3q13–q21,4q26–q31, 8p12–p22,11q22–q25,12p11–p13,12q12–q14 | – | – | ND | ||
| 13 | Spleen | – | 7q31–q36,11q22–q25 | – | Spleen | 46,XY,t(1;2)(p22;q23),del(7)(q21), add(17)(p13) 46,XY | [7] [8] |
| 14 | Spleen | – | 7q22–q32 | – | Spleen | 46,XX,del(7)(q21q31) | [12] |
| 15 | Spleen | 9q32–q34,11p12–p15,12q22–q24, 15q22–q26,16q22–24q, 18q21–q23,Xp11–p22 | – | – | ND | ||
| 16 | Spleen | 1p34–p36,3,9q31–q34,11q12–q21, 17q23q25,19,20,21 | 13q14–q31 | – | Spleen | 46,X,−Y,+346,XY | [10] [5] |
| 17 | Spleen | – | 7q31–q36 | 17q22–25 | Spleen | 46,XY,del(7)(q31)46,XY | [9] [7] |
| 18 | Spleen | 9q32–q34,16 | – | – | ND | ||
| 19 | Spleen | – | – | – | Spleen | 46,XX,add(9)(p12),add(16)(p12)46,XX | [6] [5] |
| 20 | Spleen | 2p12–p14,5q11–q23,10q21–q23, 11q12–q14,12p13–q24, 20q12–q13 | – | – | Spleen | 47,XY,+del(3)(q21q26)46,XY | [6] [9] |
| 21 | Spleen | 1p31–p36,2q32–q26,3q21–q25, 3q26–q29,5q23–q35,8q22–q24, 12p11–p13,15q21–q25,20q11–q13 | 6q21–q2413q21–q3117p11–p13 | – | LN | 47,XY,+1,add(5)(q31)47,XY | [9] [8] |
| 22 | Spleen | 5p15–q31 | – | – | Spleen | 47,XX,+del(5)(q31)46,XX | [10] [4] |
| 23 | Spleen | Xq27 | – | – | PB | 46,XY | [23] |
| 24 | PB | 1q21–q31,5p15–q21,12q12–q21 | – | – | PB | 46,XX,del(7)(q31q34),der(16)t(12;16)(q13;q24) 46,XX | [10] [19] |
| 25 | Spleen | 3q13–q29,8q11–q24,17q22–q24 | 8p11–p23 | – | Spleen | 50,XX,+5,+der(7)t(3;7)(p21;q22) x3,i(8)(q10)46,XX | [2] [6] |
| 26 | PB | 1q21–q43,3,9q32–q34 | – | 5p11–p15 | PB | 48,XY,del(1)(q42),+del(1)(p22),+3,+i(5)(p10),−9 46,XY | [4] [19] |
| 27 | BM | 2q24–q32,3q25,4q24–q28,5q12–5q21,13q22 | – | – | PB | 46,XX | [22] |
| 28 | Spleen | 6p22–p25,22q12–q13 | – | – | PB | 46,XX | [18] |
| 29 | BM | – | – | – | BM | 46,XX | [21] |
PB, peripheral blood; BM, bone marrow; LN, lymph node; ND, not done.
CGH provided more genomic imbalances than cytogenetic studies in the 21 cases analyzed with both techniques (Table 2) ▶ . A correlation between both methodologies was seen in 12 cases: two patients (cases 2 and 29) without any change in both cytogenetics and CGH; two patients (cases 14 and 22) with the same results; and the remaining eight patients (cases 9, 13, 17, 19, 23, 25, 26, and 28) with minor changes. In four patients (cases 1, 8, 16, and 21) some changes observed by cytogenetics were also showed by CGH. In the remaining five cases (cases 6, 11, 20, 24, and 27) the results were different. It should be noted that in some of these cases CGH was performed in a different tissues than cytogenetics. Interestingly losses of 7q were observed in four out of the six patients with both methodologies and in two cases only by cytogenetics.
Correlation between CGH Results and Outcome
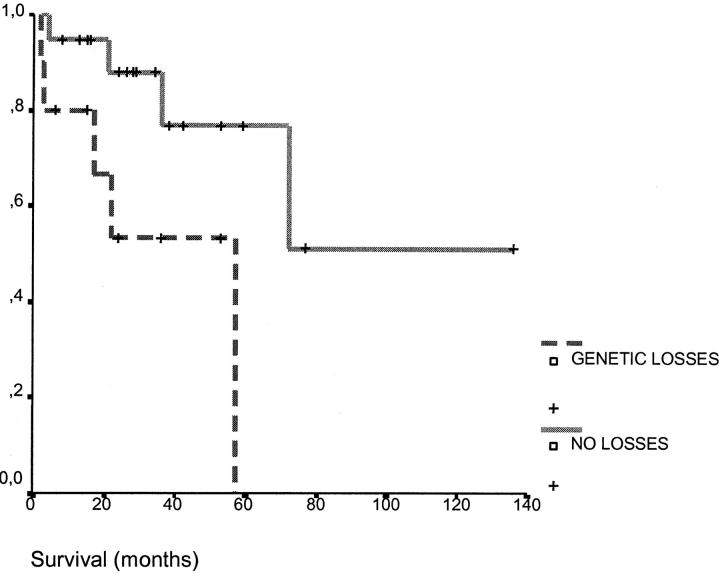
A correlation between losses and a shorter survival was found. Thus, the patients displaying a loss of chromosomal material had a survival of 36 months while the patients without losses had a longer survival (101 months, P = 0.02) (Figure 2) ▶ . By contrast, patients with deletions assessed by conventional cytogenetics did not show differences in survival in relation with patients without deletions (76 versus 46 months, P = 0.78). Patients with SMZL and more than four changes detected by CGH did not present differences in survival from the cases of SMZL with less than four changes (60 months versus 80 months, respectively, P = 0.8).
Figure 2.
Survival curves of patients with SMZL according to the presence of losses assessed by CGH (0 versus ≥ 1 loss; P = 0.02).
Discussion
In the present study, we have identified chromosomal imbalances by CGH in 83% of patients with SMZL. This incidence is similar to that previously reported in the overall setting of marginal lymphomas, 17 and higher than the overall frequency reported in indolent lymphomas in which the number of changes ranged between 48 and 68%. 14,18,24,25
Previous studies using conventional cytogenetics and fluorescence in situ hybridization have shown that interstitial deletion of the long arm of chromosome 7 is a recurrent cytogenetic abnormality in SMZL. 6,10,12,26 By CGH we have confirmed that partial loss of the 7q region is the most frequent loss in SMZL and the commonly deleted region is located at 7q31-q32. Recently, Corcoran and colleagues 27 found a deregulation of the CDK6 gene that could be involved in the pathogenesis of the small group of SMZL with translocations at 7q21. However, partial losses in 7q are more frequent than translocations in SMZL. These deletions of 7q are infrequent in other NHL 14,25,28,29 including marginal zone lymphomas. 17 Moreover, studies by loss of heterozygosity have confirmed that 7q31-q32 loss is a relatively specific genetic marker of SMZL. 11 All of the cytogenetic, fluorescence in situ hybridization, loss of heterozygosity, and CGH data support the hypothesis that a loss in 7q31-q32 plays a key role in SMZL.
In our study, gains mostly involved 3q, 5q, 12q, and 20q. Chromosomal changes in 3q have been reported in several types of B-NHL. Half of the cases of mantle cell lymphoma have gains of 3q. 30,31 This incidence is lower in patients with diffuse large cell lymphoma (24%) 15 or extranodal lymphoma (7 to 13%). 18,25 The reported overall incidence of gains of chromosome 3 in marginal zone lymphomas, including MALT (nodal and extranodal) and SMZL, is 52% and only two of the 11 cases of SMZL studied by Dierlamm and colleagues 17 showed gains on 3q. In our series of 29 SMZL patients, the incidence was 31%. This frequency of imbalances on chromosome 3 is similar to that observed using fluorescence in situ hybridization. 6-9 It should be noted that, in contrast to other marginal cell lymphomas in which trisomy is the most frequent gain, using CGH we have observed that in SMZL the gain specifically occurs at the 3q arm.
In the present series, 28% of patients with SMZL had gains in chromosome 5q, with a consensus region in 5q13-q15. Gains of 5q have only been reported by CGH in NHL sporadically, 16,18,30,32 and in MALT lymphomas the incidence is 8%. 17 Only one case of diffuse large cell lymphoma of the gastrointestinal tract has been described with an amplification of 5q33-q35. 18 Moreover, by conventional cytogenetics abnormalities of 5q are also unusual in NHL and only two chromosome translocations, t(5;14)(q11;q32) and a t(5;7)(q13;q35) have been reported, in B and T NHL, respectively. 33 In 5q13, a gene involved in CDK-activating kinase complex of cyclin H MAT1 34 and in 5q31, a gene coding a phosphatase Cdc25 associated with regulation of cell cycle have both been recently located. 34 However, the possible relationship of these genes with SMZL still needs to be clarified. Although 5q13 abnormalities have not previously been related to SMZL, the region has been involved in hairy cell leukemia. 35,36 The demonstration in the present series of a high incidence of gains on 5q in SMZL, with a consensus region located at 5q13-q15, suggests that this region could be related to SMZL.
Using conventional cytogenetics, trisomy 12 or duplication 12q are frequently observed as a secondary genomic change in lymphoid disorders. 37,38 By contrast, in marginal lymphomas, abnormalities of chromosome 12 have rarely been reported. 17 By CGH, numerical abnormalities of chromosome 12 have been detected in primary mediastinal B-cell lymphoma, 32 primary gastrointestinal large-cell lymphoma, 25 and chronic lymphocytic leukemia. 14 In our study, no cases with trisomy 12 were found. However we observed gains in the long arm of chromosome 12 in seven cases of SMZL with a consensus region in the bands 12q15-q21. In the long arm of chromosome 12 the region 12q24 has frequently been involved in mediastinal lymphomas 32 and in primary large B-cell lymphoma of the gastrointestinal tract, 18 whereas in mantle cell lymphomas, the consensus region was located at 12q13. 31 In the region 12q13-q21 several genes such as GLI, MDM2, and CDK4 have been mapped. Amplification of these genes have been observed in diffuse large B-cell lymphoma, and associated with advanced state of disease. 39 Recently, our group has observed CDK4 gene amplifications in mantle cell lymphoma. 31 However, their role in the pathogenesis of SMZL needs to be determined.
In the present study, gains of the long arm of chromosomes 9 and 20 were particularly frequent, 21 and 24%, respectively. Whereas changes observed in 9q have been reported in other lymphoma subtypes, 17,31 gains in 20q have been observed in only one case of mantle lymphoma. 30 Moreover, by conventional cytogenetics, the changes in this region are also uncommon in NHL. 40 However gains of 20q have been observed in solid tumors such as colorectal carcinoma, 41,42 breast, 43 bladder, 44 ovarian, 45 and pancreatic cancer. 46 Several candidate genes have been identified on 20q: the cellular apoptosis susceptibility (CAS) gene; 47 BTAK, a putative serine/threonine kinase gene; 48 AIB1, a steroid receptor co-activator gene; 49 PTPN1, a nonreceptor tyrosine phosphatase involved in growth regulation; 50 as well as MYB12, which encodes a transcription factor and plays an important role in cell cycle progression. 51
Regarding gene amplifications, CGH studies are detecting an increased number of high-level DNA amplifications in lymphomas, which were relatively rare in NHL by cytogenetic studies. 52 These results suggest that gene amplifications may be more frequent in NHL than initially thought. 16 We found three high-level DNA amplifications in 29 SMZL involving 3q26-q29, 5p11-p15, and 17q22-q25. Amplifications of 3q26-q29 have been observed in NHL. 16,18,30,31 Abnormalities of 5p have been reported in B-cell disorders. 17,18,31,32,39 However, to the best of our knowledge, no cases of NHL with high-level amplifications of 5p have been described. Amplifications in 17q23-25 have been previously reported in one case of mantle cell lymphoma. 31 A relationship between high-level DNA amplifications and the location of chromosome fragile sites have been suggested. 53 In fact, the three amplified regions in SMZL were located in chromosomal regions where fragile sites have been identified. These data are according to the hypothesis that fragile sites may be implicated in the amplifications of certain chromosomal regions during tumor progression. 53 The ANCA index is a measure for the number of chromosomal copy alterations in a tumor. The correlation of the ANCA index with tumor progression may reflect the tumor aggressiveness. 23 The ANCA index of SMZL in the present series was 3.8 whereas this value ranged from 4.8 in the squamous cell carcinomas of the anal canal to the 8.3 in the high-grade astrocytomas. 23
Concerning the outcome, in the present series, we have observed that the presence of genetic losses was associated with a shorter survival than in other patients without deletions (36 versus 101 months). Little data about the impact of CGH on the outcome of the NHL are available. Recently, a relationship between the presence of more than four abnormalities by CGH or losses on 9p have been related to a poor outcome in mantle cell lymphoma. 31 Our results suggest that in so-called indolent SMZL, two groups of patients based on CGH losses are present. Thus, patients with loss of chromosomal material in CGH showed a significantly poorer outcome as compared to patients without deletion or with few chromosomal changes, and therefore could be candidates for a more intensive therapeutic approach. Recently, Mateo and colleagues 11 associated the loss of 7q, assessed by loss of heterozygosity analysis, with a poor outcome.
In summary, the CGH data reported describes new regions related to SMZL (5q, 9q, 12q, and 20q). Although the high-level amplifications seem to be an uncommon mechanism in these lymphomas, three regions (3q26-q29, 5p11-p15, and 17q22-q25) were found to be amplified in SMZL. Moreover, our data confirmed that deletions of 7q31-q32 are closely associated with SMZL and showed that deletions in SMZL lead to a poor clinical outcome.
Acknowledgments
We thank Pilar Fernández, M. Ángeles Hernández, Ana Simón, and Mark Anderson for the excellent technical assistance.
Footnotes
Address reprint requests to Jesús M. Hernández Rivas, Servicio de Hematología, Hospital Universitario de Salamanca, Paseo San Vicente 58-182, 37007 Salamanca, Spain. E-mail: jmhernandezr@aehh.org.
Supported in part by grants of the Spanish Fondo de Investigaciones Sanitarias (98/1161; 98/0491, and 00/1089), and the Centro de Investigación del Cáncer, Universidad de Salamanca-CSIC, Spain (to N. C. G.).
J. M. H. and J. L. G. contributed equally to the study and should both be considered as first authors.
References
- 1.Harris NL, Jaffe ES, Diebold J, Flandrin G, Muller-Hermelink HK, Vardiman J, Lister TA, Bloomfield CD: The World Health Organization classification of neoplastic diseases of the haematopoietic and lymphoid tissues: report of the Clinical Advisory Committee Meeting, Airlie House, Virginia, November, 1997. Histopathology 2000, 36:69-86 [DOI] [PubMed] [Google Scholar]
- 2.Mollejo M, Menárguez J, Lloret E, Sánchez A, Campo E, Algara P, Cristóbal E, Sánchez E, Piris MA: Splenic marginal zone lymphoma: a distinctive type of low-grade B-cell lymphoma. A clinicopathological study of 13 cases. Am J Surg Pathol 1995, 19:1146-1157 [PubMed] [Google Scholar]
- 3.Willis TG: Bcl10 is involved in t(1;14)(p22;q32) of MALT B cell lymphoma and mutated in multiple tumor types. Cell 1999, 96:35-45 [DOI] [PubMed] [Google Scholar]
- 4.Dierlamm J, Baens M, Wlodarska I, Stefanova-Ouzounova M, Hernández JM, Hossfeld DK, Wolf-Peeters C, Hagemeijer A, Van den BH, Marynen P: The apoptosis inhibitor gene API2 and a novel 18q gene, MLT, are recurrently rearranged in the t(11;18)(q21;q21) associated with mucosa-associated lymphoid tissue lymphomas. Blood 1999, 93:3601-3609 [PubMed] [Google Scholar]
- 5.Dierlamm J, Pittaluga S, Wlodarska I, Stul M, Thomas J, Boogaerts M, Michaux L, Driessen A, Mecucci C, Cassiman JJ: Marginal zone B-cell lymphomas of different sites share similar cytogenetic and morphologic features. Blood 1996, 87:299-307 [PubMed] [Google Scholar]
- 6.Solé F, Woessner S, Florensa L, Espinet B, Mollejo M, Martín P, Piris MA: Frequent involvement of chromosomes 1, 3, 7 and 8 in splenic marginal zone B-cell lymphoma. Br J Haematol 1997, 98:446-449 [DOI] [PubMed] [Google Scholar]
- 7.Wotherspoon AC, Finn TM, Isaacson PG: Trisomy 3 in low-grade B-cell lymphomas of mucosa-associated lymphoid tissue. Blood 1995, 85:2000-2004 [PubMed] [Google Scholar]
- 8.Dierlamm J, Michaux L, Wlodarska I, Pittaluga S, Zeller W, Stul M, Criel A, Thomas J, Boogaerts M, Delaere P, Cassiman JJ, De Wolf-Peeters C, Mecucci C, Van den Berghe H: Trisomy 3 in marginal zone B-cell lymphoma: a study based on cytogenetic analysis and fluorescence in situ hybridization. Br J Haematol 1996, 93:242-249 [DOI] [PubMed] [Google Scholar]
- 9.Brynes RK, Almaguer PD, Leathery KE, McCourty A, Arber DA, Medeiros LJ, Nathwani BN: Numerical cytogenetic abnormalities of chromosomes 3, 7, and 12 in marginal zone B-cell lymphomas. Mod Pathol 1996, 9:995-1000 [PubMed] [Google Scholar]
- 10.Hernández JM, Mecucci C, Criel A, Meeus P, Michaux I, Van Hoof A, Verhoef G, Louwagie A, Scheiff JM, Michaux JL: Cytogenetic analysis of B cell chronic lymphoid leukemias classified according to morphologic and immunophenotypic (FAB) criteria. Leukemia 1995, 9:2140-2146 [PubMed] [Google Scholar]
- 11.Mateo M: 7q31-32 allelic loss is a frequent finding in splenic marginal zone lymphoma. Am J Pathol 1999, 154:1583-1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hernández JM, Schoenmakers EF, Dal Cin P, Michaux L, Van DV, Van den Berghe H: Molecular delineation of the commonly deleted segment in mature B-cell lymphoid neoplasias with deletion of 7q. Genes Chromosom Cancer 1997, 18:147-150 [PubMed] [Google Scholar]
- 13.Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D: Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 1992, 258:818-821 [DOI] [PubMed] [Google Scholar]
- 14.Bentz M, Huck K, Du Manoir S, Joos S, Werner CA, Fischer K, Dohner H, Lichter P: Comparative genomic hybridization in chronic B-cell leukemias shows a high incidence of chromosomal gains and losses. Blood 1995, 85:3610-3618 [PubMed] [Google Scholar]
- 15.Monni O, Joensuu H, Franssila K, Knuutila S: DNA copy number changes in diffuse large B-cell lymphoma—comparative genomic hybridization study. Blood 1996, 87:5269-5278 [PubMed] [Google Scholar]
- 16.Werner CA, Dohner H, Joos S, Trumper LH, Baudis M, Barth TF, Ott G, Moller P, Lichter P, Bentz M: High-level DNA amplifications are common genetic aberrations in B-cell neoplasms. Am J Pathol 1997, 151:335-342 [PMC free article] [PubMed] [Google Scholar]
- 17.Dierlamm J, Rosenberg C, Stul M, Pittaluga S, Wlodarska I, Michaux L, Dehaen M, Verhoef G, Thomas J, de Kelver W, Bakker-Schut T, Cassiman JJ, Raap AK, De Wolf-Peeters C, Van den Berghe H, Hagemeijer A: Characteristic pattern of chromosomal gains and losses in marginal zone B cell lymphoma detected by comparative genomic hybridization. Leukemia 1997, 11:747-758 [DOI] [PubMed] [Google Scholar]
- 18.Barth TF, Dohner H, Werner CA, Stilgenbauer S, Schlotter M, Pawlita M, Lichter P, Moller P, Bentz M: Characteristic pattern of chromosomal gains and losses in primary large B-cell lymphomas of the gastrointestinal tract. Blood 1998, 91:4321-4330 [PubMed] [Google Scholar]
- 19.Isaacson PG, Matutes E, Burke M, Catovsky D: The histopathology of splenic lymphoma with villous lymphocytes. Blood 1994, 84:3828-3834 [PubMed] [Google Scholar]
- 20.ISCN: Guidelines for Cancer Cytogenetics. Supplement to: an International System for Human Cytogenetic Nomenclature. Basel, Karger, 1995
- 21.Sambrook J, Fritsch EF, Maniatis T (eds): Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, Cold Spring Harbor Laboratory, 1989
- 22.Lichter P, Ried T: Molecular analysis of chromosome aberrations. In situ hybridization. Methods Mol Biol 1994, 29:449-478 [DOI] [PubMed] [Google Scholar]
- 23.Ried T, Heselmeyer-Haddad K, Blegen H, Schrock E, Auer G: Genomic changes defining the genesis, progression, and malignancy potential in solid human tumors: a phenotype/genotype correlation. Genes Chromosom Cancer 1999, 25:195-204 [DOI] [PubMed] [Google Scholar]
- 24.Karhu R, Knuutila S, Kallioniemi OP, Siltonen S, Aine R, Vilpo L, Vilpo J: Frequent loss of the 11q14-24 region in chronic lymphocytic leukemia: a study by comparative genomic hybridization. Tampere CLL Group. Genes Chromosom Cancer 1997, 19:286-290 [DOI] [PubMed] [Google Scholar]
- 25.Chan WY, Wong N, Chan AB, Chow JH, Lee JC: Consistent copy number gain in chromosome 12 in primary diffuse large cell lymphomas of the stomach. Am J Pathol 1998, 152:11-16 [PMC free article] [PubMed] [Google Scholar]
- 26.Oscier DG, Gardiner A, Mould S: Structural abnormalities of chromosome 7q in chronic lymphoproliferative disorders. Cancer Genet Cytogenet 1996, 92:24-27 [DOI] [PubMed] [Google Scholar]
- 27.Corcoran MM, Mould SJ, Orchard JA, Ibbotson RE, Chapman RM, Boright AP, Platt C, Tsui LC, Scherer SW, Oscier DG: Dysregulation of cyclin dependent kinase 6 expression in splenic marginal zone lymphoma through chromosome 7q translocations. Oncogene 1999, 18:6271-6277 [DOI] [PubMed] [Google Scholar]
- 28.Shikano T, Kaneko Y, Ishikawa Y, Niikawa N, Tono-oka T, Takeda T, Kikuchi M: Ph1-positive and Ph1-negative abnormal cell lines in a child with lymphoblastic lymphoma. Br J Haematol 1984, 58:459-464 [DOI] [PubMed] [Google Scholar]
- 29.Solé F, Woessner S, Florensa L, Montero S, Asensio A, Besses C, Sans-Sabrafen J: A new chromosomal anomaly associated with mature B-cell chronic lymphoproliferative disorders: del(7)(q32). Cancer Genet Cytogenet 1993, 65:170-172 [DOI] [PubMed] [Google Scholar]
- 30.Monni O, Oinonen R, Elonen E, Franssila K, Teerenhovi L, Joensuu H, Knuutila S: Gain of 3q and deletion of 11q22 are frequent aberrations in mantle cell lymphoma. Genes Chromosom Cancer 1998, 21:298-307 [DOI] [PubMed] [Google Scholar]
- 31.Beá S, Ribas M, Hernández JM, Bosch F, Pinyol M, Hernández L, García JL, Flores T, González M, López-Guillermo A, Piris MA, Cardesa A, Montserrat E, Miró R, Campo E: Increased number of chromosomal imbalances and high-level DNA amplifications in mantle cell lymphoma are associated with blastoid variants. Blood 1999, 93:4365-4374 [PubMed] [Google Scholar]
- 32.Joos S, Otano-Joos MI, Ziegler S, Bruderlein S, Du Manoir S, Bentz M, Moller P, Lichter P: Primary mediastinal (thymic) B-cell lymphoma is characterized by gains of chromosomal material including 9p and amplification of the REL gene. Blood 1996, 87:1571-1578 [PubMed] [Google Scholar]
- 33.Kristoffersson U, Heim S, Mandahl N, Olsson H, Akerman M, Mitelman F: Trisomy 5 and t(5;14)(q11;q32) as the sole abnormalities in two different clones from a centroblastic non-Hodgkin’s lymphoma. Cancer Genet Cytogenet 1988, 36:173-176 [DOI] [PubMed] [Google Scholar]
- 34.Dictor M, Ehinger M, Mertens F, Akervall J, Wennerberg J: Abnormal cell cycle regulation in malignancy. Am J Clin Pathol 1999, 112:S40-S52 [PubMed] [Google Scholar]
- 35.Wu X, Merup M, Juliusson G, Jansson M, Stellan B, Grander D, Zabarovsky E, Liu Y, Spasokoukotskaja T, Gahrton G, Einhorn S: Characterization of a hairy cell leukemia-associated 5q13.3 inversion breakpoint. Genes Chromosom Cancer 1997, 20:337-346 [DOI] [PubMed] [Google Scholar]
- 36.Haglund U, Juliusson G, Stellan B, Gahrton G: Hairy cell leukemia is characterized by clonal chromosome abnormalities clustered to specific regions. Blood 1994, 83:2637-2645 [PubMed] [Google Scholar]
- 37.Yunis JJ: Chromosomal translocations and gene rearrangements in non-Hodgkin lymphomas. Cancer Detect Prev 1988, 12:291-296 [PubMed] [Google Scholar]
- 38.Johansson B, Mertens F, Mitelman F: Cytogenetic evolution patterns in non-Hodgkin’s lymphoma. Blood 1995, 86:3905-3914 [PubMed] [Google Scholar]
- 39.Rao PH, Houldsworth J, Dyomina K, Parsa N, Cigudosa JC, Louie D, Popplewell L, Offit K, Jhanwar S, Chaganti RSK: Chromosomal and gene amplifications in diffuse large B-cell lymphoma. Blood 1998, 92:234-240 [PubMed] [Google Scholar]
- 40.Mitelman F: Catalog of Chromosome Aberrations in Cancer. 98 CD-ROM. 1998, Wiley-Liss, New York
- 41.Korn WM, Yasutake T, Kuo WL, Warren RS, Collins C, Tomita M, Gray J, Waldman FM: Chromosome arm 20q gains and other genomic alterations in colorectal cancer metastatic to liver, as analyzed by comparative genomic hybridization and fluorescence in situ hybridization. Genes Chromosom Cancer 1999, 25:82-90 [DOI] [PubMed] [Google Scholar]
- 42.Ried T, Knutzen R, Steinbeck R, Blegen H, Schrock E, Heselmeyer K, Du Manoir S, Auer G: Comparative genomic hybridization reveals a specific pattern of chromosomal gains and losses during the genesis of colorectal tumors. Genes Chromosom Cancer 1996, 15:234-245 [DOI] [PubMed] [Google Scholar]
- 43.Kallioniemi A, Kallioniemi OP, Piper J, Tanner M, Stokke T, Chen L, Smith HS, Pinkel D, Gray JW, Waldman FM: Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization. Proc Natl Acad Sci USA 1994, 91:2156-2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kallioniemi A, Kallioniemi OP, Citro G, Sauter G, DeVries S, Kerschmann R, Caroll P, Waldman F: Identification of gains and losses of DNA sequences in primary bladder cancer by comparative genomic hybridization. Genes Chromosom Cancer 1995, 12:213-219 [DOI] [PubMed] [Google Scholar]
- 45.Iwabuchi H, Sakamoto M, Sakunaga H, Ma YY, Carcangiu ML, Pinkel D, Yang-Feng TL, Gray JW: Genetic analysis of benign, low-grade, and high-grade ovarian tumors. Cancer Res 1995, 55:6172-6180 [PubMed] [Google Scholar]
- 46.Solinas-Toldo S, Wallrapp C, Muller-Pillasch F, Bentz M, Gress T, Lichter P: Mapping of chromosomal imbalances in pancreatic carcinoma by comparative genomic hybridization. Cancer Res 1996, 56:3803-3807 [PubMed] [Google Scholar]
- 47.Brinkmann U, Gallo M, Polymeropoulos MH, Pastan I: The human CAS (cellular apoptosis susceptibility) gene mapping on chromosome 20q13 is amplified in BT474 breast cancer cells and part of aberrant chromosomes in breast and colon cancer cell lines. Genome Res 1996, 6:187-194 [DOI] [PubMed] [Google Scholar]
- 48.Sen S, Zhou H, White RA: A putative serine/threonine kinase encoding gene BTAK on chromosome 20q13 is amplified and overexpressed in human breast cancer cell lines. Oncogene 1997, 14:2195-2200 [DOI] [PubMed] [Google Scholar]
- 49.Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan XY, Sauter G, Kallioniemi OP, Trent JM, Meltzer PS: AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science 1997, 277:965-968 [DOI] [PubMed] [Google Scholar]
- 50.Hashimoto N, Goldstein BJ: Differential regulation of mRNAs encoding three protein-tyrosine phosphatases by insulin and activation of protein kinase C. Biochem Biophys Res Commun 1992, 188:1305-1311 [DOI] [PubMed] [Google Scholar]
- 51.Noben-Trauth K, Copeland NG, Gilbert DJ, Jenkins NA, Sonoda G, Testa JR, Klempnauer KH: Mybl2 (Bmyb) maps to mouse chromosome 2 and human chromosome 20q 13.1. Genomics 1996, 35:610-612 [DOI] [PubMed] [Google Scholar]
- 52.Ben-Yehuda D, Houldsworth J, Parsa NZ, Chaganti RS: Gene amplification in non-Hodgkin’s lymphoma. Br J Haematol 1994, 86:792-797 [DOI] [PubMed] [Google Scholar]
- 53.Coquelle A, Pipiras E, Toledo F, Buttin G, Debatisse M: Expression of fragile sites triggers intrachromosomal mammalian gene amplification and sets to early amplicons. Cell 1997, 89:215-225 [DOI] [PubMed] [Google Scholar]