Abstract
To explore the association between single nucleotide polymorphisms of DNA repair genes and overall survival of patients with pancreatic cancer, we conducted a study in 378 cases of pancreatic adenocarcinoma who were treated at The University of Texas M. D. Anderson Cancer Center between February 1999 and October 2004 and were followed up to April 2006. Genotypes were determined using genomic DNA and the MassCode method. Overall survival was analyzed using the Kaplan–Meier plot, log-rank test and Cox regression. We observed a strong effect of the POLB A165G and T2133C genotypes on overall survival. The median survival time (MST) was 35.7 months for patients carrying at least 1 of the 2 homozygous variant POLB GG or CC genotypes, compared with 14.8 months for those carrying the AA/AG or TT/TC genotypes (p = 0.02, log rank test). The homozygous variants of hOGG1 G2657A, APEX1 D148E and XRCC1 R194W polymorphisms all showed a weak but significant effect on overall survival as demonstrated by either log rank test or multivariate COX regression after adjusting for other potential confounders. In combined genotype analysis, a predominant effect of the POLB homozygous variants on survival was observed. When POLB was not included in the model, a slightly better survival was observed among those carrying none of the adverse genotypes than those carrying at least one of the adverse genotypes. These observations suggest that polymorphisms of base excision repair genes signifi-cantly affect the clinical outcome of patients with pancreatic cancer. These observations need to be confirmed in a larger study of homogenous patient population.
Keywords: base excision repair, single nucleotide polymorphism, pancreatic cancer, survival
Pancreatic cancer is a rapidly fatal malignancy. The prognosis is extremely poor because this disease is usually diagnosed at a late stage and the tumors are highly aggressive and resistant to most treatments.1 Resistance to therapy leads to large individual variation in response to cytotoxic cancer therapies. To improve treatment efficacy, novel strategies are needed to help clinicians select the most suitable treatment regimen for each patient. Recent developments in pharmacogenomics and gene profiling have provided such opportunities.2 Several studies have shown that genetic variations in DNA repair and drug metabolism significantly affect the clinical outcome of patients receiving cytotoxic therapies.3–7
Gemcitabine and radiotherapy are currently the main therapeutic modalities for advanced pancreatic cancer.8 However, it is not clear what factors influence the clinical response to such treatment. Gemcitabine is a potent radiosensitizer.9 Little is known about DNA repair pathways that may alter cytotoxicity or radio-sensitivity with gemcitabine. On the other hand, it is well known that oxidative DNA damage and resulting DNA strand breaks are the most common type of radiation lesions that lead to mammalian cell death.10 The base excision repair (BER) pathway and the DNA strand break repair pathway are the major repair systems that contribute to the processing of oxidative lesions.11,12 In our previous studies we have shown a significant effect of DNA homologous recombination gene polymorphisms on overall survival of patients with pancreatic cancer.13,14 The current study focuses on the BER pathway.
Two subpathways of BER have been characterized by in vitro methods and classified according to the length of the repair patch: the short patch pathway repairs single nucleotides and the long patch pathway repairs lesions of 2–10 nucleotides.15 Short patch repair is the primary pathway for the repair of oxidative DNA damages, but long patch repair may occur when components of short patch repair are saturated or missing. The short patch repair process requires a number of enzymes: a DNA glycosylase, such as 8-oxo-guanine DNA glycosylase (hOGG1), removes the damaged base (e.g., 8-oxoguanine); apurinic/apyrimidinic endonuclease I (APEX1) recognizes and cleaves the resulting abasic (AP) site, introducing a single strand break; X-ray repair cross complementing protein 1 (XRCC1) is a scaffold protein that brings polymerase β (POLB) and ligase III (LIG3) together at the site of repair. DNA POLB adds a single nucleotide to the 3′-end of the nicked AP site and removes the resulting 5′-deoxyribose phosphate group, and a DNA LIG3-XRCC1 complex seals the generated nick.16
Polymorphisms of DNA repair genes may be capable of serving as a genetic marker for individual susceptibility to cytotoxic cancer therapy because of the role of DNA repair in protecting cells from DNA damage-mediated death. To test the hypothesis that genetic variations in BER affect clinical response to chemoradiotherapy and thus patient survival, we examined 9 single nucleotide polymorphisms of 6 major genes (hOGG1, APEX1, POLB, XRCC1, LIG3 and LIG4) that are involved in each step of the short patch BER pathway in 378 patients with a diagnosis of pancreatic ductal adenocarcinomas and who were treated at The University of Texas M. D. Anderson Cancer Center between 1999 and 2004. Overall survival was compared between patients with different genotypes.
Material and methods
Patient
A total of 378 patients with pathologically confirmed pancreatic ductal adenocarcinoma of all stages were recruited prospectively at M. D. Anderson Cancer Center (Houston, TX) between January 1999 and October 2004. These individuals participated in a larger ongoing molecular epidemiologic study in which demographic (age, sex and race) and risk factor information (smoking status, medical history, family history of cancer and exposures) were collected by personal interview and a blood sample was collected for genotyping at the time of enrollment. The study was approved by the Institutional Review Board of the M. D. Anderson Cancer Center.
For this analysis, all 378 patients were treated at M. D. Ander-son between January 1999 and October 2004 and were followed up to April 30, 2006. The minimum follow-up time was 18 months. We chose patients treated at our institute because information about patients treated elsewhere was often sparse, and sometimes patients treated outside were not given standard treatments or observed in standard fashion.
Clinical information was collected by reviewing the medical records of consenting patients. This included date of pathological diagnosis, treatment received before evaluation at M. D. Anderson, clinical tumor stage (localized, locally advanced and metastatic), performance status at first visit to M. D. Anderson, serum carbohydrate antigen 19-9 (CA19-9) values (unit/ml) at diagnosis, surgical procedure and date, pre- and postoperative chemotherapy regimen and radiation and date of death or last follow-up.
DNA extraction and genotyping
Whole blood was collected in heparinized tubes from patients at the time of enrollment. DNA was extracted from peripheral lymphocytes using the Qiagen DNA Isolation kit (Qiagen, Valencia, CA). Polymorphisms were detected using the Masscode™ technique17 by BioServe Biotechnologies (Laurel, MD). This mass spectrometry-based method uses standard PCR amplification of the region containing the polymorphism followed by hybridization with oligonucleo-tide primers that have been modified with photocleavable linkers attached to tags of variable composition (cleavable mass spectrometry tags) such that each allele-specific oligonucleotide primer is of different mass. For homozygotes, the mass spectrometer detects a single allele-specific tag, and for heterozygotes it detects 2 tags. In our previous study of other genes,18 we have observed a high concordance of data obtained from this method with those obtained from the Taqman and the PCR-RFLP methods. The reference numbers, gene locus, chromosome location, nucleotide change, amino acid change and minor allele frequency of the 9 SNPs examined in this study are described in Table I. The XRCC1, APEX1, hOGG1 and LIG4 SNPs were selected based on previous knowledge on the functional significance and their association with cancer risk or patient survival. The POLB and LIG3 SNPs were selected for their higher allele frequency (>10%) because there is no report on the functional significance of these SNPs. About 10% of the samples were analyzed in duplicates, and discrepancies were seen in less than 0.1% of the samples. Those with discordant results from 2 analyses were excluded from the final data analysis. The no call rate (samples without informative genotyping data) was 5.4% for hOGG1 C315G and POLB A165G and less than 4% for the remaining 7 polymorphisms.
TABLE I.
SNPs EXAMINED IN THE STUDY
| Gene | Chromosome | SNP | Amino acid | RS no.1 | MAF2 |
|---|---|---|---|---|---|
| hOGG1 | 3p26.2 | IVS7 + G2657A | 293794 | 0.27 | |
| 3′UTR, Ex6-C315G | S326C | 1052133 | 0.23 | ||
| APEX1 | 14q11.2-q12 | Ex5 + T5G | D148E | 3136820 | 0.51 |
| POLB | 8p11.2 | IVS10 + A165G | 2272615 | 0.27 | |
| IVS2-T2133C | 2953993 | 0.78 | |||
| XRCC1 | 19q13.2 | Ex6-C22T | R194W | 1799782 | 0.13 |
| Ex10-A4G | Q399R | 25487 | 0.35 | ||
| LIG3 | 17q11.2-q12 | IVS18-G39A | 2074522 | 0.10 | |
| LIG4 | 13q33-q34 | 5′UTR Ex2 + C54T | T9I | 1805388 | 0.13 |
Reference SNP identification number.
Minor allele frequency from the National Cancer Institute SNP500Cancer database.
Survival measurements
Our primary end point was overall survival from the time of pathologic diagnosis to date of death or last follow-up for all patients. Patients who were not deceased were censored at the last date they were known to be alive based on the date of last contact. Median follow-up time was computed among censored observations only. The minimum and maximum follow-up times were 18 and 90 months, respectively. Dates of death were obtained and crosschecked using at least 1 of the following 3 methods: Social Security Death Index, inpatient medical records and the M. D. Anderson tumor registry.
Statistical methods
The distribution of genotypes was compared by racial groups as well as by tumor stage using Pearson χ2 tests. Tests for Hardy–Weinberg equilibrium were conducted by goodness-of-fit χ2 test to compare the observed genotype frequencies with the expected genotype frequencies with 1 degree of freedom. Linkage disequilibrium of the 2 polymorphic variants of the hOGG1, POLB and XRCC1 genes was measured by using the SNPAlyze software (Dynacom, Mobara, Japan). The association between overall survival and each SNP was estimated using the method of Kaplan and Meier and assessed using the log-rank test. Because the wild-type and heterozygous variant genotypes of hOGG1, APEX1 and POLB had comparable median survival times, these 2 groups were collapsed into 1 and was used as the reference in comparison with the homozygous variant genotype. Hazard ratio and 95% confidence interval were estimated using multivariate Cox proportional regression models. The multivariate models included all factors that showed a significant association with overall survival in log rank test, i.e., stage, surgery, performance status, CA19-9 level and cytotoxic treatment. Race was also included in the model because of its association with genotype distribution. All statistical testing was conducted using the SPSS software version 12.0 (SPSS, Chicago, IL). p values of less than 0.05 were considered as statistically significant.
Results
Patient characteristics and overall survival
The demographic and clinical characteristics of the patients are described in Table II. The MST was 16.0 months (95% CI: 14–17). The median follow-up time was 34 months (95% CI: 30–39) for the living patients. Age, race, sex and cigarette smoking did not show any significant effect on overall survival (Table II). Patients with a history of diabetes lived 3 months shorter than those without diabetes (p = 0.08, log rank test). Disease stage, serum CA19-9 levels and performance status at diagnosis, and whether tumor resection was achieved were all significant predictors of survival in this patient population (Table II). During the entire disease process, 84.4% of the patients received gemcitabine, 70.4% received radiation to the tumor, 53.4% received cisplatin (or oxaliplatin) and 43.1% received 5FU (or capecitabine). In addition, about 20.6% of the patients received investigational therapy, such as bevacizumab, celecoxib, a-interferon and others. Because of the heterogeneity of the patient population and the small number of patients, treatment regimens during the entire disease process were collapsed into 3 groups: chemotherapy alone, 5FU-based chemoradiation (5FU/XRT) and gemcitabine-based chemoradiation (GEM/XRT). The 11 patients who did not receive any cytotoxic therapy were excluded from the treatment subgroup analyses. Patients who were treated with GEM/XRT did better than those who were treated with other chemotherapies (Table II). Among the 121 patients who had their tumors resected, 19 (15.7%) did not receive neoadjuvant therapy and there is no significant difference in overall survival between these patients by neo-adjuvant or adjuvant therapy modality (data not shown).
TABLE II.
PATIENT CHARACTERISTICS AND OVERALL SURVIVAL
| Variable | No. of patients | No. of death | MST* (months) | p (log rank) |
|---|---|---|---|---|
| Age (years) | ||||
| ≤50 | 50 | 39 | 18.4 | |
| 51–60 | 104 | 76 | 14.5 | |
| 61–70 | 128 | 97 | 14.4 | |
| >70 | 96 | 75 | 16.4 | 0.97 |
| Sex | ||||
| Male | 207 | 161 | 14.4 | |
| Female | 171 | 126 | 16.5 | 0.11 |
| Race | ||||
| White | 332 | 252 | 15.4 | |
| Hispanic | 22 | 18 | 12.8 | |
| African American | 19 | 12 | 18.1 | |
| Other | 5 | 5 | 16.5 | 0.51 |
| Smoking | ||||
| Never | 141 | 108 | 16.4 | |
| Ever | 237 | 179 | 14.4 | 0.68 |
| Diabetes | ||||
| No | 290 | 215 | 16.0 | |
| Yes | 88 | 72 | 13.3 | 0.08 |
| Stage | ||||
| Localized | 107 | 65 | 24.5 | |
| Locally advanced | 185 | 143 | 15.3 | |
| Metastatic | 86 | 79 | 9.8 | <0.001 |
| Performance status | ||||
| 0 | 59 | 43 | 15.3 | |
| 1 | 270 | 198 | 16.4 | |
| 2/3 | 47 | 46 | 11.5 | 0.0002 |
| Serum CA 19-9 (U/ml) | ||||
| ≤47 | 85 | 60 | 19.5 | |
| 48–1000 | 189 | 135 | 16.5 | |
| 1001–3000 | 48 | 40 | 11.7 | |
| >3000 | 55 | 51 | 9.7 | <0.001 |
| Surgical | ||||
| No | 235 | 210 | 10.7 | |
| Yes | 143 | 77 | 29.9 | <0.001 |
| Cytotoxic treatment | ||||
| Chemotherapy | 123 | 113 | 11.0 | |
| GEM/XRT | 139 | 93 | 21.4 | |
| 5FU/XRT | 105 | 71 | 18.6 | <0.001 |
p-values were from log rank test for the overall comparison within each group.
Median survival time.
Genotype and survival
The 9 SNPs were successfully amplified in 94.6–98.4% of the patients. There was a significant difference in the racial distribution of 6 SNPs (Table III). Genotype frequencies for all 9 SNPs were found to be in Hardy–Weinberg equilibrium (χ2 = 0.53–2.56, all p > 0.1). The hOGG1 G2657A, POLB A165G and POLB T2133C genotypes were not in Hardy–Weinberg equilibrium (χ2 = 4.87, 8.87 and 14.88, respectively) when data from all study subjects were analyzed together but all were in Hardy–Weinberg equilibrium (χ2 = 1.21–2.22, all p > 0.1) when data were analyzed among non-Hispanic whites only. There were no significant differences in the genotype distributions by age, sex, disease stage or surgical status (data not shown). The 2 SNPs of the hOGG1, POLB and XRCC1 genes were all in linkage disequilibrium with D′ values of –0.78, 0.91 and –0.59, respectively (all p < 0.001).
TABLE III.
GENOTYPE FREQUENCIES BY RACE
| Genotype | White N (%) | Hispanic N (%) | Black N (%) | p value (χ2 test) |
|---|---|---|---|---|
| APEX1 D148E | ||||
| TT | 85 (26.8) | 11 (52.4) | 5 (26.3) | |
| TG | 155 (48.9) | 9 (42.9) | 9 (47.4) | 0.09 |
| GG | 77 (24.3) | 1 (4.8) | 5 (26.3) | |
| hOGG1-C326G | ||||
| CC | 197 (62.9) | 10 (45.5) | 16 (88.9) | |
| CG | 104 (33.2) | 11 (50.0) | 2 (11.1) | 0.08 |
| GG | 12 (3.8) | 1 (4.5) | 0 (0) | |
| hOGG1-G2657A | ||||
| GG | 219 (66.6) | 18 (81.8) | 11 (57.9) | |
| GA | 90 (28.9) | 2 (9.1) | 5 (26.3) | 0.07 |
| AA | 12 (4.6) | 2 (9.1) | 3 (15.8) | |
| POLB-A165G | ||||
| AA | 268 (81.7) | 17 (77.3) | 8 (44.4) | |
| AG | 55 (16.8) | 5 (22.7) | 4 (22.2) | 0.00 |
| GG | 5 (1.5) | 0 (0) | 6 (33.3) | |
| POLB-T2133C | ||||
| TT | 283 (87.9) | 18 (81.8) | 5 (26.3) | |
| TC | 36 (11.2) | 4 (18.2) | 8 (42.1) | 0.00 |
| CC | 3 (0.9) | 0 (0) | 6 (31.6) | |
| XRCC1-R194W | ||||
| CC | 271 (86.6) | 16 (76.2) | 19 (100.0) | |
| CT | 39 (12.5) | 5 (23.8) | 0 (0) | 0.23 |
| TT | 3 (1.0) | 0 (0) | 0 (0) | |
| XRCC1-Q399R | ||||
| AA | 103 (32.4) | 6 (27.3) | 11 (61.1) | |
| AG | 170 (53.5) | 11 (50.0) | 7 (38.9) | 0.06 |
| GG | 45 (14.2) | 5 (22.7) | 0 (0.0) | |
| LIG3 G39A | ||||
| GG | 258 (79.4) | 21 (95.5) | 17 (89.5) | |
| GA | 66 (20.3) | 1 (4.5) | 2 (10.5) | 0.35 |
| AA | 1 (0.3) | 0 (0) | 0 (0) | |
| LIG4 C54T | ||||
| CC | 225 (70.3) | 17 (77.3) | 13 (68.4) | |
| CT | 88 (27.5) | 5 (22.7) | 5 (26.3) | 0.81 |
| TT | 7 (2.2) | 0 (0) | 1 (5.3) |
Data from 5 study subjects of other ethnicities are not shown.
A significantly reduced survival was associated with the homozygous variant alleles of the hOGG1 G2657A, APEX1 D148E and XRCC1 R194W SNPs (Table IV). The MST was 4–7 months shorter for the hOGG1 homozygous variant genotype than for the wild-type and heterozygous genotype (Fig. 1). The hOGG1 2657 AA but not the 326 GG genotype was a significant independent predictor of survival after adjusting for race and all other signifi-cant clinical factors (Table IV). When both SNPs of the hOGG1 gene were analyzed in combination, carriage of at least 1 of the homozygous variant genotypes was associated with a significantly reduced survival with MST of 11.5 vs. 16.0 months (p = 0.017, log rank test). Both homozygous variants of the APEX1 D148E and XRCC1 R194W SNPs remained as significant predictors of survival after adjusting for race and other significant clinical predictors (Table IV).
TABLE IV.
GENOTYPE FREQUENCY AND OVERALL SURVIVAL
| Genotype | No. of patients | No. of deaths | MST1 (months) | p(LR) | Multivariate hazard ratio (95% CI)2 | p value | |
|---|---|---|---|---|---|---|---|
| hOGG1-G2657A | |||||||
| GG | 253 | 93 | 14.8 | 1.0 | |||
| GA | 102 | 4 | 16.2 | 1.04 (0.79–1.37) | 0.765 | ||
| AA | 20 | 7 | 12.0 | 0.241 | 1.73 (1.05–2.85) | 0.033 | |
| GG/GA vs. AA | 0.049 | 1.35 (0.86–2.10) | 0.189 | ||||
| hOGG1-C326G | |||||||
| CC | 225 | 165 | 15.8 | 1.0 | |||
| CG | 119 | 91 | 15.4 | 0.88 (0.68–1.14) | 0.318 | ||
| GG | 13 | 12 | 8.6 | 0.159 | 1.23 (0.68–2.23) | 0.491 | |
| CC/CG vs. GG | 0.058 | 1.30 (0.72–2.33) | 0.387 | ||||
| APEX1 D148E | |||||||
| TT | 101 | 77 | 14.4 | 1.0 | |||
| TG | 178 | 128 | 16.5 | 0.94 (0.71–1.26) | 0.686 | ||
| GG | 82 | 67 | 12.9 | 0.133 | 1.42 (1.02–1.98) | 0.041 | |
| TT/TG vs. GG | 0.096 | 1.47 (1.11–1.95) | 0.007 | ||||
| POLB-A165G | |||||||
| AA | 297 | 223 | 15.4 | 1.0 | |||
| AG | 65 | 54 | 13.1 | 1.24 (0.91–1.68) | 0.172 | ||
| GG | 11 | 6 | 35.7 | 0.064 | 0.41 (0.17–0.95) | 0.037 | |
| AA/AG vs. GG | 0.039 | 0.39 (0.17–0.90) | 0.027 | ||||
| POLB-T2133C | |||||||
| TT | 311 | 236 | 15.4 | 1.0 | |||
| TC | 48 | 38 | 14.0 | 1.18 (0.83–1.67) | 0.367 | ||
| CC | 9 | 4 | 48.43 | 0.053 | 0.30 (0.11–0.85) | 0.023 | |
| TT/TC vs. CC | 0.017 | 0.29 (0.10–0.81) | 0.019 | ||||
| XRCC1-R194W | |||||||
| CC | 307 | 230 | 14.7 | 1.0 | |||
| CT | 48 | 37 | 16.5 | 0.91 (0.64–1.29) | 0.590 | ||
| TT | 3 | 3 | 5.9 | 0.023 | 4.64 (1.43–15.0) | 0.011 | |
| XRCC1-Q399R | |||||||
| AA | 123 | 90 | 16.3 | 1.0 | |||
| AG | 189 | 150 | 14.7 | 1.18 (0.91–1.54) | 0.229 | ||
| GG | 51 | 40 | 14.6 | 0.321 | 0.98 (0.67–1.43) | 0.907 | |
| LIG3 G39A | |||||||
| GG | 300 | 229 | 15.3 | 1.0 | |||
| GA | 70 | 51 | 15.3 | 0.96 (0.71–1.32) | 0.817 | ||
| AA | 1 | 1 | 22.5 | 0.956 | 0.87 (0.12–6.29) | 0.886 | |
| LIG4 C54T | |||||||
| CC | 257 | 196 | 14.7 | 1.0 | |||
| CT | 100 | 76 | 16.9 | 0.86 (0.66–1.12) | 0.270 | ||
| TT | 8 | 4 | 12.6 | 0.523 | 0.49 (0.18–1.34) | 0.165 |
MST, median survival time. The numbers of patients for each genotype do not add up to 378 because of the failure of genotyping assays in some patients.
Multivariate Cox regression with adjustment of race, stage, surgery, performance status, CA19-9 level and cytotoxic treatment.
Mean survival time is presented because MST could not be calculated.
FIGURE 1.
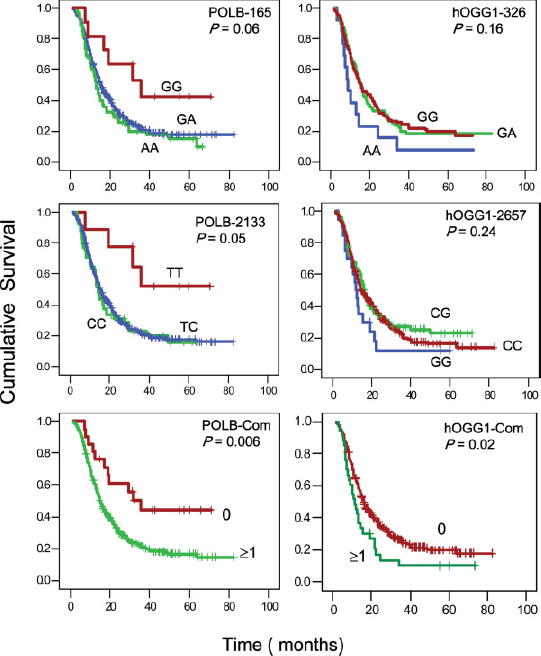
Survival plots for all patients by genotype of POLB A165G (A), POLB T2133C (B), hOGG1 G2657A (D) and hOGG1 C326G (E). Panels C and F are combined genotypes of the 2 SNPs for POLB and hOGG1, respectively. The numbers 0 and ≥1 in panels C and F indicate the number of variant alleles that are associated with significantly increased or reduced survivals. For example, in panel C, 0 indicates having POLB 165 AA/AG and 2133 TT/TC genotypes and ≥1 indicates having at least 1 of the 2 variant alleles of 165 GG and 2133 CC. In panel F, 0 indicates having hOGG1 2657GG/ GA, and 326 CC/CG genotypes and ≥1 indicates having at least 1 of the 2 variant alleles of 2657AA and 326GG. P values from log rank test, hazard ratios, and 95% confidence intervals are shown in Table IV. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
On the other hand, a significant protective effect of the POLB homozygous variant genotype on overall survival was observed. The MST was 35.7 months for those carrying at least 1 of the 2 homozygous variant POLB 165 GG and 2133 CC genotypes, compared with a MST of 14.8 months for those carrying the 165 AA/ AG and 2133 TT/TC genotypes (p = 0.006). Both individual SNPs and the combined genotype of the 2 POLB SNPs remained as significant independent predictors for survival after adjusting for race and significant clinical factors in Cox regression models (Table IV). The XRCC1 Q399R, LIG3 and LIG4 genotypes did not show any significant effect on overall survival of pancreatic cancer patients in this study (data not shown).
The effect of APEX1, XRCC1 and combined hOGG1 or POLB genotypes on survival was also confirmed when the analysis was conducted among non-Hispanic whites only. The adjusted hazard ratio (95% CI) was 1.37 (1.01–1.83, p = 0.038) for APEX1 GG and 5.08 (1.56–16.5, p = 0.007) for XRCC1 R194W TT genotypes and was 1.83 (1.03–3.25, p = 0.038) for those carrying any 1 of the 2 homozygous hOGG1 variants and 2.58 (1.21–5.50) for those with any 1 of the 2 homozygous POLB variants (p = 0.014).
Combined genotype effect
Because of hOGG1, APEX1, XRCC1 and POLB are all involved in the same DNA repair pathway; we explored the effect of combined genotypes on overall survival of the patients. We observed that patients carrying none of the adverse genotypes (hOGG1 2657 AA or 326 GG, APEX1 GG, XRCC1 194TT and POLB 165AG/GG or 2133 TC/CC) had a much better survival (mean survival time of 52.7 months) than those carrying 1, 2 or 3 adverse genotypes (MST of 16.0, 12.8 and 15.3 months, respectively) (Fig. 2, upper panel). This protective effect was mainly attributable to the POLB variant alleles. When POLB was not included in the analysis, carriage of 0, 1 or ≥2 adverse genotypes of the other 3 genes did not show a sig-nificant difference in the overall survival (data not shown). When hOGG1, APEX1, XRCC1 and LIG4 were all included in the combined genotype analysis, carriage of none of the adverse genotype had a MST of 18.1 months compared to a MST of 14.5, 13.3 and 15.3 months for those carrying 1, 2 or 3 adverse genotypes, respectively (Fig. 2, lower panel).
FIGURE 2.
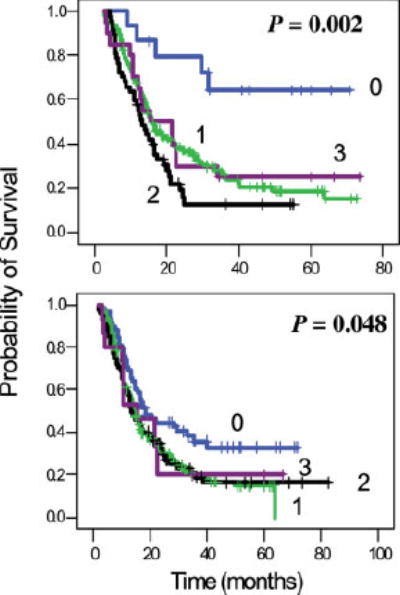
Combined genotype effects on overall survival. The numbers 0–3 indicate the number of variant alleles that are associated with significantly reduced survivals. For example, 0 indicates having none of the POLB 165 AG/AA or 2133 TC/CC, hOGG1 2657AA or 326 GG, APEX1 D148E GG and XRCC1 R194W TT genotypes and 1 indicates having at least 1 of the these variant alleles. P values are from log rank test. The hazard ratio (95% confidence interval) is 4.35 (1.74–10.9), 6.45 (2.55–16.4) and 4.25 (1.51–11.9) for patients carrying 1, 2 or 3 adverse genotypes, respectively, with adjustment of race and other clinical factors. Lower panel excluded the POLB gene but included XRCC1 Q399R AG/GG and LIG4 TT as the additional adverse genotypes. The adjusted hazard ratio (95% confidence interval) is 1.53 (1.05–2.24), 1.65 (1.08–2.52) and 1.60 (0.83–3.07), respectively, for those carrying 1, 2 or 3 adverse genotypes. [Color figure can be viewed in the online issue, which is available at www. interscience.wiley.com.]
Genotype effect by treatment
Stratified analysis by treatment or surgical status did not reveal any significant differential effect of the hOGG1, APEX1, XRCC1 and POLB genes because of the relatively low frequency of the homozygous variant alleles or reduced power associated with the small sample size in each subgroup. The combined genotype of these 4 genes showed similar effect on overall survival of patients who had their tumor resected or not resected, p = 0.04 and 0.001, respectively, by log rank test. The combined genotype effect on survival was highly significant (p = 0.005, log rank test) among patients who received radiation but not significant for patients who did not receive radiation (p = 0.11). The combined genotype effect was at borderline significance (p = 0.05) for patients who received gemcitabine/radiation but not significant for those received 5 FU/radiation (p = 0.11).
Discussion
The current study evaluated the effect of 9 SNPs of 6 major BER genes (hOGG1, APEX1, POLB, XRCC1, LIG3 and LIG4) on overall survival of patients with pancreatic adenocarcinoma. We demonstrated that the homozygous variant genotypes of the hOGG1, APEX1 and XRCC1 R194W were associated with significantly decreased overall survival and that the homozygous variant genotype of POLB was associated with a significantly increased overall survival. These observations demonstrated for the first time the sig-nificant effect of BER genes on pancreatic cancer patient survival.
Among the 6 genes examined in this study, POLB homozygous variants showed a strong protective effect on patient survival. The 2 homozygous variant genotypes were associated with a doubling of survival time compared with the wild-type and heterozygous variant genotypes. Although the frequencies of these homozygous alleles are low (2.8% and 2.4%), and the possibility that these observations were by chance alone cannot be excluded, such effect is consistent with the drug-resistant nature conferred by overex- pression of DNA POLB. POLB is expressed at a constant low level throughout the cell cycle19 and is inducible by some geno-toxic treatments.20 The primary function of POLB is thought to be in the processing of damaged bases via the BER pathway, thereby protecting cells from DNA damage-induced cytotoxicity.21,22 At the transcriptional level, POLB is overexpressed in many cancer cells.23 Overexpression of POLB has been associated with an increased chromosome instability and tumorigenesis as well as with resistance to many DNA-damaging agents, including chemotherapeutic drugs.24–26 In fact, inhibition of POLB expression has recently been used as a novel approach to increase sensitivity to cisplatin-based cancer therapy.27 The 2 SNPs examined in the current study are both localized in the intron region (intron 10 and intron 2), and the functional significance of these SNPs has not been explored. It is tempting to speculate that the homozygous variants may confer a lower expression of POLB and thus an increased sensitivity to cyto-toxic treatment and an improved survival. This hypothesis needs to be tested in experimental systems. It is also possible that these 2 SNPs are in linkage disequilibrium with other functionally signifi-cant SNPs in the same gene; therefore haplotype analysis is required to identify the responsible SNPs. These homozygous variants of the 2 SNPs were observed in a much higher frequency in African Amer-icans than in Caucasians and they were absent among Hispanics. Regardless of the mechanism underlying the observed associations, our observations suggest that POLB is an important gene influenc-ing the overall survival of patients with pancreatic cancer, especially among African Americans.
hOGG1 is a DNA repair enzyme that excises 7,8-dihydro-8-oxo-guanine (8oxoG), an oxidative DNA lesion, from DNA. Because 8oxoG is a lesion leading to a high degree of DNA mispairing, decreased hOGG1 expression level could lead to a higher frequency of mutation and could possibly increase the cancer risk of an individual under oxidative stress. There is some epidemiologic evidence supporting an association between the risk of several types of human cancers and the hOGG1 genotype.28 Although some functional studies suggest reduced repair function with variant alleles in hOGG1, the evidence is generally inconclusive.28 No study has yet been reported on the role of hOGG1 in the clinical outcome of cancer patients. In the current study, we observed a significantly reduced overall survival for patients carrying the homozygous variant genotypes of this gene. This observation would support a better DNA repair associated with the variant alleles and thus a poor response to therapy. Indeed, some epidemiological studies have shown a reduced cancer risk associated with the variant alleles of the hOGG1 gene.28 Because of the inconclusive observations on this gene in epidemiological studies and the uncertain functional effects of the variant hOGG1 genotypes, our observations need to be con-firmed in other studies as well.
APEX1 acts in base excision repair to hydrolyze abasic sites arising from enzymatic removal of damaged purine and pyrimi-dine bases. It has a 3′-phosphodiesterase activity that excises de-oxyribose fragments and phosphate groups at the 3′ terminus of DNA strand breaks caused by ionizing radiation, yielding a 3′-OH substrate for DNA repair synthesis. Nuclear expression of APEX1 protein has been related to a better survival in resected non-small cell lung cancer.29 However, overexpression of APEX1 was related to resistance to bleomycin therapy in patients with testicular cancer.30 Although the APEX1 D148E (Asp148Glu) polymor-phism does not result in reduced endonuclease activity,31 cells having the variant allele may have higher sensitivity to ionizing radiation.32 In this study, however, we observed a slightly reduced overall survival among patients carrying the homozygous variant allele, which does not support a higher sensitivity to radiotherapy. The functional significance of this SNP and its role in the clinical response to cytotoxic therapy need further investigation.
XRCC1 acts as a scaffold protein that coordinates the enzymatic activities at different steps of the BER pathway by interacting with various repair enzymes such as APEX1, polynucleotide kinase, LIG3, DNA POLB, poly (ADP-ribose) polymerase and proliferating cell nuclear antigen.33 The lack of coordination in the repair process could result in the persistence and exposure to the cellular milieu of potentially toxic DNA intermediates such as single strand breaks. The XRCC1 Q399R (Gln399Arg) polymorphism has been shown to affect the clinical response to platinum-based cancer therapy,6 and the XRCC1 R194W (Arg194Trp) SNP has not been evaluated as a predictor of clinical outcome. In the current study, we observed a significant effect of the XRCC1 R194W but not the Q399R SNP on survival in pancreatic cancer patients. However, the frequency of the XRCC1 R194W homozygote was extremely low (0.8%), our observation could be made by chance alone.
The strength of this study is its large sample size and adequate statistical power. The limitation of the study is the heterogeneity of the patients, which makes the data interpretation more difficult. Some subgroup analyses were conducted in an attempt to overcome this problem, but the small sample size in each subgroup may increase the chance of false positive results. The significant effects of combined genotype effects on overall survival among patients who received radiation but not in those without radiation were consistent with our previous observations in homologous recombination repair genes.14 It supports the hypothesis that genetic variation in the BER pathway would affect the repair of radiation-induced DNA damage and cell survival. However, patients who received no radiation were mostly with metastatic disease, therefore, the overwhelming genetic abnormalities in the tumors may confer resistance to most therapies and any subtle genetic effects can not be detected. It is therefore important to confirm these observations in other patient populations, preferably in prospectively designed trials where treatment regimens can be standardized and patient homogeneity adequately addressed.
In summary, the current study has demonstrated a statistically significant effect of hOGG1, APEX1, XRCC1 and DNA POLB gene polymorphisms on the overall survival of patients with pancreatic adenocarcinoma. Except for XRCC1, this is the first study to show that polymorphic variants of BER genes may have an important role in influencing the clinical outcome of patients with cancer. We have previously shown that DNA homologous recombination repair gene polymorphisms significantly affect the overall survival of pancreatic cancer patients.13,14 Because DNA repair is a complex process, it takes many proteins acting in concert to maintain cell viability and genome integrity. Each individual SNP of the low penetrance genes may have a weak effect at the functional level, but the combined effect of several genes may have significant clinical value in predicting response to therapy and prognosis. Genetic profiling of enzymes that are involved in drug metabolism, DNA repair or cellular response to cytotoxic agents may provide opportunities for the discovery of novel therapeutic targets and information that can direct the choice of therapy and predict the treatment tolerance, response and overall outcome.34
Abbreviations
- APEX1
apurinic/apyrimidinic endonuclease 1
- BER
base excision repair
- 95% CI
confidence interval
- 5FU
5 fluorouracil
- GEM
gemcitabine
- hOGG1
8-oxoguanine DNA glycosylase
- LIG3
ligase III
- LIG4
ligase IV
- MST
median survival time
- POLB
DNA polymerase β
- XRCC1
X-ray repair cross complementing protein 1
- XRT
radiotherapy
Footnotes
Grant sponsor: National Institutes of Health; Grant numbers: CA098380, CA101936, CA16672; Grant sponsor: Lockton Research Funds.
References
- 1.Bardeesy N, DePinho RA. Pancreatic cancer biology and genetics. Nat Rev Cancer. 2002;2:897–909. doi: 10.1038/nrc949. [DOI] [PubMed] [Google Scholar]
- 2.Phillips KA, Van Bebber SL. Measuring the value of pharmacoge-nomics. Nat Rev Drug Discov. 2005;4:500–9. doi: 10.1038/nrd1749. [DOI] [PubMed] [Google Scholar]
- 3.Bosken CH, Wei Q, Amos CI, Spitz MR. An analysis of DNA repair as a determinant of survival in patients with non small-cell lung cancer. J Natl Cancer Inst. 2002;94:1091–9. doi: 10.1093/jnci/94.14.1091. [DOI] [PubMed] [Google Scholar]
- 4.Camps C, Sarries C, Roig B, Sanchez JJ, Queralt C, Sancho E, Marti-nez N, Taron M, Rosell R. Assessment of nucleotide excision repair XPD polymorphisms in the peripheral blood of gemcitabine/cisplatin-treated advanced non-small-cell lung cancer patients. Clin Lung Cancer. 2003;4:237–41. doi: 10.3816/clc.2003.n.004. [DOI] [PubMed] [Google Scholar]
- 5.Park DJ, Stoehlmacher J, Zhang W, Tsao-Wei DD, Groshen S, Lenz HJ. A xeroderma pigmentosum group D gene polymorphism predicts clinical outcome to platinum-based chemotherapy in patients with advanced colorectal cancer. Cancer Res. 2001;61:8654–8. [PubMed] [Google Scholar]
- 6.Gurubhagavatula S, Liu G, Park S, Zhou W, Su L, Wain JC, Lynch TJ, Neuberg DS, Christiani DC. XPD and XRCC1 genetic polymorphisms are prognostic factors in advanced non-small-cell lung cancer patients treated with platinum chemotherapy. J Clin Oncol. 2004;22:2594–601. doi: 10.1200/JCO.2004.08.067. [DOI] [PubMed] [Google Scholar]
- 7.Rosell R, Taron M, Alberola V, Massuti B, Felip E. Genetic testing for chemotherapy in non-small cell lung cancer. Lung Cancer. 2003;41(Suppl 1):S97–S102. doi: 10.1016/s0169-5002(03)00151-x. [DOI] [PubMed] [Google Scholar]
- 8.Burris HA, III, Moore MJ, Andersen J, Green MR, Rothenberg ML, Modiano MR, Cripps MC, Portenoy RK, Storniolo AM, Tarassoff P, Nelson R, Dorr FA, et al. Improvements in survival and clinical bene-fit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol. 1997;15:2403–13. doi: 10.1200/JCO.1997.15.6.2403. [DOI] [PubMed] [Google Scholar]
- 9.Lawrence TS, Eisbruch A, Shewach DS. Gemcitabine-mediated radiosensitization. Semin Oncol. 1997;24:S7-24–S7-28. [PubMed] [Google Scholar]
- 10.Lawrence TS, Blackstock AW, McGinn C. The mechanism of action of radiosensitization of conventional chemotherapeutic agents. Semin Radiat Oncol. 2003;13:13–21. doi: 10.1053/srao.2003.50002. [DOI] [PubMed] [Google Scholar]
- 11.Wilson DM, III, Sofinowski TM, McNeill DR. Repair mechanisms for oxidative DNA damage. Front Biosci. 2003;8:d963–d981. doi: 10.2741/1109. [DOI] [PubMed] [Google Scholar]
- 12.Frosina G, Fortini P, Rossi O, Carrozzino F, Raspaglio G, Cox LS, Lane DP, Abbondandolo A, Dogliotti E. Two pathways for base excision repair in mammalian cells. J Biol Chem. 1996;271:9573–8. doi: 10.1074/jbc.271.16.9573. [DOI] [PubMed] [Google Scholar]
- 13.Li D, Frazier M, Evans DB, Hess KR, Crane CH, Jiao L, Abbruzzese JL. Single nucleotide polymorphisms of RecQ1, RAD54L, and ATM genes are associated with reduced survival of pancreatic cancer. J Clin Oncol. 2006;24:1720–8. doi: 10.1200/JCO.2005.04.4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li D, Liu H, Jiao L, Chang DZ, Beinart G, Wolff RA, Evans DB, Hassan MM, Abbruzzese JL. Homologous recombination DNA repair gene polymorphisms are associated with reduced survival in pancreatic cancer patients. Cancer Res. 2006;66:3323–30. doi: 10.1158/0008-5472.CAN-05-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seeberg E, Eide L, Bjoras M. The base excision repair pathway. Trends Biochem Sci. 1995;20:391–7. doi: 10.1016/s0968-0004(00)89086-6. [DOI] [PubMed] [Google Scholar]
- 16.Norbury CJ, Hickson ID. Cellular responses to DNA damage. Annu Rev Pharmacol Toxicol. 2001;41:367–401. doi: 10.1146/annurev.pharmtox.41.1.367. [DOI] [PubMed] [Google Scholar]
- 17.Kokoris M, Dix K, Moynihan K, Mathis J, Erwin B, Grass P, Hines B, Duesterhoeft A. High-throughput SNP genotyping with the Masscode system. Mol Diagn. 2000;5:329–40. doi: 10.1007/BF03262094. [DOI] [PubMed] [Google Scholar]
- 18.Li D, Jiao L, Li Y, Doll MA, Hein DW, Bondy ML, Evans DB, Wolff RA, Lenzi R, Pisters PW, Abbruzzese JL, Hassan MM. Polymorphisms of cytochrome P4501A2 and N-acetyltransferase genes, smoking, and risk of pancreatic cancer. Carcinogenesis. 2006;27:103–11. doi: 10.1093/carcin/bgi171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zmudzka BZ, Fornace A, Collins J, Wilson SH. Characterization of DNA polymerase β mRNA: cell-cycle and growth response in cultured human cells. Nucleic Acids Res. 1988;16:9587–96. doi: 10.1093/nar/16.20.9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horton JK, Srivastava DK, Zmudzka BZ, Wilson SH. Strategic down-regulation of DNA polymerase β by antisense RNA sensitizes mammalian cells to specific DNA damaging agents. Nucleic Acids Res. 1995;23:3810–15. doi: 10.1093/nar/23.19.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamath-Loeb AS, Hizi A, Kasai H, Loeb LA. Incorporation of the guanosine triphosphate analogs 8-oxo-dGTP and 8-NH2-dGTP by reverse transcriptases and mammalian DNA polymerases. J Biol Chem. 1997;272:5892–8. doi: 10.1074/jbc.272.9.5892. [DOI] [PubMed] [Google Scholar]
- 22.Hoffmann JS, Pillaire MJ, Maga G, Podust V, Hubscher U, Villani G. DNA polymerase β bypasses in vitro a single d(GpG)-cisplatin adduct placed on codon 13 of the HRAS gene. Proc Natl Acad Sci USA. 1995;92:5356–60. doi: 10.1073/pnas.92.12.5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srivastava DK, Husain I, Arteaga CL, Wilson SH. DNA polymerase β expression differences in selected human tumors and cell lines. Car-cinogenesis. 1999;20:1049–54. doi: 10.1093/carcin/20.6.1049. [DOI] [PubMed] [Google Scholar]
- 24.Bergoglio V, Pillaire MJ, Lacroix-Triki M, Raynaud-Messina B, Can-itrot Y, Bieth A, Gares M, Wright M, Delsol G, Loeb LA, Cazaux C, Hoffmann JS. Deregulated DNA polymerase β induces chromosome instability and tumorigenesis. Cancer Res. 2002;62:3511–14. [PubMed] [Google Scholar]
- 25.Yamada NA, Farber RA. Induction of a low level of microsatellite instability by overexpression of DNA polymerase β. Cancer Res. 2002;62:6061–4. [PubMed] [Google Scholar]
- 26.Canitrot Y, Cazaux C, Frechet M, Bouayadi K, Lesca C, Salles B, Hoffmann JS. Overexpression of DNA polymerase β in cell results in a mutator phenotype and a decreased sensitivity to anticancer drugs. Proc Natl Acad Sci USA. 1998;95:12586–90. doi: 10.1073/pnas.95.21.12586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boudsocq F, Benaim P, Canitrot Y, Knibiehler M, Ausseil F, Capp JP, Bieth A, Long C, David B, Shevelev I, Frierich-Heinecken E, Hubscher U, et al. Modulation of cellular response to cisplatin by a novel inhibitor of DNA polymerase β. Mol Pharmacol. 2005;67:1485–92. doi: 10.1124/mol.104.001776. [DOI] [PubMed] [Google Scholar]
- 28.Weiss JM, Goode EL, Ladiges WC, Ulrich CM. Polymorphic variation in hOGG1 and risk of cancer: a review of the functional and epi-demiologic literature. Mol Carcinog. 2005;42:127–41. doi: 10.1002/mc.20067. [DOI] [PubMed] [Google Scholar]
- 29.Kakolyris S, Giatromanolaki A, Koukourakis M, Kaklamanis L, Kanavaros P, Hickson ID, Barzilay G, Georgoulias V, Gatter KC, Harris AL. Nuclear localization of human AP endonuclease 1 (HAP1/ Ref-1) associates with prognosis in early operable non-small cell lung cancer (NSCLC) J Pathol. 1999;189:351–7. doi: 10.1002/(SICI)1096-9896(199911)189:3<351::AID-PATH435>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 30.Robertson KA, Bullock HA, Xu Y, Tritt R, Zimmerman E, Ulbright TM, Foster RS, Einhorn LH, Kelley MR. Altered expression of Ape1/ ref-1 in germ cell tumors and overexpression in NT2 cells confers resistance to bleomycin and radiation. Cancer Res. 2001;61:2220–5. [PubMed] [Google Scholar]
- 31.Hadi MZ, Coleman MA, Fidelis K, Mohrenweiser HW, Wilson DM., III Functional characterization of Ape1 variants identified in the human population. Nucleic Acids Res. 2000;28:3871–9. doi: 10.1093/nar/28.20.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu JJ, Smith TR, Miller MS, Mohrenweiser HW, Golden A, Case LD. Amino acid substitution variants of APE1 and XRCC1 genes associated with ionizing radiation sensitivity. Carcinogenesis. 2001;22:917–22. doi: 10.1093/carcin/22.6.917. [DOI] [PubMed] [Google Scholar]
- 33.Vidal AE, Boiteux S, Hickson ID, Radicella JP. XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein–protein interactions. EMBO J. 2001;20:6530–9. doi: 10.1093/emboj/20.22.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosell R, Taron M, Ariza A, Barnadas A, Mate JL, Reguart N, Margel M, Felip E, Mendez P, Garcia-Campelo R. Molecular predictors of response to chemotherapy in lung cancer. Semin Oncol. 2004;31:20–7. doi: 10.1053/j.seminoncol.2003.12.011. [DOI] [PubMed] [Google Scholar]


