Abstract
Myasthenia gravis (MG) is an autoimmune disease mainly caused by antiacetylcholine receptor autoantibodies (seropositive (SP) disease) or by Abs against unknown autoantigenic target(s) (seronegative (SN) disease). Thymectomy is usually beneficial although thymic hyperplasia with ectopic germinal centers is mainly observed in SP MG. To understand the role of thymus in the disease process, we compared the thymic transcriptome of non-MG adults to those of SP patients with a low or high degree of hyperplasia or SN patients. Surprisingly, an overexpression of MHC class II, Ig, and B cell marker genes is observed in SP but also SN MG patients. Moreover, we demonstrate an overexpression of CXCL13 in all MG thymuses leading probably to the generalized B cell infiltration. However, we find different chemotactic properties for MG subgroups and, especially, a specific overexpression of CCL21 in hyperplastic thymuses triggering most likely ectopic germinal center development. Besides, SN patients present a peculiar signature with an abnormal expression of genes involved in muscle development and synaptic transmission, but also genes implicated in host response, suggesting that viral infection might be related to SN MG. Altogether, these results underline differential pathogenic mechanisms in the thymus of SP and SN MG and propose new research areas.
Acquired myasthenia gravis (MG)4 is a neurological autoimmune disease caused by autoantibodies against components of the neuromuscular junction and leading to disabling fatigability. Seropositive (SP) MG is caused by anti-acetylcholine receptor (AChR) autoantibodies and represents 85% of patients (1). In contrast, MG patients without detectable anti-AChR Abs are named seronegative (SN). However, this distinction is misleading as these patients respond well to plasma exchange and their plasma can transfer the disease to experimental animal models (2). Moreover, in the serum of some of these patients, autoantibodies against a muscle-specific tyrosine kinase (MuSK) receptor have been found and these patients are named MuSK+ (3). For the remaining SN patients, the specificity of the autoantibodies implicated is still not known.
The thymus provides a complex environment essential for the generation of the T cell repertoire. It is composed of various cell types, essentially thymocytes and thymic epithelial cells (TECs), but also fibroblasts, macrophages, dendritic, and myoid cells (4). Differentiation of T cells occurs while they are progressing through the different thymic compartments. Successful T cell differentiation depends on the quality and the specificity of TCR/Ag-MHC interactions (positive selection). Medullary TECs, by expressing a broad panoply of tissue-specific Ags, play a crucial role in central tolerance (negative selection) and any defect in thymocyte selection could lead to autoimmune diseases (5). In MG, functional and morphological abnormalities of the thymus occur frequently and 50–60% of the SP patients exhibit thymic hyperplasia of lymphoproliferative origin with ectopic germinal center (GC) development (6). These thymic abnormalities are correlated with the anti-AChR Ab titer which decreases after thymectomy (7). The hyperplastic thymus includes all the components of the anti-AChR response: the AChR (8), B cells producing anti-AChR Abs (9), and anti-AChR autoreactive T cells (10). Thus, the thymus plays a pivotal role in the pathogenesis of SP MG and an understanding of the mechanisms leading to ectopic GC formation is expected to shed light on the pathogenesis of this disease. In contrast, there is little information on the involvement of the thymus in non-SP form of MG. The thymus of MuSK+ patients shows few or no pathological changes and the beneficial effects of thymectomy has not been proved for this subgroup (11). In SN patients, the clinical characteristics are heterogeneous and thymectomy improves some of them (11). Histological analyses of the thymus showed that SN patients can present lymph node-type infiltrates with a few GCs (12, 13). However, the pathogenic mechanisms occurring in the thymus of SN and SP patients seem to be distinct and, for example, they differently regulate Fas expression in thymocytes (14). All these observations tend to also suggest the involvement of the thymus in SN patients.
As for many autoimmune diseases, the triggering events involved in MG are not clearly defined. MG affects more women than men (4, 11). Moreover, a genetic contribution is strongly supported and the HLA-A1-B8-DR3 haplotype is associated with MG characterized by thymic hyperplasia (15). However, these susceptibility genes cannot account exclusively for MG development and other factors seem to be important triggering events. Consequently, to clarify the pathogenesis of MG, we investigated gene expressions occurring in the thymus of MG patients. By analyzing the thymic transcriptome of different MG patient subgroups, we demonstrated the existence of 1) a common gene expression signature in the thymus of all MG patients, 2) crucial thymic events associated with hyperplasia, and 3) peculiar gene expression profiles characterizing the thymus of SN from SP patients.
Materials and Methods
Samples and RNA extraction
Thymic fragments (50–100 mg) were obtained from MG patients after thymectomy or from sex-matched baby or adult females undergoing cardiovascular surgery at the Marie Lannelongue Chirurgical Center (Le Plessis-Robinson, France). We selected MG Caucasian females known to be only treated by anticholinesterase drugs and not by other therapies (corticosteroids, immunosuppressors, i.v. Igs, plasmapheresis) and with no other known disease (including thymoma). This study was approved by the local ethics committee (Comité Consultatif pour la Protection des Personnes dans la Recherche Biomédicale (CCPPRB), Kremlin-Bicêtre, France).
Total RNA was extracted from individual frozen thymic fragments at the same period of time using the FastPrep FP120 instrument (Qbiogen) followed by a TRIzol extraction (Invitrogen Life Technologies) and a DNase treatment before purification onto Qiagen columns. RNA quality was assessed on an Agilent Bioanalyser. To minimize interindividual variations, the following pools of RNA were prepared with equal amounts of total RNA extracted from thymic fragments: 1) non-MG adults, n = 4, 15–19 years old; 2) SP MG patients with low thymic hyperplasia (ML; with two or less GCs per section), n = 4; 19–25 years old; 3) SP MG patients with high thymic hyperplasia (MH; with three or more GCs per section), n = 5, 18–22 years old; 4) SN MG patients (SN: without or only a few GCs), n = 3, 16–22 years old. These MG patients were SN for anti-AChR and anti-MuSK Abs.
Each pool was hybridized to a thymic reference composed of 10 thymuses from babies aged 1 wk to 1 year old undergoing thoracic surgery.
Microarray experiments
Microarray experiments were conducted using the Human 1 cDNA arrays from Agilent (G4100A; 16,200 spots containing 12,814 unique clones) according to the manufacturer's instructions, using 20 μg of total RNA. For each array, the thymic reference was cohybridized with a given RNA pool (five arrays per pool).
Data acquisition and normalization
Labeled microarrays were scanned using the 428 Affymetrix scanner (MWG) and the images were analyzed using GenePix pro version 4.0 (Axon Instruments). Raw microarray data are available on the ArrayExpress database at 〈www.ebi.ac.uk/arrayexpress〉 (accession no. E-MEXP-518). For each array, raw data were corrected by a Lowess transformation using the TIGR Microarray Data Analysis System (〈www.tigr.org〉). To allow interarray comparisons, each condition was centered on the median calculated from the repetitions; a Lowess transformation was finally applied to each array. For each gene, a log 2 ratio (study sample/thymic reference) was calculated and the distribution by array was centered on zero.
Clustering and statistical analysis
The following analyses were applied on the probes that have successfully passed the Agilent quality control (13,572 surveyed genes). To provide a general survey of gene expression, an 8 K-median clustering analysis (Acuity software version 3.1; Axon Instruments) was applied on the median of ratios for the genes per thymic subgroups. To identify dysregulated genes in the different subgroups of MG compared with non-MG adults, log 2 ratios were analyzed using significance analysis of microarrays (SAM) software (〈www-stat.stanford.edu/∼tibs/SAM〉). To search for chromosomal susceptibility regions linked to MG, the chromosomal locations of dysregulated genes were analyzed. The gene chromosomal position was mapped using the National Center for Biotechnology Information (NCBI; 〈www.ncbi.nlm.nih.gov〉), SOURCE (〈http://genome-www5.stanford.edu/cgi-bin/source/sourceSearch〉), and Database for Annotation, Visualization, and Integrated Discovery (DAVID; 〈http://david.niaid.nih.gov/David/upload.asp〉). Concerning the precise chromosomal location, the nomenclature chromosome-arm-region band was used and the location of genes spanning several bands was arbitrarily fixed to the closest band to the centromere. We took into account the number of unique GenBanks with a known chromosomal location (10,865 genes). A χ2 test was then applied on the most highly represented chromosomal regions (>2.5%) in at least one MG subgroup for up- or down-regulated genes.
Real-time PCR
Real-time PCR was conducted for CD20, HLA-DR, CCL21, CCL19, CXCL13, CXCL10, CXCL12, and IFN-stimulated gene 12 (ISG12) (Table I). We used individual RNA samples from the donors included in the microarray study and from additional donors belonging to non-MG adult, ML, MH, and SN categories as previously described, extending the age range from 14 to 38 years old. Total mRNA was reverse transcribed using the SuperScript II RT (Invitrogen Life Technologies) according to the manufacturer's instructions. PCRs were performed on the LightCycler apparatus as previously described (16). For each PCR, a standard curve was calculated using the thymic reference and for each sample, the mean of duplicates was given. All samples were controlled by 28S rRNA amplification.
Table 1.
Primers and PCR conditions used in real-time PCR
| Gene | Primersa | Annealing Temperature | MgCl2 (mM) | |
|---|---|---|---|---|
| CD20 | GGCTGTCCAGATTATGAATGGG | (F) | 62 | 4 |
| TGGAGTTTTTCTCCGTTGCTG | (R) | |||
| HLA-DR | TGGAGCAGATTAAACACGAGTG | (F) | 68 | 4 |
| CCGCCCGGAACTTTCTGAC | (R) | |||
| CCL21 (SLC) | CCTTGCCACACTCTTTCTCCC | (F) | 65 | 3 |
| CAAGGAAGAGGTGGGGTGTA | (R) | |||
| CCL19 (ELC) | GGTGCCTGCTGTAGTGTTCA | (F) | 61 | 4 |
| GGTCCTTCCTTCTGGTCCTC | (R) | |||
| CXCL13 (BCA-1) | CTCTGCTTCTCATGCTGCTG | (F) | 64 | 4 |
| TGAGGGTCCACACACACAAT | (R) | |||
| CXCL10 (IP10) | AAGGATGGACCACACAGAGG | (F) | 62 | 4 |
| TGGAAGATGGGAAAGGTGAG | (R) | |||
| CXCL12 (SDF1-β) | GGGCTCCTGGGTTTTGTATT | (F) | 63 | 3 |
| GTCCTGAGAGTCCTTTTGCG | (R) | |||
| ISG12 | GCCTCTGCTCTCACCTCATC | (F) | 60 | 4 |
| ATCTTGGCTGCTATGGAGGA | (R) |
Forward (F) and reverse (R) primers. SLC, secondary lymphoid tissue chemokine; ELC, EBV-induced molecule I ligand chemokine; IP10, IFN-γ-inducible protein; SDF, stromal-derived factor.
Ig quantification
Proteins from individual thymic extracts from babies, non-MG adults, ML, MH, and SN patients as previously described in microarray and PCR were extracted in 1 ml of PBS containing protease inhibitors (Complete; Roche Diagnostics) using the FastPrep FP120 apparatus and centrifuged at 12,000 rpm for 20 min at 4°C to recover the supernatant. The levels of IgG and IgM in thymic extracts containing 1 mg/ml proteins were measured by immunonephelometry using the Dade Behring Nephelometer II analyzer.
Chemotaxis assay
PBMC from healthy adult donors (CCPPRB agreement no. 05-03) were recovered using a Ficoll gradient and incubated overnight in cell culture flasks with RPMI 1640-Glutamax I supplemented with 0.5% FCS (Invitrogen Life Technologies). PBL were then labeled (106/ml) for 1 h with 2 μM calcein-AM (Invitrogen Life Technologies) and washed in RPMI 1640-0.5% FCS. Chemotaxis assays were performed for 4 h at 37°C using the MultiScreen-MIC kit (3 μm pore size; Millipore): 50 μl of PBL were loaded in the upper chamber of the transwell plate. In the lower wells, we added 150 μl of RPMI 1640-0.5% FCS containing 3 mg/ml proteins from individual thymic extracts from non-MG adults, ML, MH, and SN patients as previously described. Thymic proteins were extracted in 1 ml of lysis buffer containing 50 mM Tris (pH = 7.4), 5% Triton X-100, and protease inhibitors (Complete) using the FastPrep FP120 apparatus, and centrifuged at 12,000 rpm for 30 min at 4°C to recover the supernatant. The proportion of cells migrating to the lower wells was evaluated by measuring the calcein-AM fluorescence with a microplate fluorescence reader (FL600; BioTek Instruments).
Results
To characterize gene expressions in the thymus of MG patients, the thymic transcriptome of SP MG patients with a low (ML) or a high (MH) degree of hyperplasia or SN MG was compared with that of non-MG adults.
Global characteristics of gene expression in MG thymuses
General survey of gene expression
Among the 13,572 surveyed genes, many were expressed in the thymus whatever its pathologic (MG disease) and physiologic (normal) condition, because ∼86% of the genes had a signal intensity over the background intensity. This elevated percentage is not surprising as the thymus is a complex organ composed of various cell types all contributing to the gene expression profiles analyzed. Moreover, this can also be due to the role played by the thymus in central tolerance. Indeed, Derbinski et al. (5) demonstrates that medullary TECs express a highly diverse set of genes representing all tissues of the body.
Using a clustering analysis of genes expressed in the thymus of MG and non-MG patients, four gene expression patterns were observed (Fig. 1A): one pattern corresponded to genes commonly dysregulated whatever the MG subgroup and the three others characterized a specific gene expression profile for each MG subgroup. Using SAM with a false discovery rate <5% and a fold change (FC) >1.8, 702 up- and 736 down-regulated genes were identified in MG patients compared with adults (Fig. 1B) (supplemental tables S1 and S2).5 MH patients exhibited a lower number of up-regulated genes than SP and SN patients but their expression level was higher because 18% of these genes displayed an FC >2.5 compared with 8% in ML and 7% in SN patients.
FIGURE 1.
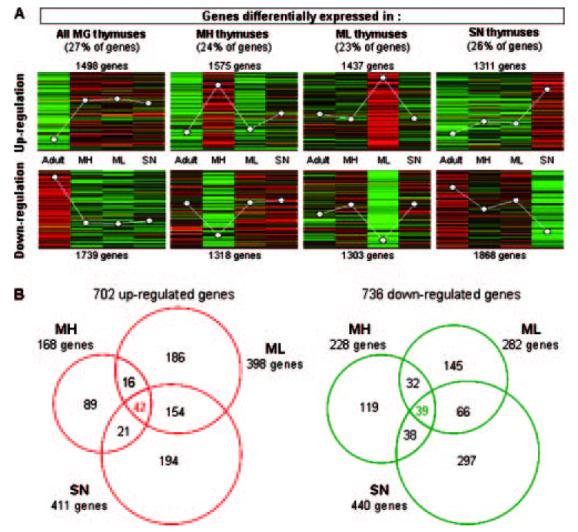
A, Clustering of gene expression in MG subgroups. Among the 13,572 surveyed genes, the Acuity software classified 12,049 GenBanks in eight expression patterns. The white dots represent the average expression level of each adult thymic category compared with the thymic reference. B, Venn diagrams of dysregulated genes in MG patients. Using SAM with a false discovery rate <5% and an FC >1.8, 702 up- (A) and 736 down-regulated (B) genes were identified in ML, MH, and SN patients compared with non-MG adults. The lists of up- and down-regulated genes are given in supplemental tables S1 and S2.
Preferential chromosomal locations of the dysregulated genes
Microarray results can be used to identify chromosomal susceptibility regions associated with pathological conditions. No particular chromosome was found overrepresented. However, we observed a significant overrepresentation of several subchromosomal locations (Fig. 2): for the down-regulated genes, the 3p21 region, and for the up-regulated genes, seven preferential regions. We observed an overrepresentation of regions linked to the Ig clusters: Ig K (2p12), Ig H (14q32), and Ig L (22q11), in all MG subgroups but especially in MH patients. However, the most heavily represented region, in the three MG subgroups, was 6p21, and more precisely 6p21.3, containing the MHC class I and MHC class II gene clusters. The overrepresentation of this region was particularly due to the expression of MHC class II genes (supplemental table S3-A). Two clusters corresponding to MHC paralogous clusters, 1q21 and 9q34, were also identified (17). The other MHC paralogous cluster, in 19p13, was not significant with the χ2 test but was nevertheless strongly represented in each MG subgroup. Although the over-representation of the MHC cluster was predictable, the overall overrepresentation of 1q21 and 9q34 MHC paralogous clusters is surprising as among the significantly up-regulated genes, only two MHC-related genes, thought to have originated by duplication, were observed. They correspond to the orphan receptor ROR γ in the 1q21 region and the bromodomain containing protein 3 in the 9q34 region (18). However, these MHC paralogous clusters might harbor susceptibility genes involved in MG. Indeed, we observed the up-regulation of the IL-6R (1q21 region) and the TNFR-associated factor 2 (9q34 region), two signaling pathways whose dysregulation has already been associated with MG susceptibility (19, 20).
FIGURE 2.
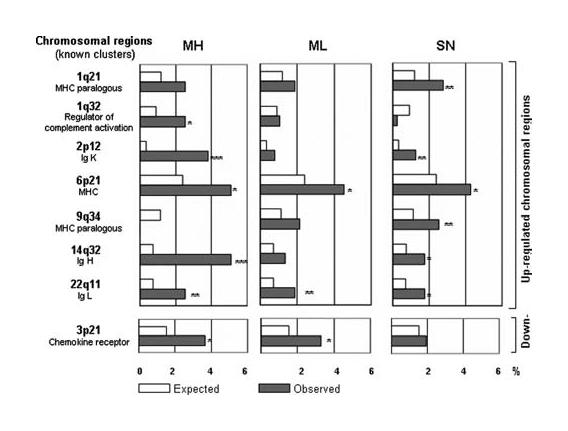
Preferential chromosomal location for up- or down-regulated genes. Among the most highly represented chromosomal regions, a χ2 test was used to search for significant differences between the number of dysregulated genes observed per chromosome or chromosomal location and the number of genes expected on the entire array (55). Eight regions were then identified as significant. Asterisks indicate the p value level: ***, <0.001; **, <0.01, and *, <0.05. The list of the genes located in the overrepresented chromosomal regions is given in supplemental table S3-A.
Gene classification in each MG condition
To characterize the thymus of MG patients, we searched for biological categories overrepresented in each MG subgroup. The up-regulated genes belonged predominantly to the immune response and, in particular, to the Ag presentation category characterized by MHC class II molecules (Table II). This was significant for each subgroup and especially for MH patients. We also found that all MG thymuses were sites of intense transcriptional activity. The up-regulation of genes associated with a chemotactic activity was observed for MH and ML patients. Interestingly, for SN patients, we specifically found genes involved in muscle development for up-regulated genes and in synaptic transmission for down-regulated genes.
Table 2.
Biological process categories overrepresented in each MG subgroupa
| MH | ML | SN | ||||
|---|---|---|---|---|---|---|
| Up-regulated genes | ||||||
| Immune response | 32 | *** | 34 | ** | 28 | NS |
| Ag presentation (MHC class II) | 5 | *** | 4 | * | 5 | ** |
| Response to stress | 19 | *** | 30 | NS | 20 | NS |
| Response to wounding | 11 | *** | 8 | NS | ||
| Inflammatory response | 8 | ** | 6 | NS | ||
| Transcription regulator activity | 15 | NS | 39 | * | 41 | * |
| RNA Pol II transcription factor activity | 7 | * | 13 | * | 11 | NS |
| RNA metabolism | 4 | NS | 15 | * | 11 | NS |
| Intracellular protein transport | 4 | NS | 18 | * | 11 | NS |
| Protein folding | 9 | * | ||||
| Cell growth | 8 | * | 4 | NS | ||
| Muscle development | 10 | ** | ||||
| Down-regulated genes | ||||||
| Cell proliferation | 27 | ** | 28 | NS | 30 | NS |
| Nucleic acid metabolism | 55 | * | ||||
| Response to stress | 25 | * | ||||
| Cell-cell signaling | NS | 25 | * | |||
| Synaptic transmission | 13 | * | ||||
| Protein kinase cascade | 11 | * | ||||
For the up- and down-regulated genes extracted with SAM, the biological process categories significantly overrepresented compared with the entire array were searched in MH, ML, and SN patients using the Expression Analysis Systematic Explorer software (EASE) on the DAVID internet site. In each MG subgroup, categories with at least three genes and with an EASE score <0.05 are listed in this table. The numbers correspond to the numbers of up- and down-regulated genes in MH, ML, or SN patients. Asterisks indicate the significance of the EASE score:
p < 0.001;
p < 0.01;
p < 0.05;
These global analyses show a striking common signature for all MG subgroups, characterized by an up-regulation of the immune response. However, the MH subgroup distinguishes itself by a heightened up-regulation of the immune response and the SN subgroup by a dysregulation of gene categories that might reflect the ability of the thymus to express various tissue-specific Ags.
Detailed analysis of dysregulated genes
Analysis of the genes dysregulated whatever the MG subgroup
As seen from the Venn diagrams (Fig. 1B), 42 up- and 39 down-regulated genes were differentially expressed in all MG subgroups compared with non-MG adults. The up-regulated genes corresponded mainly to the immune response and especially to MHC class II and Ig genes (Table III). An overexpression of B cell-related genes was also observed demonstrating B cell infiltration probably related to the expression of the B cell chemoattractant, CXCL13, whose thymic expression was increased in all MG subgroups.
Table 3.
Classification of the up- and down-regulated genes in all MG subgroupsa
| Biological Process |
Fold Change |
||||
|---|---|---|---|---|---|
| Molecular function | GenBank | MH | ML | SN | Name |
| Up-regulated genes | |||||
| Immune response (19) | |||||
| B cell related | BG759817 | 3.38 | 2.20 | 2.64 | CD79A Ag |
| B cell related | BG024663 | 2.71 | 2.04 | 2.37 | CD20 Ag |
| B cell related | M84371 | 2.63 | 1.99 | 1.88 | CD19 Ag |
| B cell related | M27394 | 1.98 | 2.23 | 2.18 | CD20 Ag |
| Chemokine | AF044197 | 2.82 | 1.97 | 1.95 | CXCL13 |
| Complement component | K02765 | 2.94 | 2.16 | 2.05 | Complement component 3 |
| IFN-induced | AF024714 | 2.09 | 2.44 | 1.99 | IFN-inducible protein (AIM2) |
| IFN-induced | X82200 | 2.06 | 2.11 | 2.26 | Staf50 |
| Ig family | AI634950 | 5.06 | 2.09 | 2.76 | Ig H constant μ |
| Ig family | BM008087 | 3.42 | 2.53 | 3.84 | Ig H constant μ |
| Ig family | Z98733 | 2.80 | 2.79 | 2.55 | Ig H chain variable region |
| Ig family | M63438 | 2.63 | 3.43 | 2.11 | Ig κ chain variable region (IGKV gene) |
| Ig family | Z46347 | 2.56 | 1.97 | 2.02 | Ig κ chain V-IV region B17 precursor |
| Ig family | Y14738 | 2.32 | 2.46 | 2.33 | Ig λ L chain |
| Ig family | M63438 | 2.17 | 2.99 | 2.29 | Ig κ chain variable region (IGKV gene) |
| Ig family | D87023 | 2.09 | 2.65 | 2.09 | Ig λ |
| MHC class II | BF795929 | 2.47 | 2.27 | 2.85 | MHC class II, DR α |
| MHC class II | BG619272 | 2.31 | 2.72 | 2.98 | MHC class II, DQ β 1 |
| MHC class II | M24364 | 2.13 | 1.97 | 1.89 | MHC class II, DQ β 1 |
| Transcription (11) | |||||
| Transcription factor activity | BG760057 | 3.72 | 2.38 | 2.14 | T-cell leukemia/lymphoma 1A |
| Transcription factor activity | AV747778 | 4.15 | 4.20 | 2.48 | v-fos FBJ murine osteosarcoma viral oncogene homolog |
| Transcription factor activity | NM_003451 | 3.63 | 3.45 | 3.70 | Zinc finger protein 177 |
| Transcription factor activity | D31716 | 3.56 | 4.03 | 2.46 | Basic transcription element-binding protein 1 |
| Transcription factor activity | U20734 | 2.94 | 2.26 | 2.16 | Transcription factor junB (junB) |
| Transcription factor activity | M92844 | 2.41 | 3.06 | 2.25 | Zinc finger transcriptional regulator (GOS24) |
| Transcription factor activity | AL549846 | 2.07 | 1.95 | 2.16 | TGF β-induced factor |
| Transcription factor activity | X63741 | 1.80 | 2.04 | 1.85 | Early growth response 3 |
| Transcriptional regulation | BF305705 | 2.07 | 2.51 | 2.43 | Histone deacetylase 5 |
| Transcriptional regulation | R56503 | 1.94 | 2.06 | 2.01 | SWI/SNF-related protein, subfamily a, member 2 |
| Transcriptional regulation | U66615 | 1.89 | 2.07 | 2.28 | SWI/SNF-related protein, subfamily c, member 1 |
| Cellular/intracellular signaling (5) | |||||
| Calcium-binding protein | AV713821 | 2.64 | 1.92 | 2.22 | S100 calcium-binding protein A4 |
| Calcium-binding protein | BC009846 | 1.96 | 2.32 | 2.10 | Hippocalcin-like 1 |
| Cell adhesion | AU124258 | 1.89 | 2.07 | 2.06 | Desmoglein 2 |
| Channel | U72245 | 1.80 | 1.96 | 1.91 | Phospholemman chloride channel |
| Receptor activity | BG325212 | 2.67 | 2.11 | 2.08 | IGF-2 receptor |
| Cellular metabolism (4) | |||||
| Nucleotide metabolism | AL535356 | 2.97 | 2.27 | 2.70 | Ribosomal protein S7 |
| Nucleotide metabolism | BI259616 | 2.73 | 3.19 | 1.99 | Uracil-DNA glycosylase 2 |
| Nucleotide metabolism | BG252637 | 1.96 | 2.56 | 3.03 | Prostatic binding protein |
| Protease | AL541945 | 2.15 | 2.39 | 2.57 | Serine proteinase inhibitor, clade F, member 1 |
| Divers (3) | |||||
| Unknown | 2.18 | 2.11 | 2.11 | Incyte EST | |
| Unknown | Z69892 | 2.02 | 1.85 | 2.16 | Clone ICRFp507I1077 |
| Unknown | AL050127 | 1.93 | 2.17 | 2.22 | Clone DKFZp586G021 |
| Down-regulated genes | |||||
| Cellular metabolism (13) | |||||
| ATP binding activity | D29641 | −2.17 | −1.93 | −2.12 | Helicase SKI2W |
| ATP binding activity | U56602 | −2.28 | −2.43 | −2.21 | Peroxisomal biogenesis factor 6 |
| ATP biosynthesis | D50371 | −2.19 | −2.16 | −1.94 | ATP synthase subunit e |
| Cell cycle | D86958 | −1.87 | −1.94 | −1.95 | RB1-inducible coiled-coil 1 |
| Protein processing | AF110647 | −1.93 | −2.12 | −2.05 | Translocon-associated protein γ subunit |
| Protein processing | BC001687 | −2.05 | −2.21 | −1.91 | Asparaginyl-tRNA synthetase |
| Protein processing | BF796127 | −4.74 | −4.34 | −3.71 | Vacuolar protein sorting 45B |
| Ribosome biogenesis | AL526336 | −2.14 | −2.42 | −1.97 | EBNA1-binding protein 2 |
| RNA binding | BE501622 | −2.20 | −2.29 | −1.85 | Small nuclear ribonucleoprotein polypeptide A |
| Transporter activity | AF176008 | −2.30 | −2.12 | −2.12 | Mitochondrial carrier homolog 2 |
| Transporter activity | BE275227 | −1.98 | −2.40 | −1.88 | Thioredoxin-like 4 |
| Ubiquitin cycle | AL136812 | −1.95 | −2.21 | −2.10 | Makorin 1 |
| Unclassified | AF017445 | −2.22 | −2.29 | −2.72 | Fucose-1-phosphate guanylyltransferase |
| Immune response (8) | |||||
| Autoantigen | NM_006029 | −2.19 | −3.00 | −2.22 | Paraneoplastic Ag MA1 |
| Cell adhesion | BG770327 | −2.00 | −2.26 | −1.98 | Galectin 3 |
| Complement component | BF338113 | −2.71 | −2.14 | −2.49 | Complement component 4B |
| Kinase activity | AF153419 | −4.36 | −4.62 | −5.15 | Iκ B kinase complex-associated protein (IKBKAP) |
| MHC related | AK027789 | −2.00 | −2.38 | −2.14 | SIMP |
| MHC related | AL050135 | −1.83 | −1.83 | −1.86 | Regulatory factor X, 5 |
| MHC related | BC008783 | −2.67 | −2.27 | −2.26 | HLA-B associated transcript 4 |
| Transcriptional regulation | T27711 | −1.84 | −2.28 | −2.32 | BCL6A |
| Cellular/Intracellular signaling (9) | |||||
| Cell adhesion | J03202 | −1.92 | −2.46 | −2.26 | Laminin B2 chain |
| Cytoskeleton | BC006141 | −1.87 | −2.18 | −2.16 | Erythrocyte protein band 4.1-like 3 |
| Cytoskeleton | M69225 | −2.14 | −2.10 | −2.11 | Dystonin |
| GTPase activity | AK027730 | −2.14 | −2.28 | −2.29 | RAB2B, member RAS oncogene family |
| Kinase activity | BC001662 | −2.07 | −1.91 | −2.02 | MAPK-activated protein kinase 3 |
| Kinase activity | BG391140 | −1.82 | −2.13 | −2.03 | c-src tyrosine kinase |
| Phosphatase | BC001095 | −1.95 | −1.92 | −2.17 | Protein phosphatase 2, regulatory subunit B, δ isoform |
| Protein interactions | BC009241 | −2.36 | −2.30 | −1.88 | TRK-fused gene |
| Receptor activity | AW020536 | −2.17 | −2.09 | −1.96 | Thyroid hormone receptor coactivating protein |
| Transcription (5) | |||||
| Transcription factor activity | AB007885 | −2.10 | −2.08 | −1.85 | Zinc finger protein 262 |
| Transcription factor activity | AB011102 | −1.88 | −2.30 | −2.39 | Zinc finger protein 292 |
| Transcription factor activity | AF230388 | −2.39 | −2.43 | −1.94 | Tripartite motif-containing 29 |
| Transcriptional regulation | BC004227 | −2.17 | −2.27 | −1.80 | Metastasis-associated family, member 3 |
| Transcriptional regulation | BE254018 | −2.12 | −2.51 | −2.16 | SWI/SNF-related protein, subfamily d, member 2 |
| Divers (4) | |||||
| Unknown | AAH04632 | −1.89 | −2.19 | −2.58 | Ubxd4-related protein |
| Unknown | AF281064 | −2.29 | −1.80 | −1.84 | CHMP1.5 protein |
| Unknown | AK026058 | −2.15 | −2.35 | −2.26 | Clone HRC08294 |
| Unknown | X75191 | −1.92 | −2.09 | −1.84 | Genomic YAC-end left arm of chromosome 15 |
The 42 up-regulated and 39 down-regulated genes in MH, ML, and SN patients were manually classified. For each gene, we give the fold change for the different MG subgroup compared to non-MG adults.
To determine whether the overexpression of Ig genes in the thymus of MG patients reflected an increased number of B cells or B cell hyperactivity, we compared the relative proportion of B cells with the global level of Ig expression. For this purpose, the intensity medians for Ig genes dysregulated in at least one MG subgroup were compared with those for specific B cell markers (reflecting the proportion of B cells within the thymus). Microarray global analyses were validated by measuring, using real-time PCR, the well-known B cell marker CD20 and, by immunonephelometry, the expression level of IgG and IgM. Of note, comparing the thymus of babies and non-MG adults, we did not observe significant difference in the expression level for the B cell markers (Fig. 3, A and B) but did observe a significant overexpression of Igs in adults with an FC of 2.9 ( p = 0.0003) for microarray data (Fig. 3C) and an FC of 2.1 (p = 0.0095) for direct Ig quantification (Fig. 3D). These analyses show the lower ability of B cells to express Igs in babies compared with adults and underlines the fact that adult B cells can respond more efficiently to an immune stress by expressing more Abs (21).
FIGURE 3.
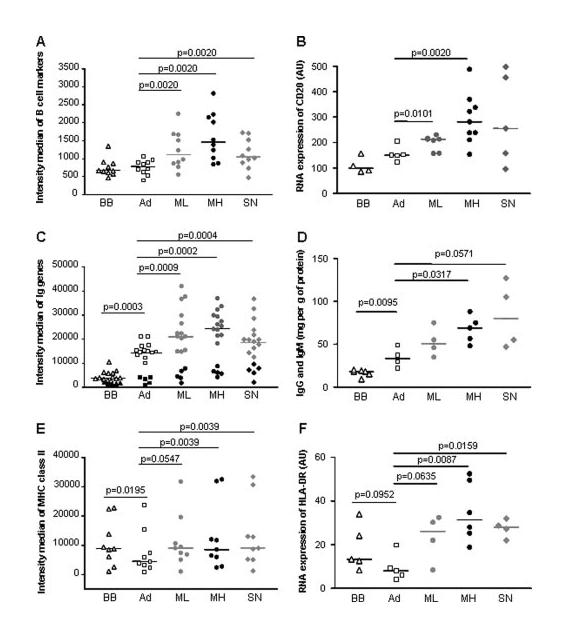
Gene expression of specific B cell markers. A, Microarray intensity medians for specific B cell markers: AF208502, BG024663, BG754034, BG759817, J03565, M27394, M80461, M84371, X66079, and Z29575. B, Analysis by real-time PCR of CD20 expression in the thymus of individual donors corresponding to babies (BB), non-MG adults (Ad), and ML, MH, SN patients. Ig expression in thymic extracts. C, Microarray intensity medians for Ig genes extracted with SAM analyses: Ig L chain genes (open or gray symbols) correspond to AF124182, AJ010442, BC005332, BF338816, D87023, M63438, M87790, S49006, S50732, X57812, Y14736, Y14738, and Z46347, and, Ig H chain genes (black symbols) to AF067420, AI634950, BF663123, BM008087, and Z98733. D, Quantification by immunonephelometry of IgG and IgM levels in the thymus of individual donors corresponding to babies (BB), non-MG adults (Ad) and ML, MH, SN patients. MHC class II expression in thymic extracts. E, Microarray intensity medians for MHC class II genes extracted with SAM analyses: AI249268, AL543515, BF795929, BG176768, BG619272, M24364, M27487, U83582, and X00033. A dot corresponds to the median of normalized intensities for each gene calculated for babies (BB) ( ), non-MG adults (Ad) (
), non-MG adults (Ad) ( ) and ML (
) and ML ( ), MH (
), MH ( ), SN (
), SN ( ) patients. The p values were obtained by the Wilcoxon matched pairs test. F, Analysis by real-time PCR of HLA-DR expression in the thymus of individual donors corresponding to babies (BB), non-MG adults (Ad) and ML, MH, SN patients.
) patients. The p values were obtained by the Wilcoxon matched pairs test. F, Analysis by real-time PCR of HLA-DR expression in the thymus of individual donors corresponding to babies (BB), non-MG adults (Ad) and ML, MH, SN patients.
In the thymus of MG patients, as expected, a significant increased expression of the B cell markers was observed for MH patients with an FC of 2.3 compared with adults (p = 0.0020) (Fig. 3A). However, we also pointed out a significantly increased B cell population in ML with an FC of 1.8 (p = 0.0020) and SN patients with an FC of 1.7 (p = 0.0020) (Fig. 3A). This increase of B cells in all MG subgroups observed with the microarray data was also validated by analyzing directly CD20 mRNA expression on individual patients (Fig. 3B). In parallel, Ig populations increased in all MG subgroups with fold changes of 1.8 and 1.6 times for ML, 2.2 and 1.7 times for MH, and 2.3 and 1.4 times for SN, respectively, for Ig H chains (p < 0.0625 for all MG categories) and Ig L chains (p < 0.0024 for all MG categories) (Fig. 3C). This overexpression of Ig in all MG patients was also observed at the protein level in thymic extracts (Fig. 3D). The similar amplification levels for B cell markers and Ig genes suggest that Ig increases were linked to the B cell increases. This observation is supported by our microarray results showing a significant correlation between B cell marker and Ig expression comparing the expression mean for the different adult categories (r2 = 0.922; p = 0.0399). This correlation was also observed between CD20 mRNA expression level and IgM-IgG production (r2 = 0.9742 p = 0.0103). Consequently, the up-regulation of Igs observed in the thymus of MG patients seems not due to an overproduction of Igs by a limited pool of B cells, but to the increased number of B cells. Moreover, taking into account all the Ig spotted on the arrays, we observed a global significant increase of their expression whatever their specificity, suggesting B cell polyclonality (our unpublished data).
In this study, a thymic overexpression of MHC class II molecules was also detected whatever the MG subgroup (Fig. 3E). HLA-DR was among the most up-regulated MHC class II genes in MG patients and we confirmed its overexpression in all MG patients by real-time PCR (Fig. 3F). The increase of MHC class II Ags could be either due to an increased number of cells expressing Ags or could be due to a proinflammatory environment, or to both. MHC class II are known to be expressed by B cells, we conducted similar analyses to that described above for Igs. We observed that hyperplastic thymuses with numerous GCs, and consequently more B cells (Fig. 3A), were not characterized by an increased expression of MHC genes compared with ML and SN patients. Moreover, comparing the expression mean for the different adult categories for either our microarray data (MHC class II vs B cell marker expression) or real-time PCR data (HLA-DR vs CD20 expression), we did not find any correlation between B cell populations and MHC class II expression. These results suggest that the increased expression of MHC components is not directly related to the increased B cell number rather supporting the inflammatory state of the MG thymus.
Analysis of the genes characterizing the hyperplastic thymus
Using SAM, 168 up- and 225 down-regulated genes were identified in the thymus of MH patients (supplemental tables S3-B and S3-C) and a quarter of them are associated with the immune response (Fig. 4). The up-regulated genes were subclassified in categories reflecting an inflammatory state and the presence of numerous GCs: Ig family, B cell-related genes, IFN-induced genes, and MHC class II molecules (Fig. 4, A and B). Moreover, we detected an up-regulation of chemokine genes and, in particular, CCL21 known to be involved in the homing of T cells to secondary lymphoid organs (22). A detailed analysis of the thymic chemokine expression showed that the number of up-regulated chemokines observed was significantly overrepresented compared with their expected number on the arrays in SP MG patients (Fig. 5A), and especially in MH patients. These results were in agreement with the chemotaxis assays, because a significant increased migration of PBL toward hyperplastic thymic extracts was found. In contrast, no overrepresentation of chemokines in the thymus of SN patients was observed. Moreover, thymic extracts from SN patients even displayed a decreased chemotactic activity (Fig. 5B).
FIGURE 4.
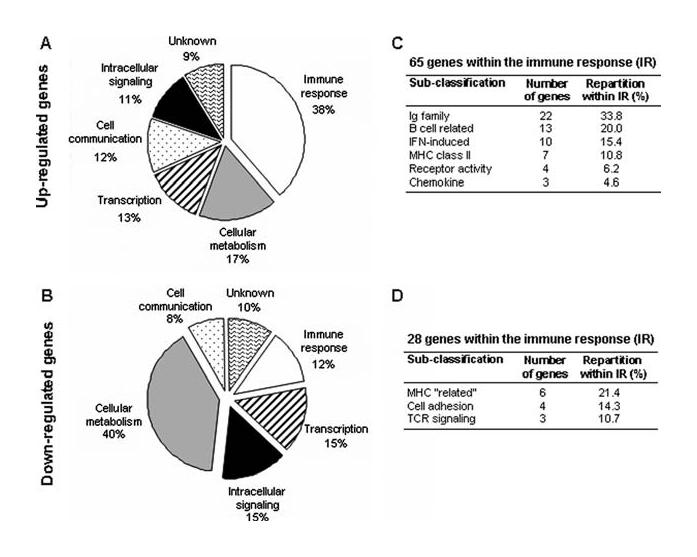
Classification of dysregulated genes in MH patients. The classification of 168 up- and 228 down-regulated genes in MH patients according to biological functions is given in A and B. The subclassification of up- and down-regulated genes belonging to the immune response category is presented in C and D. Only categories with at least three genes were indicated. The classification into functional categories was conducted using NCBI, SOURCE, and DAVID databases, and the detailed list of these genes is given in supplemental tables S3-B and S3-C.
FIGURE 5.
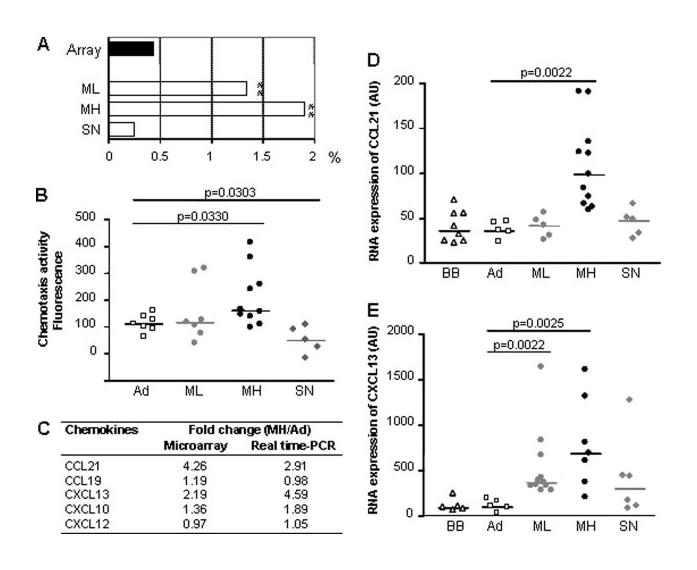
Chemokine expression analysis in MG patients. A, Percentages of chemokines observed among the up-regulated genes in ML, MH, and SN patients and present in the entire array (■). The stars (**) indicate a significant (p < 0.001) overrepresentation of the chemokine number using a χ2 test. B, Migration of PBL toward thymic extracts from individual donors corresponding to non-MG adult (Ad) or ML, MH, SN patient categories. Each dot corresponds to fluorescence mean of duplicate measures. C, Chemokine expression level in MH patients compared with non-MG adults (Ad) obtained by microarray and real-time PCR. D and E, Analysis by real-time PCR of CCL21 and CXCL13 expression in the thymus of individual donors corresponding to babies (BB), non-MG adults (Ad) and ML, MH, SN patients. The p values were obtained by the Mann-Whitney U test.
To confirm our microarray data, the expression of several chemokines, known to participate in GC formation, was analyzed by real-time PCR for MH patients (Fig. 5C). We confirmed the up-regulation of CCL21 and CXCL13. Like CCL21, CCL19 acts through CCR7, but no up-regulation of CCL19 was observed. An increased expression of CXCL10, a chemoattractant for activated T cells, was also detected, as previously described (23). However, we did not notice any MH-specific increase in the expression of CXCL12 known to contribute to the T cell homing and GC organization (22, 24).
The important up-regulation of CCL21 and CXCL13 could be directly involved in GC formation in the thymus of MG patients. We therefore investigated their expression by real-time PCR in independent MG thymus samples. A significant and specific over-expression of CCL21 was showed in the thymus of MH patients (Fig. 5D). For CXCL13, its expression was significantly up-regulated in the thymus of ML and MH patients and slightly in SN patients (Fig. 5E). Consequently, the high expression of CXCL13 could account for the increased B cell population observed in all MG thymuses and the specific overexpression of CCL21 in the thymus of MH patients could be involved in ectopic GC formation.
Analysis of the genes characterizing the thymus of SN patients
Based on the expression analysis systematic explorer (EASE) classification, the thymic transcriptome of SN patients was characterized by the up-regulation of genes belonging to the “muscle development” category and the down-regulation of genes corresponding to the “synaptic transmission” category (Table II). More interestingly, these genes were significantly dysregulated in SN compared with non-MG adults, but also to ML and MH patients (Fig. 6).
FIGURE 6.

Biological process categories overrepresented in SN patients. The list of the up-regulated muscle development genes and the down-regulated synaptic transmission genes are given in A and B, respectively. These categories were identified using the EASE software (Table II) and for each gene, the FC between each MG subgroup and non-MG adults is given. The graphs represent the gene expression ratios for non-MG adults (Ad) ( ) and ML (
) and ML ( ), MH (
), MH ( ), SN (
), SN ( ) patients compared with the thymic reference for the muscle development genes (C) and the synaptic transmission genes (D). Each dot corresponds to the median of ratios for a gene. The p values were obtained by the Wilcoxon matched pairs test.
) patients compared with the thymic reference for the muscle development genes (C) and the synaptic transmission genes (D). Each dot corresponds to the median of ratios for a gene. The p values were obtained by the Wilcoxon matched pairs test.
The synaptic transmission genes corresponded mainly to neurotransmitter receptors or protein kinases involved in the downstream signaling of these receptors (Fig. 6C). The muscle development genes are essentially cytoskeletal components implicated in muscle contraction (Fig. 6A). Their overexpression could reflect changes in a cell population in the thymus of SN patients. Because thymic myoid cells express high levels of muscle genes and could participate in autosensitization toward muscle Ags in MG (25), we investigated the myoid cell number on thymic sections. However, no increase of myoid cell number was detected, and even fewer myoid cells were enumerated in SP and SN thymuses compared with adults (our unpublished data). In a more thorough study, I. Leite (unpublished observation) also observed for these patient categories, enormous variability in the numbers of myoid cells, but some overall decrease per mm2 of medullary parenchymal tissue. It is worth noting that the decreased number of myoid cells might explain the decreased expression of the muscle AChR-α subunit detected with our microarray data (data not shown).
To further analyze the 851 genes dysregulated in SN patients (Fig. 1B), we compared their expression between SN and ML patients. In this analysis, we did not include MH patients, as their thymic transcriptome was very different due to the presence of numerous GCs. Using the Mann-Whitney U test, 42 up- and 48 down-regulated genes were identified in SN compared with ML patients (supplemental table S3-D). We regarded as interesting genes dysregulated with an FC >1.8 and found 6 up- and 3 down-regulated genes (Fig. 7A). Among the up-regulated genes were ISG12, DNAX-activating protein of 12 kDa (DAP12), and SON DNA-binding protein, all identified as involved in antiviral activity. As ISG12 has previously been associated with autoimmune diseases (26, 27), we investigated its expression by RT-PCR (Fig. 7, B and C): ISG12 was significantly up-regulated in SN patients compared with non-MG adults and also compared with ML and MH patients. To strengthen the relevance of this antiviral signature in SN patients, we analyzed the expression of a nonexhaustive list of antiviral genes clearly defined to be induced in response to pathogen infection. A significant overexpression of these genes was observed in SN patients compared with adults and ML patients (Fig. 7D). This overexpression of these genes reflects most probably an antiviral signature and not a proinflammatory response linked to IFN-α. Indeed, we previously demonstrated an up-regulation of IFN-α-regulated genes in MG patients with a high hyperplasia (MH) (16) but this was not observed for SN patients (data not shown). Moreover, among the listed genes, only 7 of 19 genes have a link more or less obvious with an IFN-α response.
FIGURE 7.

Analysis of specific genes up-regulated in SN patients. A, Details about the six genes significantly up-regulated in SN patients compared with non-MG adults and ML patients with an FC >1.8. The complete list of the genes dysregulated in SN compared with ML patients is given in supplemental table S3-D. B, Gene expression ratio of ISG12 for non-MG adult (Ad), ML, MH, or SN patient categories compared with the thymic reference by microarray. C, Analysis by real-time PCR of ISG12 expression in the thymus of individual donors corresponding to baby (BB), Ad, ML, MH, and SN categories. The p values were obtained by the Mann-Whitney U test. D, Expression of genes involved in the antiviral response: AA477235, AF019562, AF029890, AF083033, AI097512, AK024814, AL110236, AL582281, AW083130, AW328025, BC007402, BE857745, BG392389, BG506643, J04739, M30818, NM_021105, NM_033238, and X67325. A dot corresponds to the median of ratios between each thymic category and the thymic reference for each gene. The p values were obtained by the Wilcoxon matched pairs test.
Therefore, we demonstrate that thymus of SN patients differs from that of SP patients with an abnormal expression of muscle development and synaptic transmission genes but also with an antiviral signature.
Discussion
In the present study, comparing the thymic transcriptome of different MG subgroups, we demonstrate 1) a common gene expression profile associated with an important immune response suggesting an active role of the thymus in all MG subgroups studied, 2) that hyperplastic thymuses are characterized by an up-regulation of chemokines involved in ectopic GC formation, and 3) in SN patients, a dysregulation of genes involved in muscle development or synaptic transmission but also genes featuring an antiviral response.
Active role of the thymus for all MG subgroups studied
Our analyses demonstrate that the thymus of all MG patients is characterized by an intense transcriptional activity, an exacerbated immune response and, in particular, an abnormal increased B cell population, underlying originally that even nonhyperplastic thy-muses possess the characteristics of diffuse B cell infiltration. This B cell signature could be due to CXCL13, a chemokine overex-pressed in all MG thymuses and known to be involved in the homing of B cells in secondary lymphoid organs (28). Indeed, the transgenic expression of CXCL13 in β cells of the pancreatic islets mediates the recruitment of a large number of B cells (29).
The thymus of SP patients has been shown to include all the components of the anti-AChR response including B cells producing anti-AChR Abs, suggesting a possible monoclonal expansion of B cells (30). Using this microarray approach, we observe an overall increased expression of Ig genes, whatever their specificity, demonstrating the large diversity of B cells in MG thymuses. This observation is supported by Sims et al. (31) who showed that thymic GCs in MG contain a remarkably heterogeneous population of B cells, and by our work demonstrating, by in situ hybridization, the polyclonality of B cells included in individual thymic GC of MG patients (32).
The up-regulation of MHC class II components observed in the thymus of all MG subgroups was not directly related to the higher number of B cells but could be due to a generalized overexpression by various thymic cells, such as dendritic cells and TECs due to the proinflammatory state of MG thymuses. In that way, in our microarray study, CIITA (a transcription factor regulating MHC class II molecule expression (33)) is slightly overexpressed in the thymus of MG patients (our unpublished data). Consequently, MHC class II overexpression could promote an increased Ag presentation in the thymus leading to autoimmunity. This dysregulation leads to an overrepresentation of the 6p21.3 MHC cluster commonly associated with autoimmune diseases, but we also observe an up-regulation of genes from MHC paralogous clusters. These clusters correspond to disease-predisposition loci and we observe for the first time the involvement of MHC paralogous clusters altogether with the MHC cluster in an autoimmune disease. A common regulator of their transcription, conserved throughout evolution, could explain the overall up-regulation of genes from the MHC and MHC paralogous clusters (34).
These thymic transcriptome analyses underline the inflammatory state of MG thymuses, even for patients without thymic hyperplasia, demonstrating the active role of thymus in all MG subgroups studied. Thus, thymectomy might be beneficial for SN patients for whom the benefice of thymectomy has not yet been clearly defined. Considering this generalized thymic inflammatory state in MG, we can wonder why much more MG thymuses are not hyperplastic? Genes specifically dysregulated in the thymus of MH patients might be indispensable to trigger ectopic GC formation leading to hyperplasia, such as the chemokine CCL21 discussed below. However, genes specifically expressed in ML and SN thy-muses might also prevent hyperplasia and interestingly, we observed a decreased chemotaxis activity of thymic extracts from SN patients.
Involvement of chemokines in hyperplastic thymuses
Thymic hyperplasia observed in MH patients is characterized by the presence of numerous GCs (6) and a striking feature of the thymic transcriptome of these patients is the overexpression of CCL21 compared with adults and other MG subgroups. A previous analysis comparing exclusively patients with thymic hyperplasia to non-MG adults also underlines the CCL21 up-regulation (35). CCL21 and CCL19 act similarly through the same receptor, CCR7, but no up-regulation of CCL19 is observed. In the normal thymus, CCL21 and CCL19 play an important role in lymphopoiesis. They are secreted by medullary TECs and favor the migration of CD4+ and CD8+ single-positive thymocytes toward the medullary zone (24). However, these two chemokines are better known for their role in immunopoiesis. They are involved in the homing of Ag-primed dendritic cells and T lymphocytes to secondary lymphoid organs and, consequently, in the initiation of the GC formation (22). Indeed, plt mice devoid of CCL19 and CCL21 expression and CCR7-deficient mice exhibit defective GC formation with a default in the migration of lymphocytes and dendritic cells into T cell zones (36). Moreover, a model of transgenic mice expressing CCL21 from the thyroglobulin promoter demonstrates that the overexpression of this chemokine induces GC formation in the thyroid (37). Consequently, the specific up-regulation of CCL21 in the thymus of MH patients could be involved in ectopic GC formation. It is noteworthy that we previously demonstrated the significantly increased expression of IFN-inducible genes in the thymus of MH patients (16). The increased expression of inflammatory cytokines, such as IFN-γ, could contribute to the initiation of the autoimmune anti-AChR response. Moreover, the simultaneous overexpression of CCL21 and IFNs could amplify thymic hyperplasia, as IFNs has been shown to enhances the effects of CCL21 (38, 39).
Cooperation between CCL21 and CXCL13 seems indispensable to orchestrate GC formation in secondary lymphoid organs during immune responses (22). Moreover, their overexpression is observed in autoimmune diseases characterized by ectopic GC development (40, 41). We thus assume that the specific overexpression of CCL21 in hyperplastic thymuses, together with the CXCL13 up-regulation observed in all MG subgroups, could be responsible for ectopic GC formation in the thymus of MG patients. It is now important to determine the thymic cells responsible for the abnormal expression of these chemokines and the signals triggering their overproduction. We already observed that CXCL13 and CCL21 overexpression were not directly due to the presence of GCs and we even showed that CXCL13, but not CCL21, was overexpressed by TECs of MG patients (R. Le Panse, J. Bismuth, and S. Berrih-Aknin, manuscript in preparation) (42).
Peculiar gene expression signatures for the thymus of SN patients
SN MG patients are not well-characterized. This study shows that their thymus is characterized by a down-regulation of genes involved in synaptic transmission and an up-regulation of genes involved in muscle development and in the antiviral response. The dysregulation of these genes could be due to changes concerning particular cell populations but also to modifications in the ability of medullary TECs to express tissue-specific Ags. The autoantigenic target(s) involved in SN patients are not yet known. Consequently, genes dysregulated specifically in the thymus of SN patients, and known to play a role in the neuromuscular conduction, could correspond to potential autoantigenic targets. It is now well-established that neurotransmitters are expressed by thymocytes and thymic stromal cells and modulate various immune responses (43). Neurotransmitter receptors are known to be involved in autoimmune diseases leading to neurological disorders, such as MG (44). Therefore, we can thus wonder whether the neurotransmitter receptors (GABA-A, glutamate, and dopamine receptors) down-regulated in SN patients could represent antigenic targets in this category of MG patients. Drugs acting at the GABA-A receptor complex are known to have muscle-relaxant properties, but it is not clear whether this effect is due to a central or peripheral action (45). Consequently, we can hypothesize that autoantibodies targeting these receptors at the muscle endplate might lead to MG symptoms. However, further investigations are necessary to determine whether a link exists between the down-regulation of these particular neurotransmitter receptors and an unknown autoantigenic target in SN patients.
The muscle development genes up-regulated correspond to myofibrillar proteins. We are not able to clearly explain the up-regulation of these genes in SN patient thymuses. Since the 1960s, studies have demonstrated the presence of Abs against structural proteins of muscle such as tropomyosin, myosin, and actin in the sera of MG patients (46-48). The inflammatory state of MG thymuses suggested above, together with the degradation of cells expressing these Ags, such as myoid cells, could trigger their antigenic presentation. However, because these Ags are intracytoplasmic, their direct relevance in auto-immune mechanisms is not clear.
The antiviral signature detected in the thymus of SN patients was characterized by the overexpression of ISG12, DAP12, and SON protein. Thanks to diverse microarray studies, ISG12 over-expression has been observed in various pathological conditions. The exact function of ISG12 is not known, but it could possess a neuroprotective role against viral infections (49). Moreover, an IGS12 overexpression was also observed in autoimmune diseases such as Sjogren's syndrome and systemic lupus erythematosus (26, 27). DAP12 is involved in immune functions and, especially, in NK cell-mediated resistance to an infection (50). Moreover, DAP12−/− mice are resistant to induced experimental autoimmune encephalomyelitis (51). Concerning, the SON protein, this host gene represses hepatitis B virus activity (52) and it is worth noting that MG has been punctually related to infection with hepatitis C or B virus.
The antiviral signature characteristic of SN patients could be related to the etiology of this MG subgroup. Indeed, viral infections have been associated with the development of several auto-immune diseases (53). However, because the symptoms related to the autoimmune disease generally occur well after infections, it is difficult to link these two events. Nevertheless, molecular mimicry suggests that autoimmune diseases can directly be caused by a cross-reaction of an Ab against a microbial Ag that closely resembles a self-Ag. In that way, some MG patients possess Abs to the AChR which share a cross-reactive epitope with the HSV, glycoprotein D (54). Another mechanism of infection-induced autoimmunity, referred as the bystander effect, occurs secondary to the infection. The initial viral infection induces a localized inflammation of the target organ and triggers an autoantigen sensitization. In this case, an additional increased expression of cytokines during the viral response could trigger autoimmune diseases (53).
In conclusion, these thymic transcriptome analyses of MG patients suggest many novel approaches to further understand the disease. For SP patients, we are currently investigating in detail the role of CCL21 and CXCL13 in thymic hyperplasia. Drug discovery programs are already in progress to block chemokine-receptor interactions in some autoimmune diseases. We intend to evaluate whether blocking CCL21 and/or CXCL13 could represent novel therapies to reverse thymic hyperplasia development in MG patients avoiding thymectomy and the use of nonspecific glucocorticoid treatments. For SN patients in whom the antigenic target is not yet defined, this study identified genes involved in the synaptic transmission that could potentially correspond to autoimmune targets. Moreover, our results put forward that viral infection might be a key event in the triggering of SN MG.
Supplementary Material
Acknowledgments
We thank Sylvia Cohen-Kaminsky for helpful discussions, Geneviève Piétu for her assistance in setting up our microarray platform, Florent Crépineau for his computing science skills, Corinne Barthet for Ig quantification, Philippe Dartevelle and Alain Serraf for the thymic samples, and Vincent de Montpreville for histological analyses.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
This work was supported by grants from the National Institutes of Health (NS 39869), the European Community (QLG1-CT 2001-01918 and QLK3-CT 2001-00225), and the Association Française contre les Myopathies.
Abbreviations used in this paper: MG, myasthenia gravis; SP, seropositive; AChR, acetylcholine receptor; TEC, thymic epithelial cell; SN, seronegative; MuSK, muscle-specific tyrosine kinase; GC, germinal center; ML, SP MG patients with low thymic hyperplasia; MH, SP MG patients with high thymic hyperplasia; FC, fold change; ISG, IFN-stimulated gene; DAP12, DNAX-activating protein of 12 kDa.
The online version of this article contains supplemental material.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Aharonov A, Abramsky O, Tarrab-Hazdai R, Fuchs S. Humoral antibodies to acetylcholine receptor in patients with myasthenia gravis. Lancet. 1975;2:340–342. doi: 10.1016/s0140-6736(75)92779-8. [DOI] [PubMed] [Google Scholar]
- 2.Evoli A, Batocchi AP, Lo Monaco M, Servidei S, Padua L, Majolini L, Tonali P. Clinical heterogeneity of seronegative myasthenia gravis. Neuromuscul. Disord. 1996;6:155–161. doi: 10.1016/0960-8966(96)00009-0. [DOI] [PubMed] [Google Scholar]
- 3.Hoch W, McConville J, Helms S, Newsom-Davis J, Melms A, Vincent A. Auto-antibodies to the receptor tyrosine kinase MuSK in patients with myasthenia gravis without acetylcholine receptor antibodies. Nat. Med. 2001;7:365–368. doi: 10.1038/85520. [DOI] [PubMed] [Google Scholar]
- 4.Berrih-Aknin S, Eymard B. Thymus et pathologies. Biologie. 1999;5:579–585. [Google Scholar]
- 5.Derbinski J, Gabler J, Brors B, Tierling S, Jonnakuty S, Hergenhahn M, Peltonen L, Walter J, Kyewski B. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J. Exp. Med. 2005;202:33–45. doi: 10.1084/jem.20050471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berrih-Aknin S, Morel E, Raimond F, Safar D, Gaud C, Binet J, Levasseur P, Bach J. The role of the thymus in myasthenia gravis: immunohistological and immunological studies in 115 cases. Ann. N.Y. Acad. Sci. 1987;505:50–70. doi: 10.1111/j.1749-6632.1987.tb51282.x. [DOI] [PubMed] [Google Scholar]
- 7.Kuks JB, Oosterhuis HJ, Limburg PC, The TH. Anti-acetylcholine receptor antibodies decrease after thymectomy in patients with myasthenia gravis: clinical correlations. J. Autoimmun. 1991;4:197–211. doi: 10.1016/0896-8411(91)90018-8. [DOI] [PubMed] [Google Scholar]
- 8.Wakkach A, Guyon T, Bruand C, Tzartos S, Cohen-Kaminsky S, Berrih-Aknin S. Expression of acetylcholine receptor genes in human thymic epithelial cells: implications for myasthenia gravis. J. Immunol. 1996;157:3752–3760. [PubMed] [Google Scholar]
- 9.Safar D, Berrih-Aknin S, Morel E. In vitro anti-acetylcholine receptor antibody synthesis by myasthenia gravis patient lymphocytes: correlations with thymic histology and thymic epithelial-cell interactions. J Clin. Immunol. 1987;7:225–234. doi: 10.1007/BF00915728. [DOI] [PubMed] [Google Scholar]
- 10.Melms A, Schalke BC, Kirchner T, Muller-Hermelink HK, Albert E, Wekerle H. Thymus in myasthenia gravis: isolation of T-lymphocyte lines specific for the nicotinic acetylcholine receptor from thymuses of myasthenic patients. J. Clin. Invest. 1988;81:902–908. doi: 10.1172/JCI113401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evoli A, Tonali PA, Padua L, Monaco ML, Scuderi F, Batocchi AP, Marino M, Bartoccioni E. Clinical correlates with anti-MuSK antibodies in generalized seronegative myasthenia gravis. Brain. 2003;126:2304–2311. doi: 10.1093/brain/awg223. [DOI] [PubMed] [Google Scholar]
- 12.Leite MI, Strobel P, Jones M, Micklem K, Moritz R, Gold R, Niks EH, Berrih-Aknin S, Scaravilli F, Canelhas A, et al. Fewer thymic changes in MuSK antibody-positive than in MuSK antibody-negative MG. Ann. Neurol. 2005;57:444–448. doi: 10.1002/ana.20386. [DOI] [PubMed] [Google Scholar]
- 13.Lauriola L, Ranelletti F, Maggiano N, Guerriero M, Punzi C, Marsili F, Bartoccioni E, Evoli A. Thymus changes in anti-MuSK-positive and -negative myasthenia gravis. Neurology. 2005;64:536–538. doi: 10.1212/01.WNL.0000150587.71497.B6. [DOI] [PubMed] [Google Scholar]
- 14.Moulian N, Bidault J, Truffault F, Yamamoto AM, Levasseur P, Berrih-Aknin S. Thymocyte Fas expression is dysregulated in myasthenia gravis patients with anti-acetylcholine receptor antibody. Blood. 1997;89:3287–3295. [PubMed] [Google Scholar]
- 15.Price P, Witt C, Allcock R, Sayer D, Garlepp M, Kok CC, French M, Mallal S, Christiansen F. The genetic basis for the association of the 8.1 ancestral haplotype (A1, B8, DR3) with multiple immunopathological diseases. Immunol. Rev. 1999;167:257–274. doi: 10.1111/j.1600-065x.1999.tb01398.x. [DOI] [PubMed] [Google Scholar]
- 16.Poëa-Guyon S, Christadoss P, Le Panse R, Guyon T, De Baets M, Wakkach A, Bidault J, Tzartos S, Berrih-Aknin S. Effects of cytokines on acetylcholine receptor expression: implications for myasthenia gravis. J. Immunol. 2005;174:5941–5949. doi: 10.4049/jimmunol.174.10.5941. [DOI] [PubMed] [Google Scholar]
- 17.Shiina T, Ando A, Suto Y, Kasai F, Shigenari A, Takishima N, Kikkawa E, Iwata K, Kuwano Y, Kitamura Y, et al. Genomic anatomy of a premier major histocompatibility complex paralogous region on chromosome 1q21–q22. Genome Res. 2001;11:789–802. doi: 10.1101/gr.175801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kasahara M, Nakaya J, Satta Y, Takahata N. Chromosomal duplication and the emergence of the adaptive immune system. Trends Genet. 1997;13:90–92. doi: 10.1016/s0168-9525(97)01065-2. [DOI] [PubMed] [Google Scholar]
- 19.Goluszko E, Deng C, Poussin MA, Christadoss P. Tumor necrosis factor receptor p55 and p75 deficiency protects mice from developing experimental autoimmune myasthenia gravis. J. Neuroimmunol. 2002;122:85–93. doi: 10.1016/s0165-5728(01)00474-x. [DOI] [PubMed] [Google Scholar]
- 20.Deng C, Goluszko E, Tuzun E, Yang H, Christadoss P. Resistance to experimental autoimmune myasthenia gravis in IL-6-deficient mice is associated with reduced germinal center formation and C3 production. J. Immunol. 2002;169:1077–1083. doi: 10.4049/jimmunol.169.2.1077. [DOI] [PubMed] [Google Scholar]
- 21.Goidl EA, Siskind GW. Ontogeny of B-lymphocyte function. I. Restricted heterogeneity of the antibody response of B lymphocytes from neonatal and fetal mice. J. Exp. Med. 1974;140:1285–1302. doi: 10.1084/jem.140.5.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cyster JG. Chemokines, sphingosine-1-phosphate, and cell migration in secondary lymphoid organs. Annu. Rev. Immunol. 2005;23:127–159. doi: 10.1146/annurev.immunol.23.021704.115628. [DOI] [PubMed] [Google Scholar]
- 23.Feferman T, Maiti PK, Berrih-Aknin S, Bismuth J, Bidault J, Fuchs S, Souroujon MC. Overexpression of IFN-induced protein 10 and its receptor CXCR3 in myasthenia gravis. J. Immunol. 2005;174:5324–5331. doi: 10.4049/jimmunol.174.9.5324. [DOI] [PubMed] [Google Scholar]
- 24.Bleul CC, Boehm T. Chemokines define distinct microenvironments in the developing thymus. Eur. J. Immunol. 2000;30:3371–3379. doi: 10.1002/1521-4141(2000012)30:12<3371::AID-IMMU3371>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 25.Mesnard-Rouiller L, Bismuth J, Wakkach A, Poea-Guyon S, Berrih-Aknin S. Thymic myoid cells express high levels of muscle genes. J. Neuroimmunol. 2004;148:97–105. doi: 10.1016/j.jneuroim.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 26.Hjelmervik TO, Petersen K, Jonassen I, Jonsson R, Bolstad AI. Gene expression profiling of minor salivary glands clearly distinguishes primary Sjogren's syndrome patients from healthy control subjects. Arthritis Rheum. 2005;52:1534–1544. doi: 10.1002/art.21006. [DOI] [PubMed] [Google Scholar]
- 27.Ishii T, Onda H, Tanigawa A, Ohshima S, Fujiwara H, Mima T, Katada Y, Deguchi H, Suemura M, Miyake T, et al. Isolation and expression profiling of genes upregulated in the peripheral blood cells of systemic lupus erythematosus patients. DNA Res. 2005;12:429–439. doi: 10.1093/dnares/dsi020. [DOI] [PubMed] [Google Scholar]
- 28.Legler DF, Loetscher M, Roos RS, Clark-Lewis I, Baggiolini M, Moser B. B cell-attracting chemokine 1, a human CXC chemokine expressed in lymphoid tissues, selectively attracts B lymphocytes via BLR1/CXCR5. J. Exp. Med. 1998;187:655–660. doi: 10.1084/jem.187.4.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luther SA, Lopez T, Bai W, Hanahan D, Cyster JG. BLC expression in pancreatic islets causes B cell recruitment and lymphotoxin-dependent lymphoid neogenesis. Immunity. 2000;12:471–481. doi: 10.1016/s1074-7613(00)80199-5. [DOI] [PubMed] [Google Scholar]
- 30.McLachlan SM, Nicholson LV, Venables G, Mastalgia FL, Bates D, Smith BR, Hall R. Acetylcholine receptor antibody synthesis in lymphocyte cultures. J. Clin. Lab. Immunol. 1981;5:137–142. [PubMed] [Google Scholar]
- 31.Sims GP, Shiono H, Willcox N, Stott DI. Somatic hypermutation and selection of B cells in thymic germinal centers responding to acetylcholine receptor in myasthenia gravis. J. Immunol. 2001;167:1935–1944. doi: 10.4049/jimmunol.167.4.1935. [DOI] [PubMed] [Google Scholar]
- 32.Guigou V, Emilie D, Berrih-Aknin S, Fumoux F, Fougereau M, Schiff C. Individual germinal centres of myasthenia gravis human thymuses contain polyclonal activated B cells that express all the Vh and Vk families. Clin. Exp. Immunol. 1991;83:262–266. doi: 10.1111/j.1365-2249.1991.tb05625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang CH, Guerder S, Hong SC, van Ewijk W, Flavell RA. Mice lacking the MHC class II transactivator (CIITA) show tissue-specific impairment of MHC class II expression. Immunity. 1996;4:167–178. doi: 10.1016/s1074-7613(00)80681-0. [DOI] [PubMed] [Google Scholar]
- 34.Hughes AL, Nei M. Evolutionary relationships of the classes of major histocompatibility complex genes. Immunogenetics. 1993;37:337–346. doi: 10.1007/BF00216798. [DOI] [PubMed] [Google Scholar]
- 35.Nagane Y, Utsugisawa K, Obara D, Yamagata M, Tohgi H. Dendritic cells in hyperplastic thymuses from patients with myasthenia gravis. Muscle Nerve. 2003;27:582–589. doi: 10.1002/mus.10362. [DOI] [PubMed] [Google Scholar]
- 36.Gunn MD, Kyuwa S, Tam C, Kakiuchi T, Matsuzawa A, Williams LT, Nakano H. Mice lacking expression of secondary lymphoid organ chemokine have defects in lymphocyte homing and dendritic cell localization. J. Exp. Med. 1999;189:451–460. doi: 10.1084/jem.189.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin AP, Coronel EC, Sano G, Chen SC, Vassileva G, Canasto-Chibuque C, Sedgwick JD, Frenette PS, Lipp M, Furtado GC, Lira SA. A novel model for lymphocytic infiltration of the thyroid gland generated by transgenic expression of the CC chemokine CCL21. J. Immunol. 2004;173:4791–4798. doi: 10.4049/jimmunol.173.8.4791. [DOI] [PubMed] [Google Scholar]
- 38.Badr G, Borhis G, Treton D, Richard Y. IFN α enhances human B-cell chemotaxis by modulating ligand-induced chemokine receptor signaling and internalization. Int. Immunol. 2005;17:459–467. doi: 10.1093/intimm/dxh227. [DOI] [PubMed] [Google Scholar]
- 39.Sharma S, Yang SC, Hillinger S, Zhu LX, Huang M, Batra RK, Lin JF, Burdick MD, Strieter RM, Dubinett SM. SLC/CCL21-mediated anti-tumor responses require IFNγ, MIG/CXCL9 and IP-10/CXCL10. Mol. Cancer. 2003;2:22. doi: 10.1186/1476-4598-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barone F, Bombardieri M, Manzo A, Blades MC, Morgan PR, Challacombe SJ, Valesini G, Pitzalis C. Association of CXCL13 and CCL21 expression with the progressive organization of lymphoid-like structures in Sjogren's syndrome. Arthritis Rheum. 2005;52:1773–1784. doi: 10.1002/art.21062. [DOI] [PubMed] [Google Scholar]
- 41.Takemura S, Braun A, Crowson C, Kurtin PJ, Cofield RH, O'Fallon WM, Goronzy JJ, Weyand CM. Lymphoid neogenesis in rheumatoid synovitis. J. Immunol. 2001;167:1072–1080. doi: 10.4049/jimmunol.167.2.1072. [DOI] [PubMed] [Google Scholar]
- 42.Meraouna A, Cizeron-Clairac G, Panse RL, Bismuth J, Truffault F, Talaksen C, Berrih-Aknin S. The chemokine CXCL13 is a key molecule in autoimmune myasthenia gravis. Blood. 2006;108:432–440. doi: 10.1182/blood-2005-06-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Storto M, de Grazia U, Battaglia G, Felli MP, Maroder M, Gulino A, Ragona G, Nicoletti F, Screpanti I, Frati L, Calogero A. Expression of metabotropic glutamate receptors in murine thymocytes and thymic stromal cells. J. Neuroimmunol. 2000;109:112–120. doi: 10.1016/s0165-5728(00)00269-1. [DOI] [PubMed] [Google Scholar]
- 44.Vincent A, Lily O, Palace J. Pathogenic autoantibodies to neuronal proteins in neurological disorders. J. Neuroimmunol. 1999;100:169–180. doi: 10.1016/s0165-5728(99)00210-6. [DOI] [PubMed] [Google Scholar]
- 45.Green AR, Hainsworth AH, Misra A, Debens TA, Jackson DM, Murray TK, Nelson RM, Cross AJ. The interaction of ARA008055 and its enantiomers with the GABA(A) receptor complex and their sedative, muscle relaxant and anticonvulsant activity. Neuropharmacology. 2001;41:167–174. doi: 10.1016/s0028-3908(01)00053-3. [DOI] [PubMed] [Google Scholar]
- 46.van der Geld H, Oosterhuis HJ. Muscle and thymus antibodies in myasthenia gravis. Vox Sang. 1963;8:196–204. doi: 10.1111/j.1423-0410.1963.tb03293.x. [DOI] [PubMed] [Google Scholar]
- 47.Takaya M, Kawahara S, Namba T, Grob D. Antibodies against myofibrillar proteins in myasthenia gravis patients. Tokai J. Exp. Clin. Med. 1992;17:35–39. [PubMed] [Google Scholar]
- 48.Mohan S, Barohn RJ, Jackson CE, Krolick KA. Evaluation of myosin-reactive antibodies from a panel of myasthenia gravis patients. Clin. Immunol. Immunopathol. 1994;70:266–273. doi: 10.1006/clin.1994.1039. [DOI] [PubMed] [Google Scholar]
- 49.Labrada L, Liang XH, Zheng W, Johnston C, Levine B. Age-dependent resistance to lethal alphavirus encephalitis in mice: analysis of gene expression in the central nervous system and identification of a novel interferon-inducible protective gene, mouse ISG12. J. Virol. 2002;76:11688–11703. doi: 10.1128/JVI.76.22.11688-11703.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sjolin H, Tomasello E, Mousavi-Jazi M, Bartolazzi A, Karre K, Vivier E, Cerboni C. Pivotal role of KARAP/DAP12 adaptor molecule in the natural killer cell-mediated resistance to murine cytomegalovirus infection. J. Exp. Med. 2002;195:825–834. doi: 10.1084/jem.20011427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bakker AB, Hoek RM, Cerwenka A, Blom B, Lucian L, McNeil T, Murray R, Phillips LH, Sedgwick JD, Lanier LL. DAP12-deficient mice fail to develop autoimmunity due to impaired antigen priming. Immunity. 2000;13:345–353. doi: 10.1016/s1074-7613(00)00034-0. [DOI] [PubMed] [Google Scholar]
- 52.Sun CT, Lo WY, Wang IH, Lo YH, Shiou SR, Lai CK, Ting LP. Transcription repression of human hepatitis B virus genes by negative regulatory element-binding protein/SON. J. Biol. Chem. 2001;276:24059–24067. doi: 10.1074/jbc.M101330200. [DOI] [PubMed] [Google Scholar]
- 53.Bach JF. Infections and autoimmune diseases. J. Autoimmun. 2005;25(Suppl):74–80. doi: 10.1016/j.jaut.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 54.Schwimmbeck PL, Dyrberg T, Drachman DB, Oldstone MB. Molecular mimicry and myasthenia gravis: an autoantigenic site of the acetylcholine receptor α-subunit that has biologic activity and reacts immunochemically with herpes simplex virus. J. Clin. Invest. 1989;84:1174–1180. doi: 10.1172/JCI114282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yager TD, Dempsey AA, Tang H, Stamatiou D, Chao S, Marshall KW, Liew CC. First comprehensive mapping of cartilage transcripts to the human genome. Genomics. 2004;84:524–535. doi: 10.1016/j.ygeno.2004.05.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


