Abstract
Congenital hyperinsulinism (CHI), previously named persistent hyperinsulinemic hypoglycemia of infancy, is characterized by profound hypoglycemia because of excessive insulin secretion. CHI presents as two different morphological forms: a diffuse form with functional abnormality of islets throughout the pancreas and a focal form with focal islet cell adenomatous hyperplasia, which can be cured by partial pancreatectomy. Recently, we have shown that focal adenomatous hyperplasia involves the specific loss of the maternal 11p15 region and a constitutional mutation of a paternally inherited allele of the gene encoding the regulating subunit of the K+ATP channel, the sulfonylurea receptor (ABCC8 or SUR1). In the present study on a large series of 31 patients, describing both morphological features and molecular data, we report that 61% of cases (19 out of 31) carried a paternally inherited mutation not only in the ABCC8 gene as previously described but also in the second gene encoding the K+ATP channel, the inward rectifying potassium channel (KCNJ11 or KIR6.2), in 15 cases and 4 cases, respectively. Moreover our results are consistent with the presence of a duplicated paternal 11p15 allele probably because of mitotic recombination or reduplication of the paternal chromosome after somatic loss of the maternal chromosome. In agreement with the loss of the maternal chromosome, the level of expression of a maternally expressed tumor suppressor gene, H19, was greatly reduced compared to the level of expression of the paternally expressed growth promoter gene, IGF2. The expression of IGF2 was on average only moderately increased. Thus, focal forms of CHI can be considered to be a recessive somatic disease, associating an imbalance in the expression of imprinted genes in the 11p15.5 region to a somatic reduction to homozygosity of an ABCC8- or KCNJ11-recessive mutation. The former is responsible for the abnormal growth rate, as in embryonic tumors, whereas the latter leads to unregulated secretion of insulin.
Congenital hyperinsulinism (CHI), previously quoted as “persistent hyperinsulinemic hypoglycemia of infancy” (PHHI) (MIM.601820), is a glucose-metabolism disorder characterized by unregulated excess insulin secretion and profound hypoglycemia, often requiring pancreatectomy. 1 The vast majority of CHI cases (95%) seem to be sporadic, with an estimated incidence of 1 out of 50,000 live births in occidental countries. 2
Despite identical clinical presentation, two types of histological lesions are associated with CHI: a focal form, FoCHI, and a diffuse form, DiCHI. 3-5 FoCHI cases, which involve focal islet cell adenomatous hyperplasia, account for 30 to 40% of all cases for which pancreatectomy is required. 4,6 In DiCHI, all of the islets of Langerhans, throughout the pancreas, are enlarged and contain distinctly hypertrophied insulin-producing cells. These two forms can be distinguished by pancreatic venous sampling 7-9 and microscopical examination of frozen sections during surgery. 4 Focal CHI can benefit partial pancreatectomy whereas diffuse CHI requires near-total pancreatectomy. 3,8,10
Several genetic forms of CHI have been identified. Mutations in the high-affinity sulfonylurea receptor ABCC8 gene (previously SUR1 gene) (MIM.600509), 11-14 and in the inward rectifying potassium channel subunit KCNJ11 (previously KIR6.2 gene) (MIM.600937), 15-17 adjacent loci in 11p15.4, 18,19 have been found in familial autosomal-recessive forms of CHI. A distinct syndrome of dominantly transmitted familial hyperinsulinism with hyperammonemia was recently described due to mutations in the glutamate dehydrogenase gene. 20-22 More recently, a single activating mutation in the glucokinase gene causing autosomal-dominant familial hyperinsulinism has been reported. 23 Another autosomal-dominant form of familial persistent hypoglycemia of infancy has been shown not to be linked to either the ABCC8/KCNJ11 genes or to the glucokinase gene, further demonstrating the genetic heterogeneity of this disorder. 24,25
We have previously demonstrated that FoCHI is a somatic-recessive endocrine disorder caused by somatic reduction to homozygosity (or hemizygosity) of a paternally inherited mutation of the ABCC8 gene, limited to focal islet cell adenomatous hyperplasia. A specific loss of maternal alleles of the imprinted chromosome region 11p15, limited to the hyperplasia, was found in all 10 cases tested of FoCHI. 26 Germline mutations in the paternal allele of the ABBC8 gene were subsequently identified in a series of four cases. 27
Deletion mapping experiments have shown that the most commonly deleted region encompasses two regions of interest: the 11p15.5 region, subject to imprinting, and the 11p15.4 region containing the ABCC8/KCNJ11 genes, which are not imprinted. 26 Deregulation of imprinting in region 11p15.5 has been implicated in several diseases and in tumor proliferation. 28 This region contains at least 12 imprinted genes. 29 Three are clearly involved in cell growth regulation: H19 may have a growth suppressor activity 30 and the inactivation of H19 is a preneoplastic event in Wilms’ tumor formation; 31 P57KIP2 is mutated in some cases of Wiedemann-Beckwith syndrome 32 and its role as a tumor suppressor gene is still a matter of debate; 33,34 the IGF2 gene has growth promoter activity 35,36 and has also been implicated in Wiedemann-Beckwith syndrome 35,37 and in the formation of Wilms’ tumors. 33,38 The maternal loss of heterozygosity (LOH) in FoCHI necessarily leads to changes in the expression of these and other genes in the imprinted domain, suggesting that an imbalance in the amount of the corresponding gene products may result in an increase of β-cell proliferation, as already well-reported for Wilms’ tumor. 38-42
In this report, describing both morphological features and molecular data, we analyzed a series of 31 FoCHI patients by screening for ABCC8 and KCNJ11 mutations. In five FoCHI from this series, we also measured the expression of a maternally expressed tumor suppressor gene, H19, and that of a paternally expressed growth promoter gene, IGF2.
Materials and Methods
Patients
Screening for mutation in the ABCC8 and KCNJ11 genes was performed on DNA samples from 31 patients with FoCHI. For 13 patients, DNA samples from the parents were also available. Fo1, Fo2, Fo3, Fo4, Fo6, Fo7, Fo8, Fo9, and Fo10 have been reported previously in a study of LOH for the 11p15 region. 26 Fo1, Fo9, Fo11, and Fo12 have been reported previously in a study showing mutation in the ABCC8 gene. 27 In the present report, 5 of these 31 FoCHI patients were studied for the expression of 11p15.5 genes (Fo1, Fo3, Fo4, Fo6, and Fo7). LOH studies for these five patients have also been reported previously. 26 Patient Fo5 was excluded because further analysis revealed a pseudo-haploid profile suggesting that it belonged to another nosological group with a different pathogenesis. Fo18, Fo20, Fo21, and Fo33 could not be screened for ABCC8/KCNJ11 mutations because they were lost and no DNA was available for them.
Among these 31 patients, the onset of the hyperinsulinism was neonatal (within 72 hours of birth) for 23 (74%) and occurred during infancy for 8 (26%). Fifteen mg/kg/day diazoxide were given as three doses per day for at least 5 days. Diazoxide efficacy was defined as the complete normalization of blood glucose levels (>3 mmol/L), measured before and after each meal, in patients fed normally after stopping intravenous glucose and any other medication for at least 5 consecutive days. Two confirmed glucose concentrations <3 mmol/L in a 24-hour period, led us to consider the patient to be unresponsive to diazoxide. According to this criterion, all patients in this series were resistant to diazoxide. FoCHI were diagnosed by selective pancreatic venous sampling 9 combined with peroperative surgical examination of frozen sections. 4 Diagnosis was confirmed by conventional microscopy. 4,6
DNA Extraction
DNA was extracted as previously described 43 from the leukocytes of the 31 patients and, for 13 of them, from the leukocytes of their parents and from fresh pancreatic tissues for 17 patients.
Polymerase Chain Reaction (PCR)-Single-Strand Conformation Polymorphism (SSCP) and Sequence Analysis
The PCR primer pairs used to amplify all exons of the ABCC8 gene and the unique exon of the KCNJ11 gene have been described elsewhere. 14,15 PCR and SSCP analyses were performed using a modified protocol. 44 Samples containing fragments that migrated aberrantly were directly sequenced as described. 44
LOH Study
Microsatellite markers used were the same as previously and their amplification and signal detection was performed as described. 26
Quantification of Paternal Allele
To determine the number of copies of the remaining paternal chromosome 11, we used a ratio assessing the dosage of paternal allele(s) in the FoCHI lesion. To compensate for possible differences in DNA content that can wrongly enhance the signal of paternal allele in FoCHI lesion, we used, as controls, alleles generated by PCR amplification of three microsatellites from two other chromosomes, chromosome 8 (D8S262 and D8S557) and chromosome 18 (D18S57), for which no allelic imbalance was observed (data not shown). Several microsatellites were used to avoid the risk of noninformative homozygous allele. A PCR co-amplification of a microsatellite of region 11p15 and of an informative microsatellite of chromosome 8 or 18 was performed. The respective intensities of the different alleles generated were measured by quantitative autoradiography using a Phosphorimager with the Imagequant software (Molecular Dynamics-Amersham Pharmacia Biotech, Sunnyvale, CA).
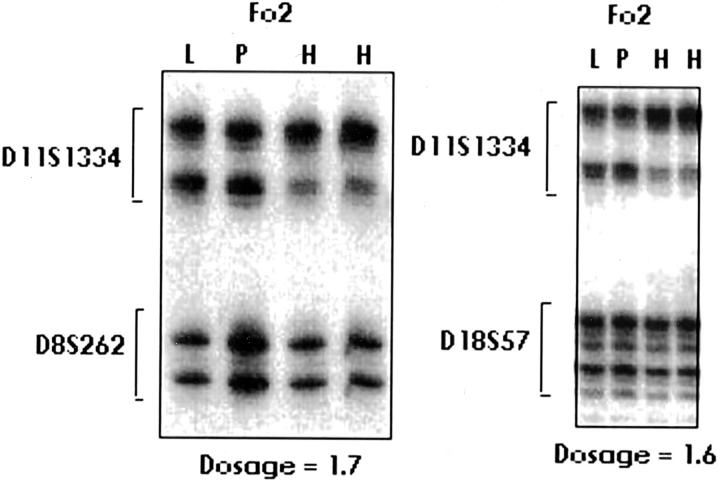
The ratio quantifying the number of copies of the paternal allele in the FoCHI lesion was calculated in two steps. First, we calculated the ratio of the signal of the paternal allele of the 11p15 microsatellite in the FoCHI lesion and in the adjacent normal pancreas. Similarly, for the control microsatellite, we calculated the ratio of the signal of the paternal allele of a chromosome 8 or 18 microsatellite in the FoCHI lesion and in adjacent normal pancreas. The number of copies of the paternal chromosome in FoCHI lesion is represented by the ratio of these two ratios (Figure 1) ▶ .
Figure 1.
Quantification of 11p15 microsatellite paternal allele in FoCHII lesion. Representative autoradiograms for gene copy number determination in patient Fo2. PCR amplification of microsatellite markers on chromosome 11 and on chromosomes 8 and 18 from focal islet cell hyperplasia (H), normal leukocytes (L), and normal pancreas (P). After quantification of the intensities of signals, the ratio indicated on the figure was calculated in two steps as described in Material and Methods.
Reverse Transcriptase (RT)-PCR and Gene Expression
Fresh tissues were frozen in liquid nitrogen and stored at −80°C until RNA isolation. For FoCHI patients, RNA was extracted from frozen samples of the FoCHI lesion and from normal pancreas after morphological control on frozen sections. Total RNA was purified using standard techniques and 1 μg was used for reverse transcription as previously described. One-fiftieth of the reaction mixture was used for PCR analysis. PCR was performed in a final volume of 50 μl of 20 mmol/L Tris-acetate, pH 9.0, 10 mmol/L ammonium sulfate, 75 mmol/L potassium acetate, 0.05% Tween-20, 10% glycerol, 0.1 mmol/L dNTP, 5 mCi α-32P dCTP, 1 mmol/L MgSO4, with 20 pmol/L of each primer for the gene analyzed. After 5 minutes at 94°C, 1 U of Tfl DNA polymerase (Promega, Charbonnières, France) was added. Amplification was performed in a Perkin-Elmer Cetus thermocycler using 26 cycles as follows: denaturation at 94°C for 1 minute, annealing at 57°C for 1 minute, and extension at 72°C for 1 minute. Aliquots were separated by electrophoresis in 10% polyacrylamide gels. Gels were fixed with acetic acid and dried. The primers used were: for IGF2 mRNA: IGF2RT-F: 5′-GCC CTC CTG GAG ACG TAC-3′, IGF2RT-R: 5′-ATC TCG GGG AAG TTG TCC-3′, for H19 mRNA: H19RT-F: 5′-ATG ACG GGT GGA GGG GCT-3′, H19RT-R: 5′-CCC ATC GTC CCC AGC-3′; for β actin-F 5′-AGA GCA AGA GAG GCA TCC-3′, actin-R 5′-TCA TTG TAG AAG GTG TGG TGC C-3′. Signal intensities were assessed by quantitative autoradiography by a Phosphorimager with the Imagequant software (Molecular Dynamics-Amersham Pharmacia Biotech, Sunnyvale, CA).
For normalization for β actin mRNA concentrations, we compared the concentration of IGF2 mRNA in the focal adenomatous hyperplasia and in normal adjacent pancreas. The ratio was obtained in two steps. In the first step, the ratio of the intensity of the signal of the IGF2 mRNA amplified by RT-PCR to the intensity of the signal of actin mRNA was calculated in the FoCHI lesion (H) and in the normal adjacent pancreas (P), respectively. In the second step, we calculated the ratio of these two ratios. This ratio permitted us to quantify the overexpression (when >1) or the underexpression (when <1) of the IGF2 gene, by comparison to the expression of the β actin gene. The same procedure was applied to the quantification of H19 by comparison with the intensities of the signal of the mRNA of IGF2.
Results
Mutations in the ABCC8 Gene
SSCP analysis of DNA from leukocyte samples of 31 patients (Table 1) ▶ detected a mobility shift on electrophoresis for the ABCC8 gene in 15 patients. All 15 patients were constitutionally heterozygous. ABCC8 mutations in focal forms were missense mutations in seven cases, splice mutations in two cases, deletion and/or insertion mutations in six cases and nonsense mutations in one case. Five of these mutations, −R842G, -R1353P, −R1421C, −R1494W, −4138del4/insGTG have been described in two previous reports from our series 4,6,27 (Table 1) ▶ . The nine other mutations have never been described previously in our reports or in the literature. 11,13,14,17,45,46 No such ABCC8 mutations were detected in the remaining hyperinsulinemic individuals belonging to a series of 131 patients, therefore precluding silent polymorphisms.
Table 1.
ABCC8 and KCNJ11 Mutations in a Series of 31 FoCHI Patients
| SUR1/ABCC8 mutation | KIR6.2/KCNJ11 mutation | Mutation origin | LOH | |
|---|---|---|---|---|
| Fo1 | R1421C* | No | Paternal | Yes |
| Fo2 | No | No | UT | Yes |
| Fo3 | No | No | UT | Yes |
| Fo4 | No | Ins405g | Paternal | Yes |
| Fo6 | No | No | UT | Yes |
| Fo7 | 3165+1g>a | No | UT | Yes |
| Fo8 | R842G** | No | Paternal | Yes |
| Fo9 | R1494W* | No | Paternal | Yes |
| Fo10 | No | No | UT | Yes |
| Fo11 | R1353P* | No | Paternal | Yes |
| Fo12 | R1494W* | No | Paternal | Yes |
| Fo13 | No | No | UT | Yes |
| Fo14 | V167L | No | UT | Yes |
| Fo15 | No | deltaF337 | Paternal | UT |
| Fo16 | 3559del4/insCGTG | No | Paternal | UT |
| Fo17 | 4202-7del12 | No | Paternal | UT |
| Fo19 | 4138del4/insGTG** | No | UT | UT |
| Fo22 | G1555S | No | UT | Yes |
| Fo23 | No | No | UT | Yes |
| Fo24 | No | No | UT | UT |
| Fo25 | No | No | UT | UT |
| Fo26 | 872fs/ter | No | Paternal | UT |
| Fo27 | No | A187V | UT | UT |
| Fo28 | No | No | UT | UT |
| Fo29 | 1580del | No | Paternal | UT |
| Fo30 | 3130del20 | No | Paternal | UT |
| Fo31 | 4153del2 | No | Paternal | UT |
| Fo32 | No | No | UT | UT |
| Fo34 | No | No | UT | UT |
| Fo35 | No | No | UT | UT |
| Fo36 | No | 813del1ins | UT | UT |
UT, Untested; LOH, loss of heterozygosity; Mutations with * or ** have already been reported. 6,27
Mutations in the KCNJ11 Gene
SSCP analysis of DNA from leukocytes of 31 patients (Table 1) ▶ detected a mobility shift on electrophoresis for the KCNJ11 gene in four patients. Sequence analysis disclosed a heterozygous pattern for four unpublished mutations: KCNJ11-405insT, KCNJ11-F337, KCNJ11-A187V, KCNJ11-813del1insTCT. As for ABCC8 mutations, KCNJ11 mutations have not been tested in a control series of normal alleles but have not been detected in the remaining hyperinsulinemic individuals belonging to a series of 121 patients, therefore precluding silent polymorphisms.
Paternal Origin of the ABCC8/KCNJ11 Mutations
In a previous paper, we have found in four cases that the mutation was of paternal origin (Fo1, Fo7, Fo8, Fo9). 27 We therefore determined the parental origin of the mutant allele in DNA samples from the parents of eight new patients with ABCC8 mutations and one with KCNJ11 mutations for which parental DNA were available. In all these nine new cases tested, the mutant allele found in the hyperplastic islet cells was already carried by the father (Table 1) ▶ . None of these mutations had occurred de novo.
Copy Number of the Paternal Chromosome
In eight patients (Fo1, Fo2, Fo3, Fo4, Fo6, Fo7, Fo8, Fo10), the copy number of the paternal chromosome 11 remaining in the focal lesion was assessed (see Quantification of Paternal Allele in Material and Methods). In patients Fo1, Fo2, Fo3, Fo4, and Fo10, we observed the ratios 2.3, 1.6, 1.8, 1.7, and 2.0, respectively. These ratios were consistent with the presence of two copies of the 11p15 paternal region in DNA from the FoCHI lesion. For patients Fo6, Fo7, and Fo8, the values obtained (1.2, 1.4, and 1.4) were inconclusive. These values probably reflect the level of contamination by normal pancreas.
Expression of IGF2 in FoPHHI
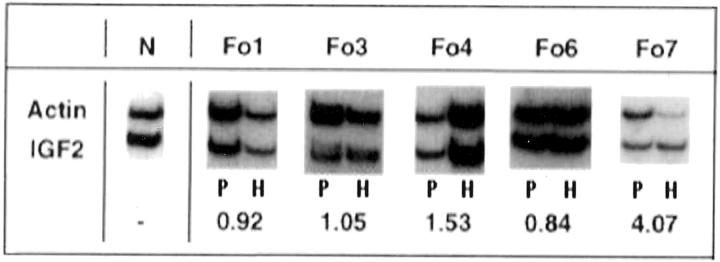
We assessed the total transcriptional activity of the IGF2 gene by determining mRNA concentrations in normal and hyperplastic pancreas samples from five FoPHHI cases (Fo1, Fo3, Fo4, Fo6, and Fo7) and an age-matched control pancreas (N). A semiquantitative RT-PCR assay was used with probes for β-actin (actin, ACTB) (Figure 2) ▶ and for glyceraldehyde phosphate dehydrogenase (data not shown) as internal controls. 47 Three independent PCR amplifications were performed for each patient.
Figure 2.
Expression of IGF2 in focal adenomatous hyperplasia. RNA was extracted from normal pancreas (left lane; P), and from hyperplastic lesions (right lane; H), from five FoPHHI patients (Fo1, Fo3, Fo4, Fo6, and Fo7) and an age-matched control pancreas (N). Expression of the IGF2 gene in hyperplastic cells was assessed by RT-PCR, normalized against β-actin mRNA. Except for case Fo7, there is no significant difference for IGF2 expression in normal pancreas and in FoCHI lesion. The numbers correspond to a ratio expressing the underexpression (when <1) or overexpression (when >1) of IGF2 in the FoCHI lesion. The ratio between the IGF2 mRNA signal and the β-actin mRNA signals in the hyperplastic lesion is divided by the same ratio in the corresponding normal pancreas.
After normalization for β-actin mRNA and for glyceraldehyde phosphate dehydrogenase concentrations, we compared the levels of expression of the mRNA for IGF2 in the focal adenomatous hyperplasia and in normal adjacent pancreas (Figure 2) ▶ . We found great variations of this ratio that did not allow us to assess an increase of IGF2 mRNA concentration in the FoCHI lesion (Figure 2) ▶ .
Imbalance Between IGF2 and H19 Expression in FoCHI Lesion
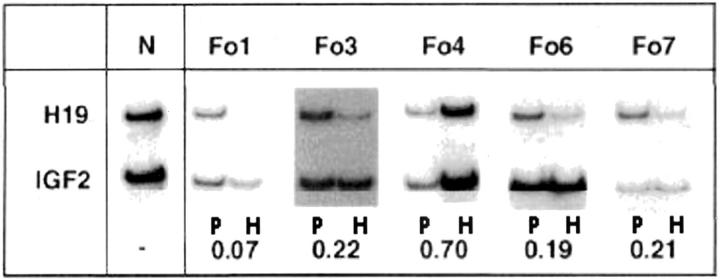
Similarly, we quantified the expression of mRNAs for H19 and IGF2. In all five cases for which sufficient amounts of mRNA were available, we found an imbalance between the expression of the IGF2 and H19 genes when comparing FoCHI lesion and normal adjacent expression (Figure 3) ▶ . In all five patients studied, we found a decrease in the expression of the H19 gene in focal adenomatous hyperplasia (H) when compared to the expression in normal adjacent pancreas (P) (Figure 3) ▶ . The mean ratio of the level of H19 mRNA compared with the level of IGF2 mRNA in FoCHI was only ∼27% of that in normal pancreatic tissue (Figure 3) ▶ . The residual expression of the H19 gene was probably because of a contamination by RNAs from normal exocrine tissue surrounding the lesion.
Figure 3.
Imbalance of the H19 and IGF2 gene expressions. Expression of the H19 and IGF2 genes was assessed by RT-PCR. The numbers correspond to the ratio of the H19 mRNA and IGF2 mRNA signals in the hyperplastic lesion divided by the same ratio calculated for the corresponding normal pancreas. They quantify the reduction of the expression of the H19 gene by comparison to the expression the IGF2 gene.
Discussion
We found that the focal form of CHI was associated with a paternal ABCC8/KCNJ11 mutation in 19 of 31 patients (61%). This rate of mutation detection seems relatively low although it is much higher than the previously reported 26.7% (23 of 86) of CHI chromosomes with an ABCC8 mutation observed in the unique screening of a previous CHI series. 14 This rate is probably due to the limited yield of the SSCP technique. Furthermore, the rate of mutation is also probably underestimated because neither the promoter region of the ABCC8/KCNJ11 genes nor the intronic regions of the genes were screened for mutations. Nevertheless this rate of mutation suggests that FoCHI is probably a more homogeneous genetic disease than the diffuse form as it is linked to a mutation of the genes coding for either one of the two subunits of the KATP channel in almost two-thirds of the cases, versus only 50% in diffuse forms of the disease (JC Fournet, MS Gross-Morand, C Mayaud, C Bellané-Chantelof, V Verkarre, et al, unpublished results). Homogeneity is further supported by the following observations: 1) all 15 cases of FoCHI who were examined for LOH had LOH for the 11p15 region (Table 1) ▶ ; 2) nine patients displayed LOH and a mutation of the ABBCC8 or KCNJ11 genes; 3) an additional set of six patients showed evidence for LOH involving the 11p15 region but not for mutations of the ABBCC8 or KCNJ11 genes; and 4) an additional set of 10 patients for whom LOH studies could not be performed carried a mutation of the ABBCC8 or KCNJ11 genes. Thus in our series of 31 patients, 25 (81%) showed evidence for the involvement of the 11p15 region, suggesting that genes contained in this region are probably involved in all cases of FoCHI.
In this series of 31 FoCHI patients, 15 different mutations of the ABCC8 gene have been identified. Six have been described in two preliminary reports. 6,27 Nine have never been described by us or in the literature. As shown in Table 1 ▶ , 7 of these 15 mutations were missense mutations. Eight mutations led to a truncated protein: one was a splice site mutation, six were insertion/deletion mutations and one was a nonsense mutation. These 15 mutations have been registered in the SUR1/ABCC8 mutation database (URL: http://umd2.necker.fr:2007).
The 3130del20 found in patient Fo30 was also found in another patient of our series with a diffuse form (JC Fournet, MS Gross-Morand, C Mayaud, C Bellané-Chantelof, V Verkarre, et al, unpublished results). Genotype analysis with several 11p15.4 region microsatellites showed that the haplotypes carrying these two mutations were identical and were therefore probably because of the same mutational event (data not shown). Thus this observation is the first clear validation of the model previously proposed 27 the same mutation of the ABCC8/KCNJ11 genes can give rise to a diffuse form, if constitutionally homozygous or compound heterozygous, and/or to a focal form if constitutionally heterozygous but somatically reduced to homozygosity.
For the first time, we also describe the involvement of KCNJ11 mutations in four cases of focal forms of PHHI. These four mutations have never been described previously. One of these four mutations was a missense mutation (KCNJ11-A187V), and one corresponded to the deletion of a phenylalanine codon without frameshift (KCNJ11-ΔF337). Two were insertion/deletion mutations leading to a truncated protein (KCNJ11-ins405G; KCNJ11–813del1insTCT). Thus, we demonstrate that mutations in either one of the genes coding for the two subunits of the K+ATP channel of β cells can equally participate in the genesis of focal forms of CHI. As previously described for the ABCC8 gene, the KCNJ11 constitutional heterozygous mutation is reduced to homozygosity by a somatic loss of a maternal allele from the 11p15 region during the pancreatic development, restricted to the focal adenomatous hyperplasia lesion. 27,48
However, the reduction to homozygosity of a paternally inherited ABCC8/KCNJ11 mutation leading to uncontrolled secretion of insulin is not the only consequence of the 11p15 somatic rearrangement. The allelic dosage experiment that we performed showed that the paternal allele is duplicated in the FoCHI lesion. It probably corresponds to a duplication of the paternal 11p15 region contemporary to the loss of maternal 11p15 alleles, as observed in 11p15.5 related tumors (Wilms’ tumor, rhabdomyosarcoma) or diseases, such as Wiedemann-Beckwith syndrome. However, despite this allelic paternal duplication, we did not observe a consistent increase of the expression of the paternally expressed IGF2 gene in the focal lesion. Such wide variations in IGF2 expression have also been reported in various series of Wilms’ tumors with or without LOH or loss of imprinting and in adjacent normal kidney samples. 33,41,42 These variations can be because of the instability of the IGF2 mRNA, but can also be linked to different expression levels of IGF2 in the FoCHI lesion.
The somatic loss of maternal 11p15 region during pancreatic development also necessarily alters the expression of maternally imprinted genes of region 11p15.5 for which only the maternal allele is expressed. Among them, the H19 gene has a tumor suppressor activity and is involved in several tumor models with rearrangements, mostly LOH, of the 11p15.5 region. We showed that the expression of the tumor suppressor gene H19, monoallelically expressed from the maternal allele, was greatly reduced, in agreement with the loss of the maternal 11p15 region. This alteration of the expression of imprinted genes probably explains why inherited mutations are always of paternal origin.
The pattern of expression of IGF2 and H19 observed in FoPHHI was similar to that in embryonic tumors such as Wilms’ tumor and rhabdomyosarcoma. 33,40,49-51 In these tumors, the lack of H19 was not necessarily coupled to higher levels of IGF2 expression. 33,40 Recent data from Wilms’ tumor patients suggested that the H19 gene is generally inactivated in association with an overgrowth lesion. Although in Wilms’ tumor it is generally assumed that silencing of H19 leads to biallelic IGF2 expression, most of the tumors analyzed have displayed a silenced H19 but monoallelic IGF2 expression. These observations suggest that H19 may play a role by itself, as suggested by reversion of the malignant phenotype of a rhabdomyosarcoma cell line transfected with the H19 gene. 30 Although these results could not be reproduced, the role of H19 may be less enigmatic. Indeed, the H19 transcript has been shown to be associated with polysomes in a variety of cell types in both mice and humans. 52 H19 seems directly or indirectly to modulate cytoplasmic levels of the product of the IGF2 allele. Thus the H19 gene seems to be an antagonist to IGF2 in trans.
The imbalance between IGF2 and H19 rather than the up-regulation of IGF2 alone may therefore participate to pancreatic hyperplasia in FoPHHI, as previously proposed in a Wilms’ tumor model. 33 Analysis of this model led to the suggestion that normal cell growth requires coordinated expression of both H19, a putative tumor suppressor gene, and IGF2, an autocrine growth factor gene. Abolition of H19 expression and/or up-regulation of IGF2 may therefore increase cell growth. Indeed, inactivation of H19 has been shown to be a preneoplastic event in Wilms’ tumor formation. 31 Alternatively, the expression of other genes of the region may be dysregulated and may be involved in the formation of this tumor. These genes include P57KIP2 and unknown genes.
The somatic event of 11p15 LOH is probably a rare phenomenon during fetal development: in our series of 31 cases of FoCHI, we found no case of recurrence in the same patient or in the same family. We report for the first time one instance of an haplo-identical mutation (3130del20) in a patient with a diffuse form and in a patient with a focal form in a large series of 131 patients thus further supporting the rarity of this somatic event. It may take place during a narrow window in fetal life: the event is probably late because the lesion is tiny (a few millimeters) and involves few pancreatic lobules. It probably occurs during fetal life because most cases (74% in our series) are neonatal and, as far as we know, no recurrence of FoPHHI on the pancreatic remnant after a complete removal of the focal adenomatous hyperplasia by partial pancreatectomy has ever been described. 6 At this stage, we can conclude that the probability for this somatic chromosomal event to occur in a fetus carrying a heterozygous mutation of ABCC8/KCNJ11 of paternal origin is rare, probably <1%. However, this question is critical for genetic counseling purposes and epidemiological studies are needed to provide a more precise figure.
Acknowledgments
We thank Nicole Brousse, Chantal Duros, Geneviève Gourdon, Isabelle Henry, Francis Jaubert, and Myriam Le Strat for their help and contribution to this work.
Footnotes
Address reprint requests to Pr. Claudine Junien, INSERM UR 383, Hôpital Necker-Enfants Malades, Clinique Maurice Lamy, 149, rue de Sèvres, 75743 PARIS Cedex 15, France. E-mail: junien@necker.fr.
Supported by grants from the Mutuelle Générale de l’Education Nationale, the Délégation à la Recherche Clinique, Assistance Publique-Hôpitaux de Paris, the Ligue Nationale Contre le Cancer and the Association pour la Recherche contre le Cancer, and INSERM “PROGRES n° 4P010D”. P. de L. was supported by a grant from the Evian Foundation.
References
- 1.Stanley C: Hyperinsulinism in infants and children. Pediatr Clin North Am 1997, 44:363-374 [DOI] [PubMed] [Google Scholar]
- 2.Bruining G: Recent advances in hyperinsulinism and the pathogenesis of diabetes mellitus. Curr Opin Pediatr 1990, 2:758-765 [Google Scholar]
- 3.Goossens A, Gepts W, Saudubray J, Bonnefont J, Nihoul-Fekete C, Heitz P, Kloppel G: Diffuse and focal nesidioblastosis. : A clinicopathological study of 24 patients with persistent neonatal hyperinsulinemic hypoglycemia. Am J Surg Pathol 1989, 13:766-775 [PubMed] [Google Scholar]
- 4.Rahier J, Sempoux C, Fournet JC, Poggi F, Brunelle F, Nihoul-Fekete C, Saudubray JM, Jaubert F: Partial or near-total pancreatectomy for persistent neonatal hyperinsulinaemic hypoglycaemia: the pathologist’s role. Histopathology 1998, 32:15-19 [DOI] [PubMed] [Google Scholar]
- 5.Jaffé R, Hashida Y, Yunis E: Pancreatic pathology in hyperinsulinemic hypoglycemia of infancy. Lab Invest 1980, 42:356-365 [PubMed] [Google Scholar]
- 6.de Lonlay-Debeney P, Poggi-Travert F, Fournet JC, Sempoux C, Vici CD, Brunelle F, Touati G, Rahier J, Junien C, Nihoul-Fekete C, Robert JJ, Saudubray JM: Clinical features of 52 neonates with hyperinsulinism. N Engl J Med 1999, 340:1169-1175 [DOI] [PubMed] [Google Scholar]
- 7.Brunelle F, Negre V, Barth M, Fekete C, Czernichow P, Saudubray J, Kuntz F, Tach T, Lallemand D: Pancreatic venous samplings in infants and children with primary hyperinsulinism. Pediatr Radiol 1989, 19:100-103 [DOI] [PubMed] [Google Scholar]
- 8.Lyonnet S, Bonnefont J-P, Saudubray J-M, Nihoul-Fékété C, Brunelle F: Localization of focal lesion permitting partial pancreatectomy in infants. Lancet 1989, 2:671. [DOI] [PubMed] [Google Scholar]
- 9.Dubois J, Brunnelle F, Touati G, Sebag G, Nuttin C, Thach T, Nihoul-Fekete C, Rahier J, Saudubray J: Hyperinsulinism in children: diagnosis value of pancreatic venous sampling correlated with clinical, pathological and surgical outcome in 25 cases. Pediatr Radiol 1995, 25:512-516 [DOI] [PubMed] [Google Scholar]
- 10.Craver R, Hill C: Cure of hypoglycemic hyperinsulinism by enucleation of a focal islet cell adenomatous hyperplasia. J Pediatr Surg 1997, 32:1526-1527 [DOI] [PubMed] [Google Scholar]
- 11.Thomas PM, Cote GJ, Wohllk N, Haddad B, Mathew PM, Rabl W, Aguilar Bryan L, Gagel RF, Bryan J: Mutations in the sulfonylurea receptor gene in familial persistent hyperinsulinemic hypoglycemia of infancy. Science 1995, 268:426-429 [DOI] [PubMed] [Google Scholar]
- 12.Thomas PM, Wohllk N, Huang E, Kuhnle U, Rabl W, Gagel RF, Cote GJ: Inactivation of the first nucleotide-binding fold of the sulfonylurea receptor, and familial persistent hyperinsulinemic hypoglycemia of infancy. Am J Hum Genet 1996, 59:510-518 [PMC free article] [PubMed] [Google Scholar]
- 13.Nestorowicz A, Wilson BA, Schoor KP, Inoue H, Glaser B, Landau H, Stanley CA, Thornton PS, Clement JP, IV, Bryan J, Aguilar-Bryan L, Permutt MA: Mutations in the sulfonylurea receptor gene are associated with familial hyperinsulism in Ashkenazi Jews. Hum Mol Genet 1996, 5:1813-1822 [DOI] [PubMed] [Google Scholar]
- 14.Nestorowicz A, Glaser B, Wilson BA, Shyng SL, Nichols CG, Stanley CA, Thornton PS, Permutt MA: Genetic heterogeneity in familial hyperinsulinism. Hum Mol Genet 1998, 7:1119-1128 [DOI] [PubMed] [Google Scholar]
- 15.Nestorowicz A, Inagaki N, Gonoi T, Schoor K, Wilson B, Glaser B, Landau H, Stanley C, Thornton P, Seino S, Permutt M: A nonsense mutation in the inward rectifier potassium channel gene, Kir6.2, is associated with familial hyperinsulinism. Diabetes 1997, 46:1743-1748 [DOI] [PubMed] [Google Scholar]
- 16.Inoue H, Ferrer J, Warren-Perry M, Zhang Y, Millns H, Turner RC, Elbein SC, Hampe CL, Suarez BK, Inagaki N, Seino S, Permutt MA: Sequence variants in the pancreatic islet beta-cell inwardly rectifying K+ channel Kir6.2 (Bir) gene: : identification and lack of role in Caucasian patients with NIDDM. Diabetes 1997, 46:502-507 [DOI] [PubMed] [Google Scholar]
- 17.Thomas P, Ye Y, Lightner E: Mutation of the pancreatic islet inward rectifier Kir6.2 also leads to familial persistent hyperinsulinemic hypoglycemia of infancy. Hum Mol Genet 1996, 5:1809-1812 [DOI] [PubMed] [Google Scholar]
- 18.Aguilar-Bryan L, Nichols C, Wechsler S, Clement J, Boyd A, Gonzalez G, Herrera Sosa H, Nguy K, Bryan J, Nelson D: Cloning of the beta cell high-affinity sulfonylurea receptor: : a regulator of insulin secretion. Science 1995, 268:423-426 [DOI] [PubMed] [Google Scholar]
- 19.Inagaki N, Gonoi T, Clement J, Namba N, Inazawa J, Gonzalez G, Aguilar-Bryan L, Seino S, Bryan J: Reconstitution of IKATP: an inward rectifier subunit plus the sulfonylurea receptor. Science 1995, 270:1166-1170 [DOI] [PubMed] [Google Scholar]
- 20.Zammarchi E, Filippi L, Novembre E, Donati M: Biochemical evaluation of a patient with a familial form of leucine-sensitive hypoglycemia and concomitant hyperammoniema. Metabolism 1996, 45:957-960 [DOI] [PubMed] [Google Scholar]
- 21.Weinzimer S, Stanley C, Berry G, Yudkoff M, Tuchman M, Thornton P: A syndrome of congenital hyperinsulinism and hyperammoniema. J Pediatr 1997, 130:661-664 [DOI] [PubMed] [Google Scholar]
- 22.Stanley C, Lieu Y, Hsu B, Ponez M: Hypoglycemia in infants with hyperinsulinism and hyperammoniema: gain of function mutations in the pathway of leucine-mediated insulin secretion. Diabetes 1997, 46(Suppl 1):217A [Google Scholar]
- 23.Glaser B, Kesavan P, Heyman M, Davis E, Cuesta A, Buchs A, Stanley C, Thornton P, Permutt A, Matschinsky F, Herold K: Familial hyperinsulinism caused by an activating glucokinase mutation. N Engl J Med 1998, 338:226-230 [DOI] [PubMed] [Google Scholar]
- 24.Kukuvitis A, Deal C, Arbour L, Polychronakos C: An autosomal dominant form of familial persistent hyperinsulinemic hypoglycemia of infancy, not linked to the sulfonylurea receptor locus. J Clin Endocrinol Metab 1997, 82:1192-1194 [DOI] [PubMed] [Google Scholar]
- 25.Thornton PS, Satin-Smith MS, Herold K, Glaser B, Chiu KC, Nestorowicz A, Permutt MA, Baker L, Stanley CA: Familial hyperinsulinism with apparent autosomal dominant inheritance: clinical and genetic differences from the autosomal recessive variant. J Pediatr 1998, 132:9-14 [DOI] [PubMed] [Google Scholar]
- 26.de Lonlay P, Fournet JC, Rahier J, Gross-Morand MS, Poggi-Travert F, Foussier V, Bonnefont JP, Brusset MC, Brunelle F, Robert JJ, Nihoul-Fekete C, Saudubray JM, Junien C: Somatic deletion of the imprinted 11p15 region in sporadic persistent hyperinsulinemic hypoglycemia of infancy is specific of focal adenomatous hyperplasia and endorses partial pancreatectomy. J Clin Invest 1997, 100:802-807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verkarre V, Fournet JC, de Lonlay P, Gross-Morand MS, Devillers M, Rahier J, Brunelle F, Robert JJ, Nihoul-Fekete C, Saudubray JM, Junien C: Paternal mutation of the sulfonylurea receptor (SUR1) gene and maternal loss of 11p15 imprinted genes lead to persistent hyperinsulinism in focal adenomatous hyperplasia. J Clin Invest 1998, 102:1286-1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feinberg AP: Imprinting of a genomic domain of 11p15 and loss of imprinting in cancer: an introduction. Cancer Res 1999, 59 (Suppl 7):1743s-1746s [PubMed] [Google Scholar]
- 29.Engemann S, Strodicke M, Paulsen M, Franck O, Reinhardt R, Lane N, Reik W, Walter J: Sequence and functional comparison in the Beckwith-Wiedemann region: implications for a novel imprinting centre and extended imprinting. Hum Mol Genet 2000, 9:2691-2706 [DOI] [PubMed] [Google Scholar]
- 30.Hao Y, Crenshaw T, Moulton T, Newcomb E, Tycko B: Tumour-suppressor activity of H19 RNA. Nature 1993, 365:764-767 [DOI] [PubMed] [Google Scholar]
- 31.Cui H, Hedborg F, He L, Nordenskjold A, Sandstedt B, Pfeifer-Ohlsson S, Ohlsson R: Inactivation of H19, an imprinted and putative tumor repressor gene, is a preneoplastic event during Wilms’ tumorigenesis. Cancer Res 1997, 57:4469-4473 [PubMed] [Google Scholar]
- 32.Hatada I, Ohashi H, Fukushima Y, Kaneko Y, Inoue M, Komoto Y, Okada A, Ohishi S, Nabetani A, Morisaki H, Nakayama M, Niikawa N, Mukai T: An imprinted gene P57KIP2 is mutated in Beckwith-Wiedemann syndrome. Nat Genet 1996, 14:171-173 [DOI] [PubMed] [Google Scholar]
- 33.Steenman M, Rainier S, Dobry C, Grundy P, Horon I, Feinberg A: Loss of imprinting of IGF2 is linked to reduced expression and abnormal methylation of H19 in Wilms’ tumour. Nat Genet 1994, 7:433-439 [DOI] [PubMed] [Google Scholar]
- 34.Matsuoka S, Edwards M, Bai C, Parker S, Zhang P, Baldini A, Harper J, Elledge S: p57KIP2, a structurally distinct member of the p21CIP1 Cdk inhibitor family, is a candidate tumor suppressor gene. Genes Dev 1995, 9:650-662 [DOI] [PubMed] [Google Scholar]
- 35.Ohlsson R, Nystrom A, Pfeifer-Ohlsson S, Tohonen V, Hedborg F, Schofield P, Flam F, Ekstrom T: IGF2 is parentally imprinted during human embryogenesis and in the Beckwith-Wiedemann syndrome. Nat Genet 1993, 4:94-97 [DOI] [PubMed] [Google Scholar]
- 36.Giannoukakis N, Deal C, Paquette J, Goodyer C, Polychronakos C: Parental genomic imprinting of the human IGF2 gene. Nat Genet 1993, 4:98-101 [DOI] [PubMed] [Google Scholar]
- 37.Junien C: Beckwith-Wiedemann syndrome, tumourigenesis and imprinting. Curr Opin Genet Dev 1992, 2:431-438 [DOI] [PubMed] [Google Scholar]
- 38.Ogawa O, Eccles M, Szeto J, McNoe L, Yun K, Maw M, Smith P, Reeve A: Relaxation of insulin-like growth factor II gene imprinting implicated in Wilms’ tumour. Nature 1993, 362:749-751 [DOI] [PubMed] [Google Scholar]
- 39.Ogawa O, Becroft D, Morison I, Eccles M, Skeen J, Mauger D, Reeve A: Constitutional relaxation of insulin-like growth factor II gene imprinting associated with Wilms’ tumour and gigantism. Nat Genet 1993, 5:408-412 [DOI] [PubMed] [Google Scholar]
- 40.Moulton T, Crenshaw T, Hao Y, Moosikasuwan J, Lin N, Dembitzer F, Hensle T, Weiss L, McMorrow L, Loew T, Draus W, Gerald W, Tycko B: Epigenetic lesions at the H19 locus in Wilms’ tumour patients. Nat Genet 1994, 7:440-447 [DOI] [PubMed] [Google Scholar]
- 41.Reeve AE, Eccles MR, Wilkins RJ, Bell GI, Millow LJ: Expression of insulin-like growth factor-II transcripts in Wilms’ tumour. Nature 1985, 317:258-260 [DOI] [PubMed] [Google Scholar]
- 42.Scott J, Cowell J, Robertson ME, Priestley LM, Wadey R, Hopkins B, Pritchard J, Bell GI, Rall LB, Graham CF, Knott T: Insulin-like growth factor-II gene expression in Wilms’ tumour and embryonic tissues. Nature 1985, 317:260-262 [DOI] [PubMed] [Google Scholar]
- 43.Béroud C, Fournet J-C, Froger D, Jeanpierre C, Droz D, Chretien Y, Bouvier R, Marechal J, Weissenbach J, Junien C: Correlations of allelic imbalance of chromosome 14 with tumor progression and prognostic parameters in renal cell carcinoma. Genes Chromosom Cancer 1996, 17:215-224 [DOI] [PubMed] [Google Scholar]
- 44.Blanquet V, Turleau C, Gross-Morand M-S, Sénamaud-Beaufort C, Doz F, Besmond C: Spectrum of germline mutations in the RB1 gene: a study of 232 patients with hereditary and non-hereditary retinoblastoma. Hum Mol Genet 1995, 4:383-388 [DOI] [PubMed] [Google Scholar]
- 45.Meissner T, Beinbrech B, Mayatepek E: Congenital hyperinsulinism: molecular basis of a heterogeneous disease. Hum Mutat 1999, 13:351-361 [DOI] [PubMed] [Google Scholar]
- 46.Otonkoski T, Ammala C, Huopio H, Cote GJ, Chapman J, Cosgrove K, Ashfield R, Huang E, Komulainen J, Ashcroft FM, Dunne MJ, Kere J, Thomas PM: A point mutation inactivating the sulfonylurea receptor causes the severe form of persistent hyperinsulinemic hypoglycemia of infancy in Finland. Diabetes 1999, 48:408-415 [DOI] [PubMed] [Google Scholar]
- 47.Radvanyi F, Christgau S, Baekkeskov S, Jolicoeur C, Hanahan D: Pancreatic beta cells cultured from individual preneoplastic foci in a multistage tumorigenesis pathway: a potentially general technique for isolating physiologically representative cell lines. Mol Cell Biol 1993, 13:4223-4232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Glaser B, Ryan F, Donath M, Landau H, Stanley CA, Baker L, Barton DE, Thornton PS: Hyperinsulinism caused by paternal-specific inheritance of a recessive mutation in the sulfonylurea-receptor gene. Diabetes 1999, 48:1652-1657 [DOI] [PubMed] [Google Scholar]
- 49.Zhan S, Shapiro DN, Helman LJ: Activation of an imprinted allele of the insulin-like growth factor II gene implicated in rhabdomyosarcoma. J Clin Invest 1994, 94:445-448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhan S, Shapiro DN, Helman LJ: Loss of imprinting of IGF2 in Ewing’s sarcoma. Oncogene 1995, 11:2503-2507 [PubMed] [Google Scholar]
- 51.Taniguchi T, Sullivan MJ, Ogawa O, Reeve AE: Epigenetic changes encompassing the IGF2/H19 locus associated with relaxation of IGF2 imprinting and silencing of H19 in Wilms tumor. Proc Natl Acad Sci USA 1995, 92:2159-2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li YM, Franklin G, Cui HM, Svensson K, He XB, Adam G, Ohlsson R, Pfeifer S: The H19 transcript is associated with polysomes and may regulate IGF2 expression in trans. J Biol Chem 1998, 273:28247-28252 [DOI] [PubMed] [Google Scholar]