Abstract
Regulation of the lymphoid enhancer factor 1 (Lef-1) transcription factor is important for the inductive formation of many epithelial-derived organs including airway submucosal glands (SMGs). Although Wnts have been linked to developmental processes involving transcriptional activation of the Lef-1 protein, there is little in vivo information directly linking Wnts with the transcriptional regulation of the Lef-1 promoter. In the present study, we hypothesized that Wnt3a directly regulates Lef-1 gene expression required for SMG morphogenesis in mice. In support of this hypothesis, TOPGAL reporter mice demonstrated activation of β-catenin/Tcf complexes during early phases of SMG development and immunolocalization studies confirmed abundant expression of Tcf4, but not Tcf1 or Tcf3, at this stage. ChIP analysis in primary airway epithelial cells revealed that Tcf4 associates with a known Wnt Responsive Region in the Lef-1 promoter and transfection of Cos-1 cells with dominant active β-catenin and Tcf4 synergistically activated the Lef-1 promoter. Using Wnt3a deficient and Lef-1 promoter-GFP reporter mice, we also demonstrate that Wnt3a induces Lef-1 gene expression in newly forming SMG buds of mice and the maintenance of gland bud growth. These findings provide the first in vivo evidence that Wnt3a can transcriptionally regulate the Lef-1 gene.
Keywords: Wnt3a, TOPGAL, Lef-1, Development, Airway, Glands, Tcf4
Introduction
Submucosal glands (SMGs) are epithelial secretory structures that reside beneath the airway epithelium that play important roles in hypersecretory lung diseases such as asthma, chronic bronchitis, and cystic fibrosis (CF) (Verkman et al., 2003). In humans, SMG hypertrophy and hyperplasia are thought to aggravate the pathoprogression of this disease through excessive production of airway mucus (Salinas et al., 2005). Hence, an understanding of the mechanisms of gland growth and differentiation will allow for the development of improved treatments of diseases that result in gland expansion and hypersecretion. In humans, SMGs reside throughout the cartilaginous airways of the tracheobronchial tree and in the submucosa of the nasal passages. In mice, the development of these structures begins as early as E15.5 for nasal submucosal glands, while tracheal glands begin to develop during the first few days of birth.
Epithelial appendages, like hair follicles, mammary glands, and teeth, rely heavily on Wnt signaling pathways for proper development. To this end, inhibiting Wnt signaling through ectopic expression of Dkk1, a Wnt inhibitor, results in the loss of formation of these structures (Andl et al., 2002; Chu et al., 2004). The canonical Wnt signaling pathway consists of a secreted Wnt signaling molecule that binds to its receptor Frizzled and co-receptor LRP (Logan and Nusse, 2004). Wnts binding with their cellular receptors signal a cascade of events that leads to the inhibition of β-catenin degradation, which is normally found associated with E-cadherin or other similar membrane proteins, keeping free cytoplasmic and nuclear β-catenin at low levels (Nelson and Nusse, 2004). Once stabilized β-catenin is translocated to the nucleus, it can associate with transcription factors and invoke transcriptional changes by recruiting chromatin remodeling proteins (Nusse, 2005). Target genes of the Wnt signaling pathway include: 1) structural proteins, such as E-cadherin or, cytokeratins (Jamora et al., 2003; Zhou et al., 1995); 2) cellular proliferation factors, such as cyclin D1 (Shtutman et al., 1999; Tetsu and McCormick, 1999); and 3) transcription factors, such as Lef-1 (Driskell et al., 2004; Filali et al., 2002; Hovanes et al., 2001; Hovanes et al., 2000; Li et al., 2006).
Lef-1 is a Tcf factor that mediates Wnt signaling by binding with β-catenin at specific sites in the promoters of genes such as Cyclin D1 and keratin-14 (K14) to influence cellular division and migration (Nusse, 2005). Lef-1 knock out mice lack fully formed hair follicles, mammary glands, teeth, and airway SMGs (Duan et al., 1999; van Genderen et al., 1994). Misregulated expression of this gene has also been implicated in colon cancer in conjunction with hyperactive Wnt signaling (Hovanes et al., 2000; Li et al., 2006). Consequently, dissecting the regulatory mechanisms of Lef-1 expression has become imperative to understanding a diversity of developmental processes in both the normal and diseased states.
Attempts to elucidate the transcriptional regulation of the Lef-1 gene have involved both in vitro and in vivo studies. In vitro studies have revealed that transcriptional activity of the Lef-1 gene can be modulated by the Wnt signaling pathway and also by the transcription factor Pitx2 (Hovanes et al., 2000; Vadlamudi et al., 2005). Early studies investigating the potential for the Wnt pathway to induce Lef-1 gene transcription have identified two Tcf binding sites in the Lef-1 promoter [−1000 bp (TCTTTGCT) and −921 bp (AACAAAGA)]. These Tcf sites were shown to bind Tcf factors using in vitro binding assays and to be necessary for upregulation of Lef-1 transcriptional activity by ectopically expressed β-catenin/Tcf complexes (Hovanes et al., 2000). Additional studies of this region of the Lef-1 promoter identified a separate 110 bp region in the Lef-1 promoter, which was named the Wnt Response Element (WRE) (−879 to −769 bp), because deletion of this region resulted in a loss of Wnt3a inducible transcription (Filali et al., 2002). The WRE also contains a candidate Tcf binding site at −785 bp (ACTTTATT). The deletion of the WRE increased the baseline transcriptional activity of the promoter, which is consistent with this region of the promoter being inhibitory in the absence of β-catenin (Filali et al., 2002). Recent studies have investigated the association of Tcf factors to these regions of the Lef-1 promoter using Chromatin Immunoprecipitations with pan-Tcf antibodies (Li et al., 2006). These studies suggested that Tcf factors associate preferentially with either the WRE or the previously identified Tcf binding sites depending on the cell type used for analysis. However, the specific Tcf factors that associate with this area have not been identified. In this report, we refer to both the Tcf binding sites (−1000 and −921 bp) and the WRE (−879 to −769 bp) collectively as the Wnt Response Region (WRR).
The in vivo importance of the Lef-1 promoter WRE in regulating Lef-1 gene expression has also been investigated in transgenic Lef-1 promoter/reporter mice (Driskell et al., 2004; Liu et al., 2004). The results of these studies suggested that the WRE is important for controlling epithelial and mesenchymal expression patterns of the Lef-1 promoter in hair follicles and mammary glands (Liu et al., 2004). Studies demonstrating that reciprocal Wnt-signals in the epithelia and mesenchyme of the developing mammary gland are in part controlled by Lef-1, support the potential importance of Wnt regulated Lef-1 gene expression (Boras-Granic et al., 2006). Additionally, the Lef-1 promoter WRE was required for the inductive expression during airway gland morphogenesis in mice (Driskell et al., 2004). However, unlike other models of epithelial appendages, no studies have evaluated the importance of Wnt signaling in SMG morphogenesis. Furthermore, evidence for whether Wnts can directly regulate the Lef-1 promoter in vivo in any organ system has not been reported.
In the present study, we have begun to define the in vivo regulatory mechanism that controls the induction of the Lef-1 gene during airway gland development. To this end, we have demonstrated that the canonical Wnt signaling pathway is active during gland bud formation using β-catenin/Tcf-βgal reporter (TOPGAL) mice. Furthermore, using Wnt3a KO mice, we demonstrate that Wnt3a regulates Lef-1 protein and promoter expression during airway SMG morphogenesis. Molecular studies attempting to define the Wnt-regulated transcription factors required for induction of the Lef-1 promoter demonstrated that Tcf4 is the likely candidate in gland buds and binds to the Lef-1 promoter WRR in primary airway cells and transformed airway cell lines. In summary, these studies demonstrate that Lef-1 gene transcription in vivo can be regulated by Wnt3a, and places a physiologic context to this regulatory mechanism in airway gland morphogenesis.
Materials and Methods
Transgenic mice
Lef-1 deficient mice were previously described (van Genderen et al., 1994) and Wnt3a deficient mice were kindly provided by Dr. Andy McMahon at Harvard University (Takada et al., 1994) and were on a mixed C57BL/6J:BALB/c background. Genotyping for these mice was performed as previously described (Sasaki et al., 2005; Takada et al., 1994). TOPGAL mice were purchased from Jackson Laboratories and were genotyped according to their protocol. β-galactosidase staining of TOPGAL mouse tissues was performed as previously described (Driskell et al., 2004). Lef-1/GFP reporter mice were generated using the previously described LF-2700/-200 human Lef-1 promoter fragment (−2700 to −200 bp relative to the translational start codon ATG at +1) (Filali et al., 2002) controlling the expression of nuclear-targeted EGFP fusion protein. The transgene cassette lacking all plasmid sequences was excised, gel purified, and then injected into the pronucleus of fertilized oocytes of C57BL/6JxSJL/J F2 lineage. ICR pseudopregnant females were implanted with the transgenic embryos and transgenic founders and progeny were screened by PCR analysis of genomic DNA with the following primers: LFGFP-Fwd: 5′-CAC-GTT-ATT-TAC-CCT-GTT-TCG-GGC-3′ and LFGFP-Rev: 5′-CAG-ATG-AAC-TTC-AGG-GTC-AGC-TTG-3′. Transgenic mice were maintained by successive backcross breeding to C57BL/6J. Timed pregnancies were performed as previously described (Driskell et al., 2004) with vaginal plugging on the morning following evening mating defined as E0.5 days. Mice were sacrificed and tissue was immediately frozen in Optimum Cutting Temperature Media (OCT) and sectioned at 10 μm for immunostaining.
In Vitro Analysis of the Lef-1 Promoter
The Cos1 cell line was transfected using Lipofectamine according to standard protocols. A previously described expression construct (LF-2700/-200-Luciferase) containing 2.5 kb of the human Lef-1 promoter upstream of the firefly luciferase reporter (Vadlamudi et al., 2005) gene was used to study Tcf4/β-catenin responsiveness of the Lef-1 promoter. LF-2700/-200-Luciferase contains sequences in the promoter that span from −2700 to −200 bp. Cells were grown to 70% confluency on 60-mm dishes prior to co-transfection with 1 μg LF-2700/-200-Luciferase construct and 0.5 μg of renilla luciferase plasmid (Promega), which encoded the renilla luciferase gene under the control of the SV40 promoter. Cells were harvested for luciferase assays at 24hrs post-transfection. Harvesting and analysis of lysates was performed as previously described (Vadlamudi et al., 2005). Normalization of transfection efficiencies for each experimental point was performed by dividing the firefly luciferase activity units by renilla luciferase activity. The resulting value was then used to compare the expression level of a given construct to the promoterless luciferase plasmid backbone for the same experiment. For Tcf4 and (S37)β-catenin induction assays, 4 μg of expression plasmids encoding Tcf4 and/or (S37)β-catenin was cotransfected with the Lef-1 promoter-reporter and renilla luciferase constructs, as described above. The total amount of DNA transfected in all experimental comparisons was always normalized by the inclusion of an empty vector plasmid control (pcDNA).
In vivo analysis of the Lef-1 promoter activity in gland buds
Wnt3a +/− male mice were bred to Lef-1 promoter-EGFP reporter female transgenic mice. Double heterozygous females were then bred to Wnt3a +/− males and embryos were harvested at E15.5. 10 μm sections of the snouts were obtained from littermates of Lef-1 promoter-EGFP transgene positive embryos with either Wnt3a-KO or Wnt3a-WT genotypes (defined by abnormalities in lower torso development and confirmed by PCR in independent studies). Breedings typically yielded only one double transgenic Wnt3a-KO:Lef-1-EGFP embryo and five independent breedings of this type were evaluated using matched littermate embryos for analysis. Sections were stained with an anti-EGFP antibody and FITC-labeled secondary antibody to localize the nuclear targeted EGFP transgene. Serial sections of the nasal cavity were analyzed where Lef-1 promoter activity has been shown to express in nasal SMG buds (Driskell et al., 2004). The number of EGFP positive nuclei in each nasal SMG bud was then quantified for matched littermate embryos. Only regions of the glandular placode residing below the basal lamina of the surface airway epithelium were quantified. Statistical differences were evaluated by ANOVA followed by the Mann-Whitney test.
Western Blotting of newborn mouse tracheas and skin
Tracheas and skin were dissected from C57 postnatal day 1 (P1) mice and immediately flash frozen in liquid nitrogen and minced in RIPA buffer. Protein concentrations were quantified and 75μg of protein was run on a 10.5% SDS-PAGE gel, transferred to Nylon membranes, and probed with antibodies against Tcf1 (SantaCruz H-18), Lef-1 (Exalpha T-100M), Tcf3 (SantaCruz M-20), and Tcf4 (Exalpha X1070M). 293 cells were also transfected with control plasmids overexpressing Tcf1 (Upstate 21–172), Lef-1, Tcf3 (Upstate 21–173), and Tcf4 (kind gift of Dr. James Wells). 75μg of lysates were used as positive controls for Western blots and demonstrated strong reactivity with the antibodies.
Chromatin Immunoprecipitation
Cos1 cells were transfected in triplicate in 150 mm plates with 6 μg of LF-2700/-200-Luciferase plasmid and either 36 μg of GFP or Tcf4V5His (kind gift of Dr. James Wells) expression plasmids. Cells were then allowed to incubate for 24 hours before harvesting the DNA for chromatin immunoprecipitation. DNA harvesting and shearing was performed according to the Active Motif DNA shearing kit (53005); 10 min incubations with the DNA shearing cocktail resulted in consistent 300–600 bp DNA fragmentation for Cos1 cells. Precipitation and purification of DNA fragments was performed according to the EZChIP Kit (#17–371) protocol from Upstate Biologicals. Antibodies used for precipitation of protein/DNA complexes included control Mouse IgG, anti-Tcf4 (Exalpha X1070M), and anti-V5 (Invitrogen R960-25). Final analysis of the precipitated DNA was performed using Real Time PCR on 3 μl of the final DNA precipitate. Primers used to amplify the precipitated DNA fragments included: 1) WRE Fwd: 5′-CTC-GAG-CCG-GGA-ACA-AAG-A-3′ and WRE Rev: 5′-GGG-AAG-AGA-AAG-AGA-AGT-TTG-CC-3′ which amplified a 181 bp fragment from −932 bp to −751 bp of the human Lef-1 promoter and 2) Cont Fwd: 5-CAG-CGG-AGC-GGA-GAT-TAC-AG-3′ and Cont Rev: 5′-TCT-CTG-AGT-TTC-CCA-GGG-ACC-3′ which amplified a 302 bp fragment from −30 bp to +272 bp of the human Lef-1 gene. The base pair numbering originates from our previous work with the Lef-1 promoter (NCBI accession number AY129650) (Filali et al., 2002). The PCR protocol used 45 cycles of denaturation at 95°C for 15 sec followed by annealing at 60°C for 45 seconds. Analysis of the Real Time data was performed by calculating relative copies of the target sequence in the immunoprecipitated fraction and dividing this number by the copy number detected in the Input Control. Assays were done with both specific capture antibodies and control IgG for each sample. The above protocol was also used to analyze Tcf4 association at the endogenous Lef-1 promoter in A549 and primary airway epithelial cells.
Immunostaining and Apoptosis Assays
Immunostaining was performed on 10 μm fresh frozen sections. The tissue was then fixed in 4% paraformaldehyde for 15 minutes and stained with primary antibodies for 1 hour followed by incubation with the appropriate secondary antibody for an additional hour. Control reactions were stained with secondary antibodies only. Antibodies used for immunostaining included anti-Lef-1, anti-Tcf4 (Exalpha X1070M), anti-GFP (Abcam ab6673), Phospho-H3 (Cell Signaling 9701), Cyclin D1 (Abcam ab15196), and anti-K14 (Labvision RB-2090). The rabbit polyclonal anti-human Lef-1 was generated to a His-tagged N-terminal fusion Lef-1 protein. A BamHI and SalI fragment containing N-terminal nucleotides 3–718 of human Lef-1 cDNA was subcloned in-frame into the BamHI and XhoI site of the pTrcHisA vector (Invitrogen). The fusion protein containing 238aa of Lef-1 sequence was expressed and purified from bacteria and rabbit antisera were generated. The antibody was subsequently affinity purified by chromatography on immobilized His-Lef-1-N238aa fusion protein. Apoptosis was detected using the TUNEL Assay Detection kit from R&D Systems (TA4626) on 10 μm sections from wild type and Lef-1 deficient mice.
Results
Lef-1 expression in nasal and tracheal submucosal gland buds
Previous studies have demonstrated that Lef-1 mRNA is induced in gland buds of developing tracheal and nasal submucosal glands in mice and ferrets (Driskell et al., 2004; Duan et al., 1998). Additionally, transgenic mice harboring a 2.5 kb human Lef-1 promoter-LacZ reporter express the reporter gene selectively in gland buds of mouse trachea and nasal mucosa (Driskell et al., 2004). These studies underscore the tight regulation of Lef-1 expression during the development of airway glands in a variety of species and implicate transcriptional regulation of the Lef-1 gene in gland morphogenesis. Although Lef-1 mRNA and promoter expression have been demonstrated during gland morphogenesis, confirmation of Lef-1 protein expression has been lacking. This has hindered characterization of the phenotype of Lef-1 expressing cells that give rise to SMGs. Previous studies using bronchial xenografts and retroviral vectors to track lineage fates in developing SMGs have suggested that a subset of basal cells in the airway are the progenitors of this region (Engelhardt et al., 1995). To this end, we sought to characterize Lef-1 protein expression in developing airway glands and to determine whether they expressed the K14 basal cell marker.
A Lef-1 antibody was generated against an N-terminal His-tagged Lef-1 fusion protein and was analyzed for its capability to recognize Lef-1 expression in developing airway submucosal glands and hair follicles (as a control for well-defined patterns of Lef-1 protein staining). Nuclear Lef-1 staining was seen in both the nasal and tracheal epithelial gland buds (Fig. 1A–B, 1E–F), while no background staining was seen in samples treated only with secondary antibody (Fig. 1C, 1G). Additionally, Lef-1 protein staining was also seen in basal cells of the surface airway nasal epithelium, a finding that mirrored mRNA expression patterns observed in nasal mucosal epithelium (Driskell et al., 2004). These findings demonstrate a close correlation between Lef-1 protein expression patterns in developing SMGs and those previously observed for Lef-1 mRNA. To confirm the specificity of our antibody, we evaluated the Lef-1 staining pattern in the well-characterized hair follicle development model using Lef-1 deficient P1 mouse tissue (van Genderen et al., 1994). Wild type hair buds showed typical Lef-1 staining patterns in the basal layer of the epidermis, the hair placode, and the dermal condensate (Fig. 1D). In contrast, Lef-1 deficient hair buds showed no staining in the hair follicle buds or the basal epidermal layer (Fig. 1H). With the hypothesis that glandular progenitors expressing Lef-1 were derived from a subset of basal cells (Engelhardt et al., 1995), we evaluated co-localization of the K14 basal cell marker in Lef-1 expressing glandular buds (Fig. 1I). Results from these studies demonstrated that indeed Lef-1 expressing cells within the glandular bud are K14 positive and support the hypothesis that glandular progenitors are a subset of Lef-1/K14 co-expressing cells.
Fig. 1. Lef-1 protein expression is upregulated in K14 positive mouse epithelial cells that form the developing airway SMG placode.
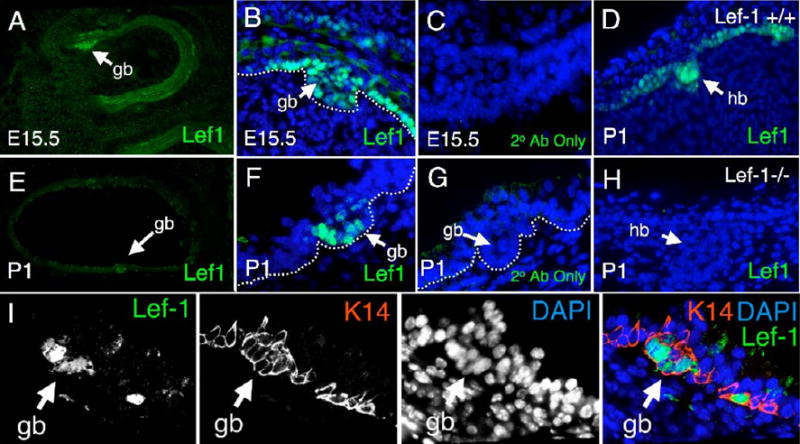
(A–H) Lef-1 expression was evaluated in frozen sections of developing nasal (E15.5) and tracheal (P1) SMG buds (gb) and hair follicles (P1) by immunostaining with a (A, B, E, F, D, and H) Lef-1 primary antibody and FITC-labeled secondary antibody or (C, G) FITC-labeled secondary antibody alone. Panels B–D and F–H are stained with DAPI to label nuclei. Panels depict (A–C) cranial sections of E15.5 day embryos and (E–G) tracheal sections P1 newborn mice. (D and H) The specificity of the Lef-1 antibody was evaluated by comparing staining patterns in wild type and Lef-1 deficient P1 developing hair follicle buds (hb). (I) Coimmunofluorescent staining for K14 and Lef-1 in a tracheal gland bud of newborn mouse trachea (P1). Black and white panels depict the single fluorescent channels as labeled with the merged image on the far right.
Active β-catenin/Tcf complexes are detected in developing airway SMGs
Based on previous in vitro studies demonstrating that Wnt signals can induce transcription from the Lef-1 promoter (Filali et al., 2002; Hovanes et al., 2001; Hovanes et al., 2000), we hypothesized that Wnts may also upregulate Lef-1 expression during airway gland morphogenesis. We evaluated the in vivo status of active β-catenin/Tcf complexes in developing airway SMGs using the TOPGAL reporter mice commonly used to assess involvement of Wnt signaling (DasGupta and Fuchs, 1999). TOPGAL mice were analyzed for β-catenin/Tcf-responsive β-galactosidase expression in their developing nasal and tracheal SMGs. Tracheal gland buds, which begin to form at post-natal days P1-3, demonstrated the highest levels of TOPGAL reporter expression in the trachea with lower basal levels of expression throughout the surface airway epithelium (Fig. 2A and B). Similarly, nasal gland buds, which begin to develop at the E15.5 embryo stage, also demonstrated high levels of TOPGAL expression, but little expression was seen in the surface airway epithelium (Fig. 2D and E). No β-gal staining was seen in either of these two regions of the airway in TOPGAL negative littermate control mice (Fig. 2C and E). These results support the notion of canonical Wnt pathway involvement during airway gland development and are consistent with the hypothesis that β-catenin/Tcf activation regulates the Lef-1 promoter in vivo.
Fig. 2. β-catenin/Tcf pathway is activated in developing airway SMGs of TOPGAL mice.
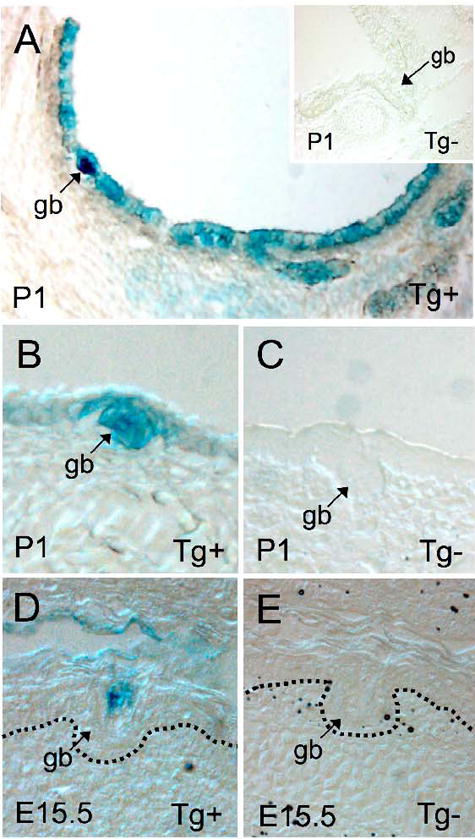
(A–C) Tracheas were dissected from newborn TOPGAL transgene positive (Tg+) and negative (Tg−) littermate mice (P1), stained for β-galactosidase activity, and histologic 10 μm frozen sections were evaluated. Tracheal SMG buds (gb) are shown for (A, B) Tg+ and (C, inset A) Tg− littermates. (D, E) TOPGAL activity in nasal SMG buds (gb) was also analyzed in frozen section of E15.5 embryos for (D) Tg+ and (E) Tg− littermates following whole mount staining for β-galactosidase.
Wnt3a is required for the induction of Lef-1 expression in developing airway SMG placodes
Multiple studies have shown that Lef-1 promoter activity in cell lines is induced by Wnts through the mobilization of nuclear β-catenin (Driskell et al., 2004; Filali et al., 2002; Hovanes et al., 2001; Hovanes et al., 2000; Li et al., 2006). However, conclusive evidence that Wnts upregulate Lef-1 expression in vivo has not been demonstrated. These previous in vitro studies have implicated Wnt signaling in the regulation of the Lef-1 promoter by identifying Tcf sites that are necessary for the induction of Lef-1 transcription using dominant active β-catenin/Tcf and/or Wnt signals. Sequences responsible for Wnt3a responsiveness of the Lef-1 promoter in vitro are also necessary for transcriptional activity of the Lef-1 promoter during airway submucosal gland morphogenesis (Driskell et al., 2004). In addition, it has been shown that overexpression of a dominant active form of Lef-1, when transiently expressed, can rescue the vestigal tail (vt) Wnt3a mutation in mice (Galceran et al., 2001). Hence, we hypothesized that Wnt3a is required for Lef-1 expression during airway SMG morphogenesis and that Wnt3a deficient mice would likely lack airway SMGs, since 12 day old Lef-1 deficient mice lack nasal and tracheal glands (Duan et al., 1999). Wnt3a knockout mice die in utero around E16.5, so only nasal gland development could be studied (Takada et al., 1994). To our surprise, Wnt3a deficient mice contained properly formed nasal SMG placodes. However, as hypothesized Wnt3a deficient mice lacked Lef-1 protein expression in SMG placodes seen in wild type littermates (Fig. 3A and B). These results demonstrated that Lef-1 expression in developing SMGs was indeed under the control of Wnt3a, but also conflicted with earlier reports demonstrating the lack of SMG development in 12 day old Lef-1 deficient mice (Duan et al., 1999). In contrast to SMGs, Wnt3a was not necessary for Lef-1 protein expression in developing vibrissa at E15.5 (Fig. 3C and D).
Fig. 3. Wnt3a regulates Lef-1 gene expression during airway SMG morphogenesis.
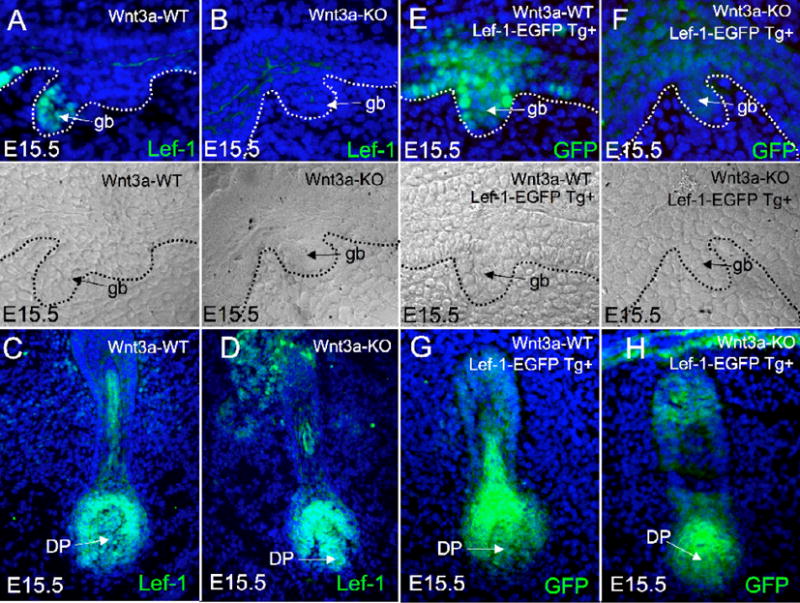
(A–D) Immunofluorescent detection of Lef-1 protein was evaluated in (A, B) SMG buds and (C and D) whisker follicles from E15.5 Wnt3a deficient and wild type littermate mice. 10 μm frozen sections for each genotype were stained with a rabbit polyclonal antibody against the N-terminal 238aa of Lef-1 and a FITC-labeled secondary antibody. (E–H) Transgenic mice harboring a 2.5 kb Lef-1 promoter-EGFP transgene were used to assess whether Wnt3a was required for the transcriptional activation of the Lef-1 promoter in (E, F) SMG buds and (G and H) whisker follicles from E15.5 Wnt3a deficient and wild type littermate mice harboring the transgene reporter. 10 μm frozen sections for each genotype were stained with a goat polyclonal antibody against EGFP and a FITC-labeled secondary antibody. Nomarski images are given in the bottom of panels A, B, E, and F with the basement membrane marked by a dotted line to demarcate the glandular buds (gb). All sections were counter stained with DAPI to label nuclei (blue).
To determine whether Wnt3a acted to control Lef-1 expression at the transcriptional level, we evaluated the expression patterns of a Lef-1 promoter-EGFP reporter cassette in transgenic mice crossed onto the Wnt3a deficient background. This same promoter segment was previously shown to express in airway SMG buds and developing hair follicles at endogenous sites of Lef-1 mRNA (Driskell et al., 2004; Liu et al., 2004). These newly generated Lef-1 promoter-EGFP reporter transgenic mice express a nuclear-targeted EGFP fusion protein. As previously described for the Lef-1 promoter LacZ reporter transgenic mice (Driskell et al., 2004), this Lef-1 promoter expressed nuclear targeted EGFP in the nasal gland placode cells and sporadically in basal cells of the surface airway epithelium of wt mice (Fig. 3E), mirroring the endogenous Lef-1 protein expression patterns (Fig. 1A, B). In the five Wnt3a-KO mice analyzed, sporadic expression of the Lef-1 promoter-EGFP transgene in basal cells of the surface airway epithelium was always observed, suggesting that control of the Lef-1 promoter is not under Wnt3a control. In contrast, Wnt3a deficient mice had significantly reduced expression of the Lef-1 promoter-EGFP cassette in nasal gland placodes (Fig. 3F and Fig. 4). However, this glandular phenotype demonstrated variable penetrance. Quantitative assessment of five paired E15.5 double transgenic Wnt3a-KO and Wnt3a-WT embryos demonstrated that one Wnt3a-KO mice had normal levels of Lef-1 promoter EGFP nuclear staining in SMG buds in comparison to its matched littermate (Fig. 4E, group 4). The remaining four pairs all demonstrated reduction in nuclear EGFP expression in gland buds on the Wnt3a-KO background, with two pairs (group 3 and 5) demonstrating no detectable nuclear expression (Fig. 4E). The reason for this variability is currently unclear, but may be due to the outbred genetic background established by crossing the Wnt3a KO (with a C57BL/6JxBALB/c mixed lineage) and Lef-1 reporter mice (with a C57BL/6JxSJL/J mixed lineage). Genetic variability in modifier genes introduced into these experiment may influence either the penetrance of the Wnt3a phenotype and/or the regulation of the heterologous Lef-1 promoter. Despite the variable penetrance, statistical analysis of the combined data set revealed a significant (p<0.016) decline in the number of glandular bud cells that expressed the Lef-1 promoter in the absence of Wnt3a. These findings strongly suggest that Wnt3a can regulate transcriptional activation of the Lef-1 promoter during gland morphogenesis and support findings in TOPGAL mice demonstrating active β-catenin/Tcf complexes at sites of Wnt3a action. In contrast to findings in SMGs, Lef-1 promoter-GFP reporter expression patterns in both the dermal papillae and the matrix cells of whisker follicles were similar in wild type and Wnt3a deficient littermates (Fig. 3G and H). These findings are consistent with the fact that Lef-1 protein expression in vibrissa was unaltered on the Wnt3a deficient background and suggests that Wnt3a does not play a critical role in activating Lef-1 expression during this stage of whisker development.
Fig 4. Wnt3a influences Lef-1 promoter activation during airway SMG morphogenesis.
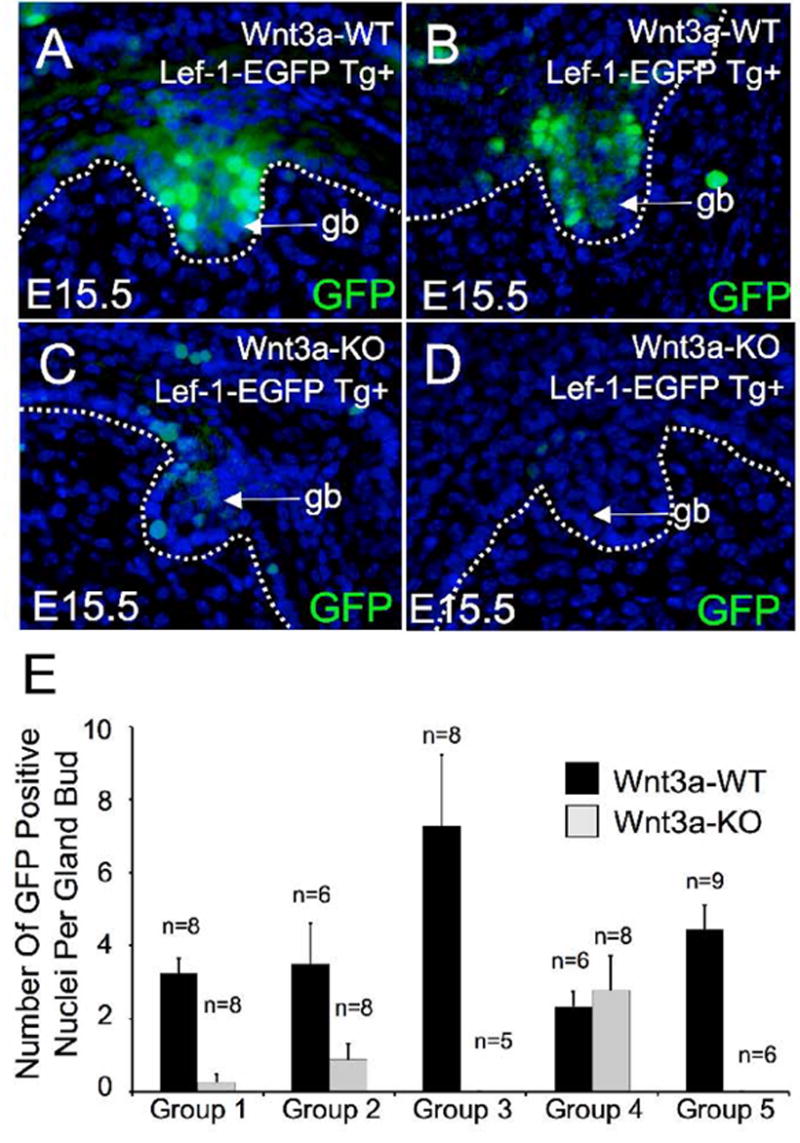
(A–D) Immunofluorescent detection of nuclear-targeted EGFP under the control of a 2.5 kb Lef-1 promoter was evaluated in SMG buds from Wnt3a deficient and wild type littermate mice. 10 μm frozen sections for each genotype were stained with a goat polyclonal antibody against EGFP and a FITC-labeled secondary antibody. All sections were counter stained with DAPI to label nuclei (blue). (A, C) and (B, D) are two examples of studies evaluating matched littermates with (A, B) Wnt3a-WT and (C, D) Wnt3a-KO genotypes. (E) Morphometric quantification of nuclear EGFP expression in nasal SMG buds from five independent litters. The graph depicts the mean (+/−SEM) number of nuclei that expressed detectable levels of nuclear EGFP in SMG placodes for a given genotype and experiment group. The number of placodes (n) evaluated for each animal is given above the mean value. Each group consisted of Wnt3a-WT and Wnt3a-KO littermate embryos that were immunostained at on the same day. All photomicrogaphs were captured at identical time exposures and quantification was performed in the combined data set at one time representing a total of 37 Wnt3a-Wt and 35 Wnt3a-KO gland buds. Panels A and C were taken from group 2 and panels B and D were taken from group 3. Statistical differences between Wnt3a-Wt and Wnt3a-KO mice were significant as assessed by ANOVA followed by the Mann-Whitney test (p<0.016).
Lef-1 expression is required for maturation of SMGs but not initiation of glandular placode formation
It has been previously reported that 12 day post-natal Lef-1 knockout mice do not have nasal or tracheal submucosal glands (Duan et al., 1999). These previous results appeared inconsistent with our present findings that nasal gland placodes formed normally in Wnt3a deficient mice despite the lack of Lef-1 protein expression. Lef-1 is a necessary transcription factor required for the proper formation of many epithelial appendages (van Genderen et al., 1994). For example, hair follicles, teeth, and mammary glands do not form properly in Lef-1 knockout mice. Hair follicle morphogenesis occurs, but continued development of de novo follicle formation after E18.5 is inhibited (van Genderen et al., 1994). In addition, mammary glands do not develop past the initial epithelial bud stages in Lef-1 deficient mice (van Genderen et al., 1994). Given these previous findings in other model epithelial appendages, we hypothesized that Lef-1 may be required for the continued maturation of SMGs, but not the initial stages of placode formation. To address this hypothesis, we reevaluated maturation of nasal and tracheal glands at earlier stages than previously investigated. Consistent with this hypothesis, newborn (P1) Lef-1 deficient mice did demonstrate limited nasal submucosal gland development compared to wild type controls (Fig. 5A–B, 5E–F). Nasal glands in Lef-1 deficient mice were clearly abnormal and failed to fully develop, reminiscent of finding in hair follicles of Lef-1 deficient mice (van Genderen et al., 1994). In contrast, tracheal gland buds were not detected (Fig. 5C–D, 5G–H), confirming previous findings (Duan et al., 1999). These results are consistent with the presence of nasal glandular placodes in Wnt3a deficient mice lacking glandular Lef-1 expression and suggest that Lef-1 is required for the maintenance of SMG growth but not initiation.
Fig. 5. Lef-1 expression is not required for nasal SMG placode formation but is required for maturation of glands.
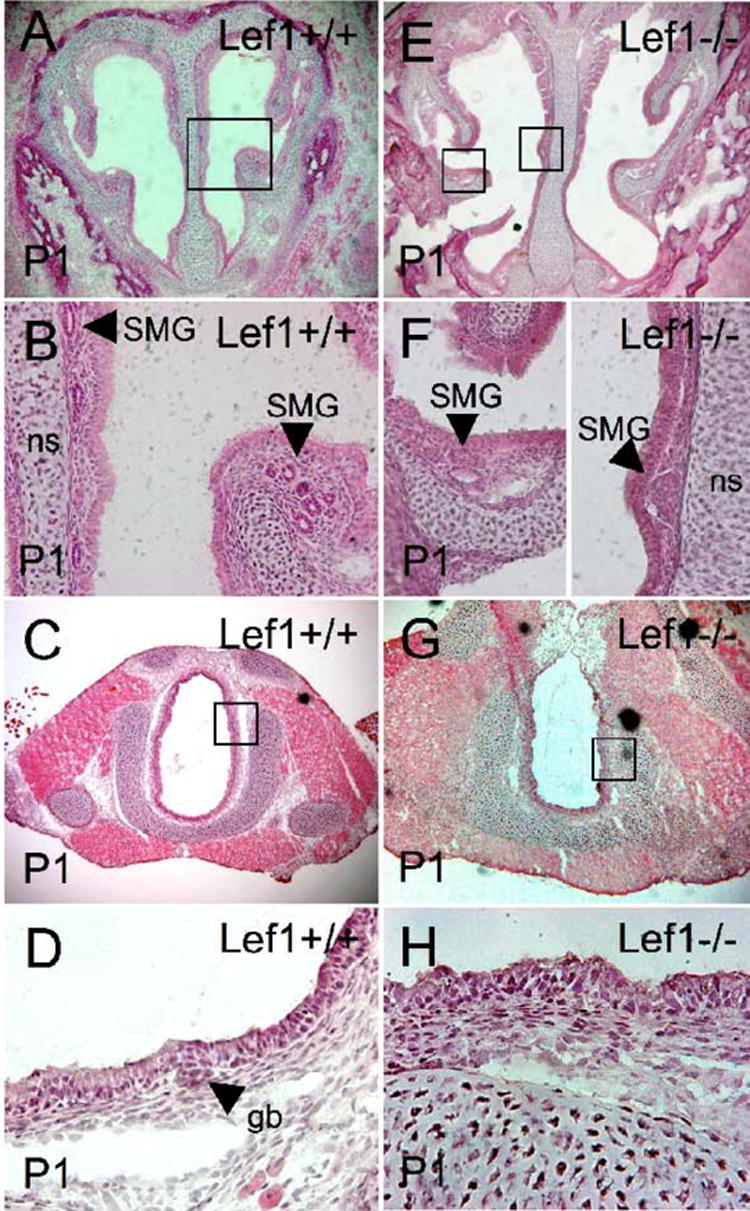
The presence of nasal and tracheal glands were evaluated in P1 (A–D) wild type and (E–H) Lef-1 deficient mice by evaluating 10 μm H & E stained frozen sections. Nasal SMGs with normal morphology were seen in the submucosa of (A, B) wild type animals, while SMGs in the mucosa of (E, F) Lef-1 deficient mice failed to undergo normal tubulogenesis. Boxed regions in A and E are enlarged in panels B and F, respectively. Tracheal SMG buds were found in the proximal trachea of (C, D) wild type animals, while no SMG buds were found in (G, H) Lef-1 deficient mice. Boxed regions in C and G are enlarged in panels D and H, respectively. SMG, submucosal glands; gb, gland bud; ns, nasal septum.
Lef-1 expression has been shown to regulate cellular replication and differentiation during the morphogenesis of epithelial appendages by influencing the expression of genes associated with cellular proliferation and migration (Fuchs et al., 2001; Waterman, 2004; Zhou et al., 1995). We hypothesized that Lef-1 might also play a role in cellular proliferation during SMG growth and development. Our analysis of phosphorylated Histone H3 (PO4H3) expression in nasal SMGs, as a marker for S-phase of the cell cycle, revealed a marked decrease in cellular replication during the initial stages (E15.5) of nasal SMG morphogenesis in Lef-1 knockout mice as compared to control wild type littermates (Fig. 6A and B). Further analysis at later stage of gland development (E17.5 and P1) revealed similar findings of decreased glandular epithelial cell replication in Lef-1 deficient mice (Fig. 6C–F). Interestingly, mesenchymally derived cells surrounding glandular tubules also demonstrated decreased PO4H3 expression on the Lef-1 knockout background, which was most significant at E17.5 and P1 time points (Fig. 6C–F). These findings suggest that Lef-1 is required for normal cell cycle progression of glandular epithelial cells. Consistent with these findings, a loss of Cyclin D1 expression was also observed in Lef-1 deficient mice at later stages (P1) of SMG development as compared to wild type controls (Fig. 6H and J). Interestingly, Cyclin D1 was similarly detected in E15.5 SMG buds of both Lef-1 knockout wild type mice (Fig. 6G and I). These finding cumulatively suggests that later stages of glandular proliferation are most significantly affected by Lef-1 expression. Such findings are consistent with the fact that both Lef-1 and Wnt3a are not required for the initiation of gland bud formation.
Fig. 6. SMG buds and tubules fail to develop in the absence of Lef-1 due to impaired cellular proliferation.
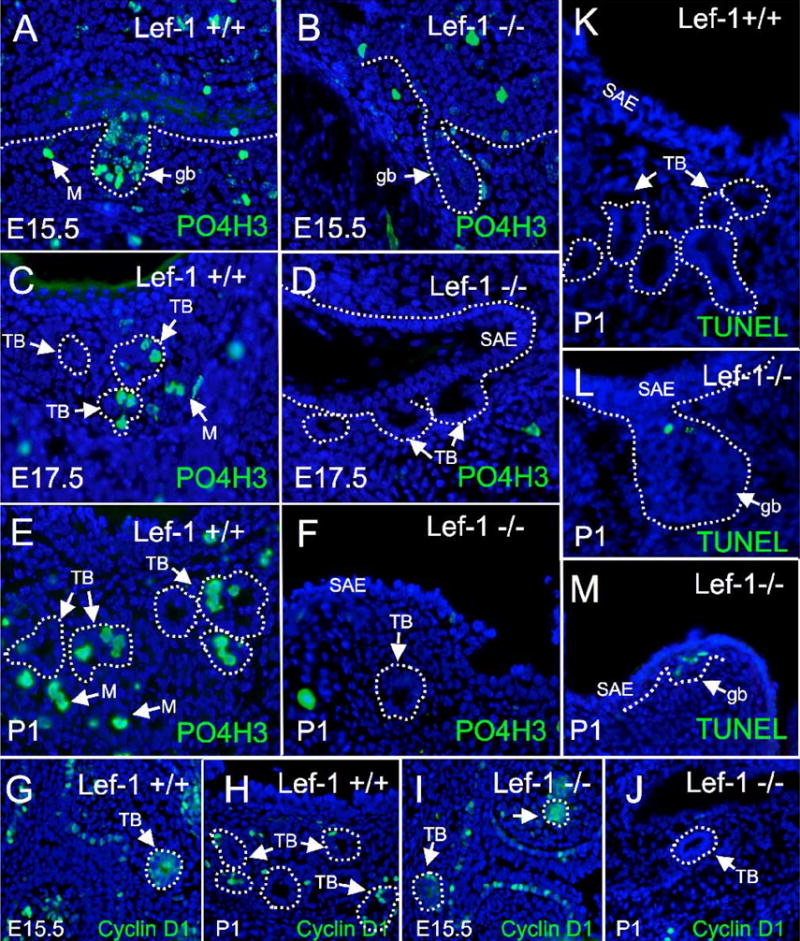
To investigate potential mechanisms responsible for improper SMG maturation in Lef-1 deficient mice, we evaluated expression of two markers for cellular proliferation and the extent of apoptosis in Lef-1 deficient and wild type littermate nasal SMGs at E15.5, E17.5, and P1 developmental time points. Immunostaining for phosphorylated histone H3 (PO4H3) and Cyclin D1 revealed difference in the extent of cellular replication in developing nasal SMG buds (gb), tubules (TB), and mesenchymal cells (M) between Lef-1 deficient and wild type mice. (A–J) Indirect immunofluorescent staining with FITC-labeled secondary antibodies for (A–F) PO4H3 or (G–J) Cyclin D1 in wild type (+/+) and Lef-1 deficient (−/−) littermate nasal tissue sections for the indicated developmental time points and genotypes. (K–M) TUNEL assays were also performed on P1 nasal SMGs of (K) wild type and (L, M) Lef-1 deficient littermates. No TUNEL labeling was observed in wild type SMGs, however, TUNEL positive nuclei (green) were observed in Lef-1 deficient glandular bud epithelial cells (gb) and also in cells below the basement membrane no longer associated with visible buds in regions where glandular structures normally form (data not shown). Merged FITC images with DAPI nuclear counter stain are given in all panels. The basal lamina of epithelia in the surface airway (SAE), glandular buds, and glandular tubules is marked by a dashed white line when appropriate.
Similar to SMGs, abnormal growth of mammary glands in Lef-1 deficient mice has also been associated with a failure of mammary epithelial placodes to progress past the initial stages of development (van Genderen et al., 1994). Recent studies have implicated reciprocal Wnt signals between the epithelia of the placode and surrounding mesenchyme that are defected in the absence of Lef-1 (Boras-Granic et al., 2006). In these studies the progressive loss of mammary placodes in Lef-1 deficient mice was associated with increased apoptosis of mesenchymal cells surrounding the epithelial placodes leading to abortive degeneration of the mammary epithelial placode. In light of these recent results, we hypothesized that loss of cellular proliferation in forming gland tubules caused by Lef-1 deficiency may lead to reabsorption of SMG through a similar apoptosis mechanism. To investigate this hypothesis, TUNEL assays were performed on E15.5, E17.5 and P1 nasal glands. Results from this analysis demonstrated TUNEL positive cells in P1 nasal glands of Lef-1 deficient mice that were never seen in wild type littermates (Fig. 6K–M). Interestingly, no TUNEL labeling was observed in E15.5 or E17.5 nasal buds of Lef-1 deficient mice (data not shown). These findings are consistent with apoptotic cell death as a mechanism for SMG clearance when Lef-1 is absent and cellular proliferation is abrogated.
Tcf4 mediates Wnt activation of the Lef-1 promoter in airway epithelia
Tcfs have been shown to bind the Lef-1 promoter in vitro (Hovanes et al., 2001) and in vivo (Li et al., 2006) through association at the WRR. Tcf factors can bind to similar DNA sequences and regulate context-specific activation and/or repression of diverse promoters (Waterman, 2004). An example of these opposing effects is seen in the contrasting phenotypes found in Tcf1 or Tcf4 null mice. Tcf4 null mice lack intestinal crypt progenitor cells due to a loss of proliferation in the crypt stem cell compartment, indicating that the expression of Tcf4 can play a important in vivo role in cellular proliferation (Korinek et al., 1998; Roose et al., 1999). In contrast, Tcf1 null mice exhibited increased susceptibility to neoplasms in their intestines and mammary glands, suggesting an inhibitory role in proliferation (Roose et al., 1999; Verbeek et al., 1995). Consequently, it is important to define the Tcf factor(s) that bind to the Lef-1 promoter WRR to fully understand Wnt involvement in SMG morphogenesis. To better define the transcription factors involved in Wnt3a activation of the Lef-1 promoter within airway epithelial cells, we sought to identify candidate Tcf factors present in airway epithelia and test whether these factors associate with the Lef-1 promoter WRR, which contains known Tcf bindings sites as well as the WRE required for Wnt3a activation in vitro. As a first assessment, Western blots were performed to detect levels of Tcf1, 3, 4 and Lef-1 in whole cell lysates from trachea and skin. In addition to Lef-1, Tcf4 was detected in newborn (P1) skin and tracheal lysates, but Tcf1 and 3 were absent (Fig. 7A). Immunostaining of P1 newborn tracheas confirmed that indeed Tcf4 expression localized to the surface airway epithelium (SAE) as well as the airway SMG placodes (Fig. 7D), while Tcf1, and Tcf3 were not expressed (Fig. 7B and C). Immunostaining of E15.5 embryos also demonstrated Tcf4 expression in nasal gland buds (Fig. 7F), while control experiments omitting primary antibody gave no detectable staining (Fig. 7E). These results identify Tcf4 as a potential candidate for Wnt3a mediated activation of the Lef-1 promoter during airway gland development. Tcf4 expression in SMG buds and SAE was unaffected by deletion of Lef-1 or Wnt3a (Fig. 7G and H). Given that Lef-1 is required for normal glandular development and its expression is dependent on Wnt3a, these findings indicating that Tcf4 cannot compensate for Lef-1 specific function during the morphogenesis of SMGs.
Fig. 7. Tcf4 is highly expressed in developing airway epithelium and SMGs and is unaltered by Wnt3a or Lef-1 deficiency.
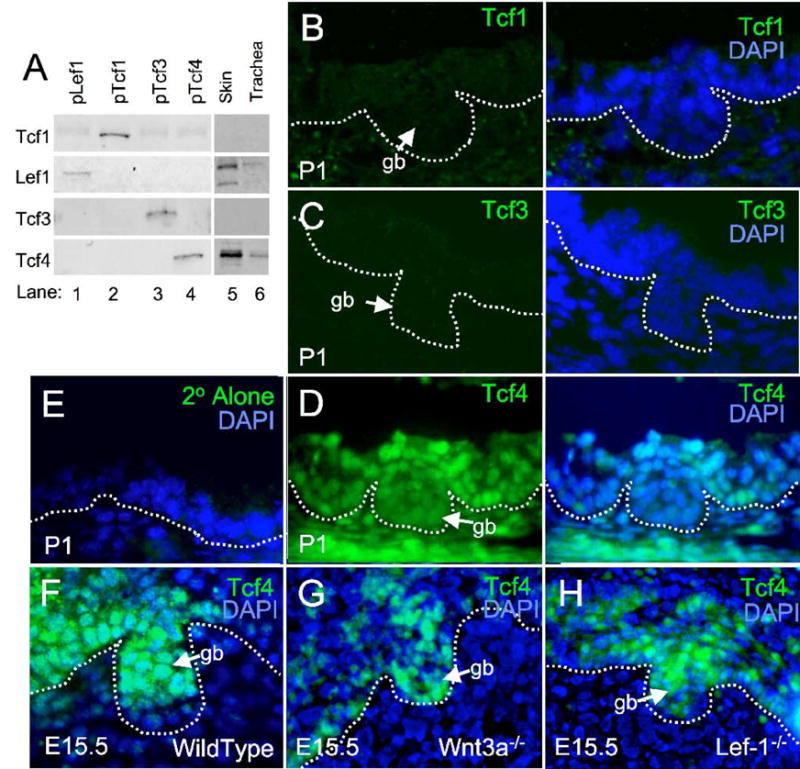
To investigate candidate Tcfs that might be responsible for Wnt-mediated activation of the Lef-1 gene during glandular development, we evaluated the expression patterns of Tcf1, 3, and 4 in developing trachea and nasal epithelium. (A) Western blot analysis for Lef-1, Tcf1, 3 and 4 was evaluated using 75 μg of protein lysate from 293 cells transiently transfected with the expression constructs indicated above each lane (lane 1–4) and also in tissue harvested from skin and trachea of newborn mice (P1) (lane 5 and 6). (B–D) Immunostaining for the indicated Tcf proteins was performed on P1 tracheal gland buds using primary Tcf antibodies as indicated in the methods and FITC-labeled secondary antibody (left panel). Merged FITC images with DAPI counter stain are given to the right of each panel. (E) P1 tracheal epithelium was stained with only the FITC secondary antibody used for Tcf4 detection. (F–H) Nasal SMGs from wild Type, Wnt3a deficient, and Lef-1 deficient E15.5 embryos were immunostained for Tcf4 expression and demonstrated expression in the nasal epithelium and nasal SMG bud (gb). The basal lamina of the surface airway epithelium and glandular bud epithelium is marked by a dashed white line.
Given that Tcf4 is expressed in developing airway SMGs, and Tcf4 in conjunction with nuclear β-catenin has been shown to activate Lef-1 promoter activity, we hypothesized that Tcf4 associates with the WRR in the Lef-1 promoter. Indeed, recently studies have demonstrated that Tcf factors bind to the WRR in a cellular context-specific fashion using Chromatin Immunoprecipitation (ChIP) (Li et al., 2006). However, the specific Tcf factors that associated with this region of the Lef-1 promoter were not elucidated. Furthermore, the specific 2.5 kb promoter fragment used in our studies has not been previously evaluated for Tcf responsiveness. (Driskell et al., 2004; Filali et al., 2002; Hovanes et al., 2001)To confirm that previous findings of TCF/β-catenin-dependent Lef-1 promoter transcription (Hovanes et al., 2001) also applied to the larger 2.5 kb Lef-1 promoter studied in our transgenic models, we investigated the ability of Tcf4 and a dominant active mutant of β-catenin (S37) to induce the Lef-1 promoter in transient transfection assays in Cos1 cells (the same cell line previously used to study Tcf/β-catenin-dependent Lef-1 promoter transcription). Indeed, Tcf4 and β-catenin-S37 synergistically induced transcription from the 2.5 kb Lef-1 promoter (Fig. 8B), consistent with previous observation with a shorter promoter (Hovanes et al., 2001).
Fig. 8. Tcf4 mediates Wnt signaling at the WRR in the Lef-1 promoter.
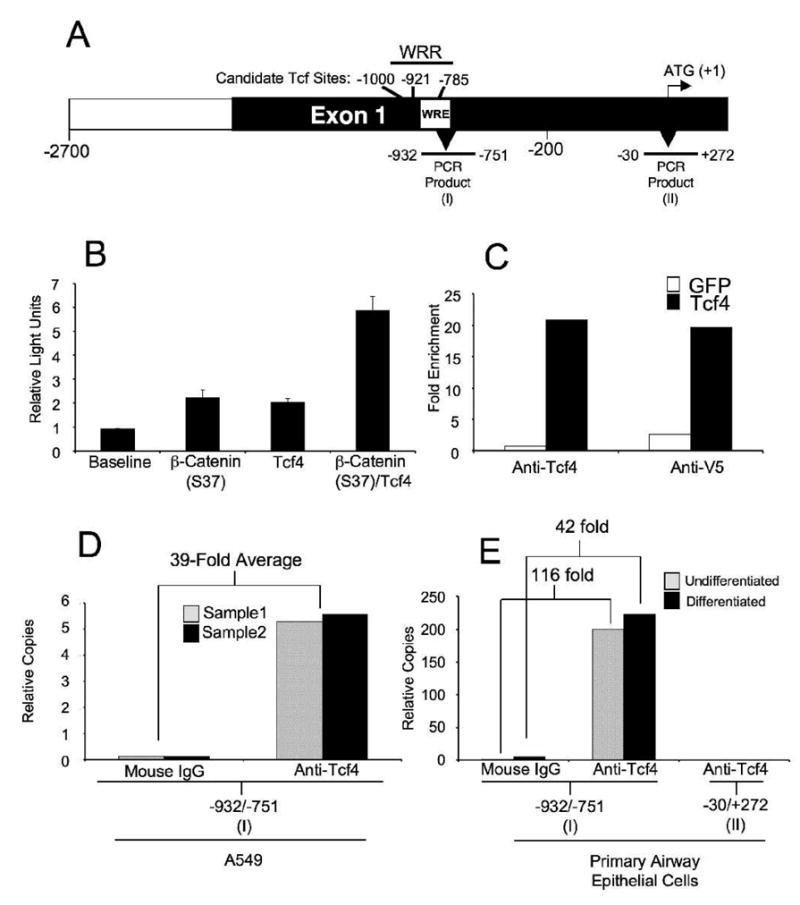
(A) A schematic of the Lef-1 promoter (not drawn to scale) with candidate Tcf binding sites [−1000 bp (TCTTTGCT), −921 bp (AACAAAGA), and −785 bp (ACTTTATT)] and WRE illustrated. Regions of the Lef-1 promoter evaluated in ChIP assays by real time PCR are marked below the promoter diagram with bp locations of the end primers. PCR product (I) amplifies the WRR region and PCR product (II) amplifies a region downstream of Tcf binding sites. (B) To determine if Tcf4/beta-catenin can induce the transcriptional activity of the 2.5 kb Lef-1, Cos1 cells were cotransfected with the LF-2700/-200-Luciferase plasmid and dominant active β-catenin (S37) and/or Tcf4 expression plasmids. Whole cell lysates were collected 24 hours post transfection and analyzed for luciferase activity. Results depict the mean +/−SEM for N=3 transfections. (C) To determine if Tcf4 associates with the WRR of the LF-2700/-200 Lef-1 promoter, Cos1 cells were cotransfected with the LF-2700/-200-Luciferase plasmid together with either a Tcf4-V5-His-tagged or GFP expression plasmid. Nuclear cell lysates were collected 24hr after transfection and ChIP assays were performed using either anti-Tcf4 or anti-V5 antibodies. Mouse IgG was used as a negative control for immunoprecipitation in the ChIP assay. PCR product (I) primers were used for Real Time PCR analysis of immunoprecipitates. The results are presented as fold enrichment of relative PCR product (I) copies detected in anti-Tcf4 or anti-V5 precipitates over the relative PCR product (I) copies detected in Mouse IgG precipitates. (D) Tcf4 association at the WRR of the endogenous Lef-1 promoter was analyzed in A549 cells by performing ChIP assays on A549 nuclear cell lysates with either anti-Tcf4 antibody or Mouse IgG control. Real Time PCR analysis was performed using probes for PCR product (I) and is presented as relative copies. (E) Tcf4 association at the WRR in the Lef-1 promoter in undifferentiated and differentiated primary airway epithelial was assessed by ChIP. Real Time PCR analysis was preformed using probes to PCR product (I) and (II) and using the indicated antibodies for precipitation.
Although purified Tcf proteins have been shown to bind and associate with the Lef-1 promoter at the WRR, identification of specific Tcf factors that associate in vivo has not been investigated. Using ChIP, we sought to demonstrate that Tcf4 associates with the WRR of the Lef-1 promoter in vivo. Because of the low resolution of ChIP assays caused by random DNA shearing, and the close proximity of known Tcf binding sites with the WRE, these studies can not distinguish between binding at these two regions and can only determine association of the overall WRR. ChIP was performed in Cos1 cells transiently transfected with a Lef-1 promoter/reporter plasmid and Tcf4 expression plasmids that expressed either V5-tagged Tcf4. Using capture antibodies against either Tcf4 or V5, results demonstrated a ~19–20 fold enrichment of Tcf4 binding to the WRR of the Lef-1 promoter in cells overexpressing either of the Tcf4 constructs in comparison to cells transfected with a GFP construct (Fig. 8C). These findings demonstrate a correlation between Tcf4/β-catenin transcriptional induction of the 2.5 kb Lef-1 promoter and Tcf4 binding to the promoter’s WRR. Furthermore, given that Tcf4 levels are extremely low in Cos1 cells (Hovanes et al., 2001), these findings demonstrate the specificity of our Tcf4 antibody directed ChIP assay. Using this validated ChIP assay, we then sought to determine if endogenous Tcf4 in airway epithelial cells also associated with the endogenous Lef-1 promoter. Indeed results from these studies demonstrated significant enrichment of Tcf4 binding to the endogenous Lef-1 promoter WRR in both A549 transformed airway cells and primary airway epithelial cells as compared to controls omitting the primary Tcf4 antibody in the ChIP assay (Fig. 8D and E). In contrast, no detectable Tcf4 association was observed on a segment of the Lef-1 gene ~700 bp 3′ to the WRR (Fig. 8E) These results confirm that Tcf4 does bind to the Lef-1 promoter in airway epithelial cells in vivo within the vicinity of the WRR. Since this WRE is known to be required for Lef-1 promoter expression during SMG morphogenesis (Driskell et al., 2004), these findings support the notion that Tcf4 is involved in regulating Lef-1 gene expression in airway cells during SMG development.
Discussion
Lef-1 plays a dynamic role in many developmental processes, of which the best studied are those involved in regulating epithelial skin appendages including hair follicles and mammary glands. Pathways that control Lef-1 activation have been best studied at the level of post-transcriptional regulation. In this context, Wnt signals play a major role in mobilizing the co-activator β-catenin to the nucleus to facilitate Lef-1 activation as a transcription factor. A second less characterized pathway for Lef-1 activation during organogenesis in vivo includes transcriptional activation of the Lef-1 promoter (Widelitz, 2004). In this context, transgenic mice harboring Lef-1 promoter reporters have demonstrated that transcriptional regulation of the Lef-1 gene can be highly regulated during hair follicle, mammary gland, and SMG development (Driskell et al., 2004; Liu et al., 2004). Additionally, Lef-1 transcriptional regulation appears to play an important role in the development of colon cancers (Hovanes et al., 2001). These studies evaluating Lef-1 promoter activation in transgenic mice and cancers have implicated Wnt/Tcf/β-catenin signaling pathways as important in the regulation of Lef-1 gene transcription during development. However, to date, there has been no direct evidence demonstrating Wnts can indeed control Lef-1 gene transcription in vivo in the context of a developmental process. In the current study, we provide this evidence by demonstrating that Lef-1 gene expression is regulated by Wnt3a in the context of SMG development. Such information may also lead to an enhanced understanding of other developmental systems involving Wnt signaling and the regulation of Lef-1 dependent processes.
Although Lef-1 is required for skin appendage and airway SMG development, notable differences can be found between the types of cells that express Lef-1 during the morphogenesis of these structures. In hair follicles and mammary glands, Lef-1 is expressed in a dynamic fashion in both the epithelial placode and surrounding mesenchymal cells leading to reciprocal interactions that control organ development. In contrast, Lef-1 expression during SMG development is limited to epithelial cells of the placode and developing glandular tubules, suggesting that fundamental differences exist in the developmental biology of SMGs and skin appendages. Despite these differences, it appears that Lef-1 may have a similar functional role in both hair follicle and SMG morphogenesis, since like hair follicle (Fuchs et al., 2001; Millar et al., 1999), SMG formation is also inhibited due to the repressed ability of cells in the forming appendage to proliferate.
Our findings assessing SMG development in Wnt3a deficient mice implicate Wnt signaling in the regulation of Lef-1 gene transcription. Although the loss of Wnt3a did not lead to the inhibition of placode formation as we originally hypothesized, the loss of Lef-1 expression led to the eventual regression of SMGs through the lack of cellular proliferation. A multitude of Wnts are expressed during the formation of epithelial appendages and the compensatory nature of these proteins are thought to contribute to the lack of epithelial appendage phenotypes during embryogenesis when any one Wnt gene is deleted. For example, the hair follicle expresses at least 3 Wnts during placode formation and as many as 6 different Wnts during maturation and cycling, including Wnt3a (Reddy et al., 2001). Consequently, deletion of a single Wnt gene often does not result in a grossly abnormal epithelial appendage phenotype. These findings may also help to explain why variable penentrance in Wnt3a regulation of the Lef-1 promoter was seen on a mixed outbred genetic background.
It has been previously reported that 12 day old Lef-1 knockout mice lack airway SMGs in the nasal mucosa and trachea (Duan et al., 1999). In the present study, we have demonstrated that nasal gland buds do indeed form in Lef-1 deficient mice, however they fail to undergo normal tubulogenesis since cellular proliferation is abrogated by the absence of Lef-1. This observation mirrors (Sasaki et al., 2005)the loss of proliferative markers during hair follicle development of Lef-1 knockout mice (Millar et al., 1999; Zhou et al., 1995). The observation of SMG apoptosis in Lef-1 deficient mice, which is most likely a downstream consequence caused by a lack cellular proliferation, is shared with several other development pathways. For example, the progressive loss of mammary placodes in Lef-1 knockout mice leads to increased apoptosis of mesenchymal cells surrounding the epithelial placodes (Boras-Granic et al., 2006). Similarly, during tooth development in Lef-1 knockout mice at E16.5 is accompanied by apoptosis in the dental epithelium (Sasaki et al., 2005). In B cells, Lef-1 functions to also promote cell survival, with increased apoptosis observed in Lef-1 deficient mice (Reya et al., 2000). (Boras-Granic et al., 2006)However, these observations differ from what is seen in hair follicles, where no additional apoptosis was seen in Lef-1 knockout mice compared to wild type controls (data not shown). These findings suggest that Lef-1 plays organ specific roles in the maintenance of morphogenesis of diverse developmental programs.
Wnt activation of Lef-1 promoter reporter constructs in vitro is dependent on Tcf/β-catenin signaling and Wnt responsive sequences in the promoter that contain Tcf binding sites (Filali et al., 2002; Hovanes et al., 2001; Hovanes et al., 2000; Li et al., 2006). Our studies also demonstrate that Tcf4/β-catenin complexes can activate Lef-1 transcriptional activity and that Tcf4 also associates with regions in the Lef-1 promoter that have been assigned to Wnt signaling. The WRE in the Lef-1 promoter, which may work in conjunction with Tcf binding sites, is required for transcriptional activation of a heterologous Lef-1 promoter in SMGs (Driskell et al., 2004; Liu et al., 2004). Hence, our studies demonstrate for the first time Tcf4 association with the endogenous Lef-1 promoter at sites responsible for transcriptional activation of the promoter in nasal and tracheal SMGs and Wnt3a activation of the promoter in vitro (Filali et al., 2002). In vivo supportive evidence for the importance of Wnt3a in activating the Lef-1 promoter in epithelial cells of the SMG placode is three-fold. First, endogenous Lef-1 protein fails to be induced in SMG placodes in Wnt3a deficient mice. Second, the TOPGAL Tcf/β-catenin reporter is induced at highest levels in SMG placodes of mice. Finally, Wnt3a influenced transcriptional activation of the Lef-1 promoter in epithelial cells of the SMG placode. Tcf4 appears to be the likely candidate for Wnt3a-mediated activation of the Lef-1 gene during SMG morphogenesis since it is expressed at high levels in gland buds and binds to the Lef-1 promoter in the vicinity of the WRE. Given that Tcf4 is ubiquitously expressed in the airway epithelial cells and is unaltered on the Wnt3a deficient background (Fig. 7G), it is likely that Wnt3a regulates Lef-1 transcription through the local activation of β-catenin in the forming SMG placode. This hypothesis is supported by the fact that co-expression of dominant active β-catenin and Tcf4 synergistically enhance transcription from the Lef-1 promoter. Given that Tcf4 expression is also unaltered on the Lef-1 deficient background (Fig. 7H), it also appears that Tcf4 expression cannot compensate for Lef-1 during SMG morphogenesis. However, further investigation is needed to determine if Tcf4 and β-catenin are absolutely required for Wnt3a induction of the Lef-1 promoter in vivo during airway SMG development.
In conclusion, these studies describe the first report of Wnt-mediated transcriptional activation of the Lef-1 promoter in vivo. Such findings provide further insights into the diverse mechanisms by which Lef-1 regulation controls developmental processes. Further elucidation of these mechanisms may uncover additional regulatory systems that control Lef-1 transcription in response to diverse Wnt signals in both development and cancer.
Acknowledgments
This work was supported by NIH RO1 DK47967 (J.F.E.), the University of Iowa Gene Therapy Center Animal Models Core (DK54759), and the Roy J. Carver Chair in Molecular Medicine (J.F.E.). We also greatly appreciate the technical assistance of Norma Sinclair, Trish Yarolem, and JoAnne Schwarting in the Transgenic Core Facility at University of Iowa. We also thank Dr. James Wells for providing Tcf4 expression constructs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andl T, Reddy ST, Gaddapara T, Millar SE. WNT signals are required for the initiation of hair follicle development. Dev Cell. 2002;2:643–53. doi: 10.1016/s1534-5807(02)00167-3. [DOI] [PubMed] [Google Scholar]
- Boras-Granic K, Chang H, Grosschedl R, Hamel PA. Lef1 is required for the transition of Wnt signaling from mesenchymal to epithelial cells in the mouse embryonic mammary gland. Dev Biol. 2006 doi: 10.1016/j.ydbio.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Chu EY, Hens J, Andl T, Kairo A, Yamaguchi TP, Brisken C, Glick A, Wysolmerski JJ, Millar SE. Canonical WNT signaling promotes mammary placode development and is essential for initiation of mammary gland morphogenesis. Development. 2004;131:4819–29. doi: 10.1242/dev.01347. [DOI] [PubMed] [Google Scholar]
- DasGupta R, Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126:4557–68. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- Driskell RR, Liu X, Luo M, Filali M, Zhou W, Abbott D, Cheng N, Moothart C, Sigmund CD, Engelhardt JF. Wnt-responsive element controls Lef-1 promoter expression during submucosal gland morphogenesis. Am J Physiol Lung Cell Mol Physiol. 2004;287:L752–63. doi: 10.1152/ajplung.00026.2004. [DOI] [PubMed] [Google Scholar]
- Duan D, Sehgal A, Yao J, Engelhardt JF. Lef1 transcription factor expression defines airway progenitor cell targets for in utero gene therapy of submucosal gland in cystic fibrosis. Am J Respir Cell Mol Biol. 1998;18:750–8. doi: 10.1165/ajrcmb.18.6.2987. [DOI] [PubMed] [Google Scholar]
- Duan D, Yue Y, Zhou W, Labed B, Ritchie TC, Grosschedl R, Engelhardt JF. Submucosal gland development in the airway is controlled by lymphoid enhancer binding factor 1 (LEF1) Development. 1999;126:4441–53. doi: 10.1242/dev.126.20.4441. [DOI] [PubMed] [Google Scholar]
- Engelhardt JF, Schlossberg H, Yankaskas JR, Dudus L. Progenitor cells of the adult human airway involved in submucosal gland development. Development. 1995;121:2031–46. doi: 10.1242/dev.121.7.2031. [DOI] [PubMed] [Google Scholar]
- Filali M, Cheng N, Abbott D, Leontiev V, Engelhardt JF. Wnt-3A/beta-catenin signaling induces transcription from the LEF-1 promoter. J Biol Chem. 2002;277:33398–410. doi: 10.1074/jbc.M107977200. [DOI] [PubMed] [Google Scholar]
- Fuchs E, Merrill BJ, Jamora C, DasGupta R. At the roots of a never-ending cycle. Dev Cell. 2001;1:13–25. doi: 10.1016/s1534-5807(01)00022-3. [DOI] [PubMed] [Google Scholar]
- Galceran J, Hsu SC, Grosschedl R. Rescue of a Wnt mutation by an activated form of LEF-1: regulation of maintenance but not initiation of Brachyury expression. Proc Natl Acad Sci U S A. 2001;98:8668–73. doi: 10.1073/pnas.151258098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovanes K, Li TW, Munguia JE, Truong T, Milovanovic T, Lawrence Marsh J, Holcombe RF, Waterman ML. Beta-catenin-sensitive isoforms of lymphoid enhancer factor-1 are selectively expressed in colon cancer. Nat Genet. 2001;28:53–7. doi: 10.1038/ng0501-53. [DOI] [PubMed] [Google Scholar]
- Hovanes K, Li TW, Waterman ML. The human LEF-1 gene contains a promoter preferentially active in lymphocytes and encodes multiple isoforms derived from alternative splicing. Nucleic Acids Res. 2000;28:1994–2003. doi: 10.1093/nar/28.9.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamora C, DasGupta R, Kocieniewski P, Fuchs E. Links between signal transduction, transcription and adhesion in epithelial bud development. Nature. 2003;422:317–22. doi: 10.1038/nature01458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19:379–83. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- Li TW, Ting JH, Yokoyama NN, Bernstein A, van de Wetering M, Waterman ML. Wnt activation and alternative promoter repression of LEF1 in colon cancer. Mol Cell Biol. 2006;26:5284–99. doi: 10.1128/MCB.00105-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Driskell RR, Luo M, Abbott D, Filali M, Cheng N, Sigmund CD, Engelhardt JF. Characterization of Lef-1 promoter segments that facilitate inductive developmental expression in skin. J Invest Dermatol. 2004;123:264–74. doi: 10.1111/j.0022-202X.2004.23201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- Millar SE, Willert K, Salinas PC, Roelink H, Nusse R, Sussman DJ, Barsh GS. WNT signaling in the control of hair growth and structure. Dev Biol. 1999;207:133–49. doi: 10.1006/dbio.1998.9140. [DOI] [PubMed] [Google Scholar]
- Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–7. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse R. Wnt signaling in disease and in development. Cell Res. 2005;15:28–32. doi: 10.1038/sj.cr.7290260. [DOI] [PubMed] [Google Scholar]
- Reddy S, Andl T, Bagasra A, Lu MM, Epstein DJ, Morrisey EE, Millar SE. Characterization of Wnt gene expression in developing and postnatal hair follicles and identification of Wnt5a as a target of Sonic hedgehog in hair follicle morphogenesis. Mech Dev. 2001;107:69–82. doi: 10.1016/s0925-4773(01)00452-x. [DOI] [PubMed] [Google Scholar]
- Reya T, O'Riordan M, Okamura R, Devaney E, Willert K, Nusse R, Grosschedl R. Wnt signaling regulates B lymphocyte proliferation through a LEF-1 dependent mechanism. Immunity. 2000;13:15–24. doi: 10.1016/s1074-7613(00)00004-2. [DOI] [PubMed] [Google Scholar]
- Roose J, Huls G, van Beest M, Moerer P, van der Horn K, Goldschmeding R, Logtenberg T, Clevers H. Synergy between tumor suppressor APC and the beta-catenin-Tcf4 target Tcf1. Science. 1999;285:1923–6. doi: 10.1126/science.285.5435.1923. [DOI] [PubMed] [Google Scholar]
- Salinas D, Haggie PM, Thiagarajah JR, Song Y, Rosbe K, Finkbeiner WE, Nielson DW, Verkman AS. Submucosal gland dysfunction as a primary defect in cystic fibrosis. Faseb J. 2005;19:431–3. doi: 10.1096/fj.04-2879fje. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Ito Y, Xu X, Han J, Bringas P, Jr, Maeda T, Slavkin HC, Grosschedl R, Chai Y. LEF1 is a critical epithelial survival factor during tooth morphogenesis. Dev Biol. 2005;278:130–43. doi: 10.1016/j.ydbio.2004.10.021. [DOI] [PubMed] [Google Scholar]
- Shtutman M, Zhurinsky J, Simcha I, Albanese C, D'Amico M, Pestell R, Ben-Ze'ev A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad Sci U S A. 1999;96:5522–7. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S, Stark KL, Shea MJ, Vassileva G, McMahon JA, McMahon AP. Wnt-3a regulates somite and tailbud formation in the mouse embryo. Genes Dev. 1994;8:174–89. doi: 10.1101/gad.8.2.174. [DOI] [PubMed] [Google Scholar]
- Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–6. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- Vadlamudi U, Espinoza HM, Ganga M, Martin DM, Liu X, Engelhardt JF, Amendt BA. PITX2, beta-catenin and LEF-1 interact to synergistically regulate the LEF-1 promoter. J Cell Sci. 2005;118:1129–37. doi: 10.1242/jcs.01706. [DOI] [PubMed] [Google Scholar]
- van Genderen C, Okamura RM, Farinas I, Quo RG, Parslow TG, Bruhn L, Grosschedl R. Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-deficient mice. Genes Dev. 1994;8:2691–703. doi: 10.1101/gad.8.22.2691. [DOI] [PubMed] [Google Scholar]
- Verbeek S, Izon D, Hofhuis F, Robanus-Maandag E, te Riele H, van de Wetering M, Oosterwegel M, Wilson A, MacDonald HR, Clevers H. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature. 1995;374:70–4. doi: 10.1038/374070a0. [DOI] [PubMed] [Google Scholar]
- Verkman AS, Song Y, Thiagarajah JR. Role of airway surface liquid and submucosal glands in cystic fibrosis lung disease. Am J Physiol Cell Physiol. 2003;284:C2–15. doi: 10.1152/ajpcell.00417.2002. [DOI] [PubMed] [Google Scholar]
- Waterman ML. Lymphoid enhancer factor/T cell factor expression in colorectal cancer. Cancer Metastasis Rev. 2004;23:41–52. doi: 10.1023/a:1025858928620. [DOI] [PubMed] [Google Scholar]
- Widelitz RB. Regulating the regulators: routing the Wnt-beta-catenin--Lef signals. J Invest Dermatol. 2004;123:VIII–X. doi: 10.1111/j.0022-202X.2004.23239.x. [DOI] [PubMed] [Google Scholar]
- Zhou P, Byrne C, Jacobs J, Fuchs E. Lymphoid enhancer factor 1 directs hair follicle patterning and epithelial cell fate. Genes Dev. 1995;9:700–13. doi: 10.1101/gad.9.6.700. [DOI] [PubMed] [Google Scholar]


