Abstract
Human immunodeficiency virus type 1 (HIV-1) infection can be inhibited by small molecules that target the CCR5 coreceptor. Here, we describe some properties of clonal viruses resistant to one such inhibitor, SCH-D, using both chimeric, infectious molecular clones and Env-pseudotypes. Studies using combinations of CCR5 ligands, including small molecule inhibitors, monoclonal antibodies (MAbs) and chemokine derivatives such as PSC-RANTES show that the fully SCH-D-resistant viruses enter target cells by using the SCH-D-bound form of CCR5. However, the way resistance to SCH-D and other small molecule CCR5 inhibitors is manifested depends on the target cell and the nature of the assay (single- vs multi-cycle). In multi-cycle assays using primary lymphocytes, SCH-D does not inhibit resistant molecular clones, and it can even enhance their infectivity modestly. In contrast, the same viruses (as Env-pseudotypes) are significantly inhibited by SCH-D in single-cycle entry assays using U87-CD4/CCR5 cells, resistance being manifested by incomplete inhibition at high SCH-D concentrations. When a single-cycle, Env-pseudotype entry assay was performed using either U87-CD4/CCR5 cells or PBMC under comparable conditions, entry was inhibited by up to 88% in the former cells but by only 28% in the PBMC. Hence, there are both cell- and assay-dependent influences on how resistance is manifested. We also take this opportunity to correct our previous report that SCH-D-resistant isolates are also substantially cross-resistant to PSC-RANTES (Marozsan, A. J., Kuhmann, S. E., Morgan, T., Herrera, C., Rivera-Troche, E., Xu, S., Baroudy, B. M., Strizki, J., and Moore, J. P. (2005). Generation and properties of a human immunodeficiency virus type 1 isolate resistant to the small molecule CCR5 inhibitor, vicriviroc (SCH-D; SCH-417690). Virology 338, 182-199). A substantial element of this resistance was attributable to the unappreciated carry-over of SCH-D from the selection cultures into analytical assays.
Keywords: HIV-1, CCR5, entry inhibition, resistance, escape mutants
Introduction
A new class of drugs to treat infection with human immunodeficiency virus type 1 (HIV-1) is now in clinical development: the CCR5 inhibitors (Kazmierski et al., 2003; Seibert and Sakmar, 2004; Westby and van der Ryst, 2005). These compounds, small molecules or monoclonal antibodies (MAbs), bind to the CCR5 coreceptor that is used in conjunction with CD4 to mediate the fusion of HIV-1 with its target cells. By doing so, they impede the ability of the HIV-1 Env complex to interact efficiently with CCR5; the fusion process is blocked and the viral replication cycle interrupted. The small molecule CCR5 inhibitors are considered to act via an allosteric mechanism involving the stabilization or induction of a CCR5 conformation that is not efficiently recognized by the HIV-1 Env complex (Billick et al., 2004; Dragic et al., 2000; Kazmierski et al., 2003; Kenakin, 2004; Seibert et al., 2006; Tsamis et al., 2003; Watson et al., 2005). Two different small molecule CCR5 inhibitors, maraviroc (UK-427,857) and vicriviroc (SCH-D) are now in Phase II or Phase III clinical trials. Both compounds cause significant (∼1.5 log10) reductions in plasma viral load when administered orally to HIV-1-infected people (Fatkenheuer et al., 2005; Schuermann et al., 2005).
As would be expected from its propensity to mutate under selection pressure, HIV-1 develops resistance to the CCR5 small molecule inhibitors in vitro. We have shown previously that an R5 primary isolate, CC1/85, when cultured in peripheral blood mononuclear cells (PBMC) with either AD101 (a pre-clinical precursor of SCH-D) or with SCH-D itself, gradually becomes resistant to the inhibitors (see Table 1 for nomenclature of isolates and env clones used in this study) (Marozsan et al., 2005; Trkola et al., 2002). In general, the resistant viruses retain the R5 phenotype, in that they continue to be dependent on CCR5 for entry into primary CD4+ T-cells, in the presence or absence of the inhibitor. Specifically, the replication of the resistant viruses was efficiently inhibited by CCR5-specific MAbs such as PA14 and 2D7 and replication of the resistant viruses in PBMC from CCR5-Δ32 homozygotes did not occur (Marozsan et al., 2005; Trkola et al., 2002). However, when we studied the sensitivities of the escape mutants to the chemokine ligands of CCR5, a more complex set of data emerged. Thus, the AD101 escape mutant isolate, CC101.19, was only modestly resistant to inhibition by RANTES, and the clonal viruses bearing env genes derived from the isolate were fully sensitive to it (Kuhmann et al., 2004; Trkola et al., 2002). In contrast, two different SCH-D resistant isolates were highly cross-resistant to the chemically modified, more potent RANTES derivative, PSC-RANTES (Marozsan et al., 2005). This finding was unexpected because PSC-RANTES and SCH-D bind to distinct sites on CCR5, and because PSC-RANTES is known to down-regulate a substantial fraction of CCR5 from the cell surface (Hartley et al., 2004). One of the virus isolates resistant to SCH-D (D1/85.16) was able to use CXCR4 in a cell line, but not in PBMC (Marozsan et al., 2005). However, in general, CCR5 inhibitor escape mutants do not switch to using CXCR4, or any other coreceptor, despite the presence of these alternative receptors on the target cells (Marozsan et al., 2005; Trkola et al., 2002). CCR5 use must therefore be favored, even if an inhibitory CCR5 ligand is present in the cultures.
TABLE 1.
Nomenclature and properties of viruses and env genes used in this study.
| virus isolate | selecting compound |
representative env clone |
SCH-D resistant |
|---|---|---|---|
| CC1/85 parental isolate |
none | CC1/85 cl.7 | no |
| CC101.19 (Trkola et al., 2002) |
AD101 | CC101.19 cl.7 | yes |
| D1/85.16 (Marozsan et al., 2005) |
SCH-D | D1/85.16 cl.23 | yes |
The genetics of CCR5 inhibitor resistance are complex. The amino acid substitutions associated with, and in some cases proven to be causative of, resistance development are in the gp120 subunit of the Env complex (Kuhmann et al., 2004; Marozsan et al., 2005), which is logical given that gp120 contains the CCR5 binding site (Hartley et al., 2005). In the case of the AD101-resistant isolate CC101.19, the amino acid changes shown to be responsible for resistance are in the V3 region of gp120 (Kuhmann et al., 2004), an element that is likely to form part of the CCR5 binding site (Hartley et al., 2005; Huang et al., 2005). However, an Env-chimeric virus, D1/85.16 cl.23, derived from the D1/85.16 isolate and resistant to SCH-D, has no sequence changes in V3 (Marozsan et al., 2005). Overall, then, much remains to be learned about how CCR5 inhibitor resistance develops under in vitro conditions. Moreover, there is now preliminary evidence for the evolution of escape mutants during clinical trials of SCH-D, the resistant viruses having phenotypic and genotypic properties that appear to be consistent with the ones generated in vitro (Landovitz et al., 2006).
In this study, we have investigated how the CCR5 inhibitor-resistant viruses produced in the aforementioned studies continue to be CCR5-dependent for entry. We studied the properties of three clonal viruses that are isogenic outside of their env genes. The env clone designated CC1/85 cl.7 was isolated from the genomic DNA of cells infected with the parental, CCR5 inhibitor-sensitive isolate CC1/85 (Table 1). Likewise, the CC101.19 cl.7 and D1/85.16 cl.23 env genes are from the CC101.19 and D1/85.16 isolates that were selected for resistance to AD101 and SCH-D, respectively (Marozsan et al., 2005; Trkola et al., 2002). By using combinations of CCR5 ligands - small molecule inhibitors, MAbs and chemokines - we show here that the resistant viruses must have mutated to recognize, as an entry coreceptor, the complex formed between CCR5 and the small molecule ligands. In addition, we show that the efficiency with which this complex promotes entry varies with cell type, being more efficient in PBMC than in U87-CD4/CCR5 cells. This observation has implications for the use of engineered cell lines such as U87-CD4/CCR5 in testing for resistance to CCR5 inhibitors (Coakley et al., 2005).
Results
Interactions between multiple CCR5 ligands
One way to study the properties of viruses resistant to the small molecule CCR5 inhibitors is to assess their interactions with other CCR5 ligands, such as chemokines and MAbs. The various CCR5 ligands used in these studies are listed in Table 2, together with a summary of their properties, with emphasis on whether each of them interferes with the binding of the others.
TABLE 2.
Properties of selected CCR5 ligands.
| CCR5 ligand |
Type | Binding site on CCR5 |
Probable mode of HIV-1 inhibition |
Interaction with SCH-D |
Interaction with PSC- RANTES |
|---|---|---|---|---|---|
| PSC- RANTES |
chemically modified chemokine |
extracellular loops and amino terminus |
Down- regulation of CCR5 from cell surface |
SCH-D inhibits PSC- RANTES induced down regulation |
N/A |
| SCH-D | small molecule (Mr = 533) |
TM domains | allosteric blockade of gp120- CCR5 interaction |
N/A | SCH-D inhibits PSC- RANTES induced down regulation |
| PRO140 | humanized IgG4 MAb |
extracellular loop 2 and amino terminus |
steric blockade of gp120- CCR5 interaction |
SCH-D partially inhibits PRO140 binding |
PSC- RANTES inhibits PRO140 binding |
| PA12 | mouse IgG1 MAb |
Amino terminus |
steric blockade of gp120- CCR5 interaction |
SCH-D does not inhibit PA12 binding |
PSC- RANTES does not inhibit PA12 binding |
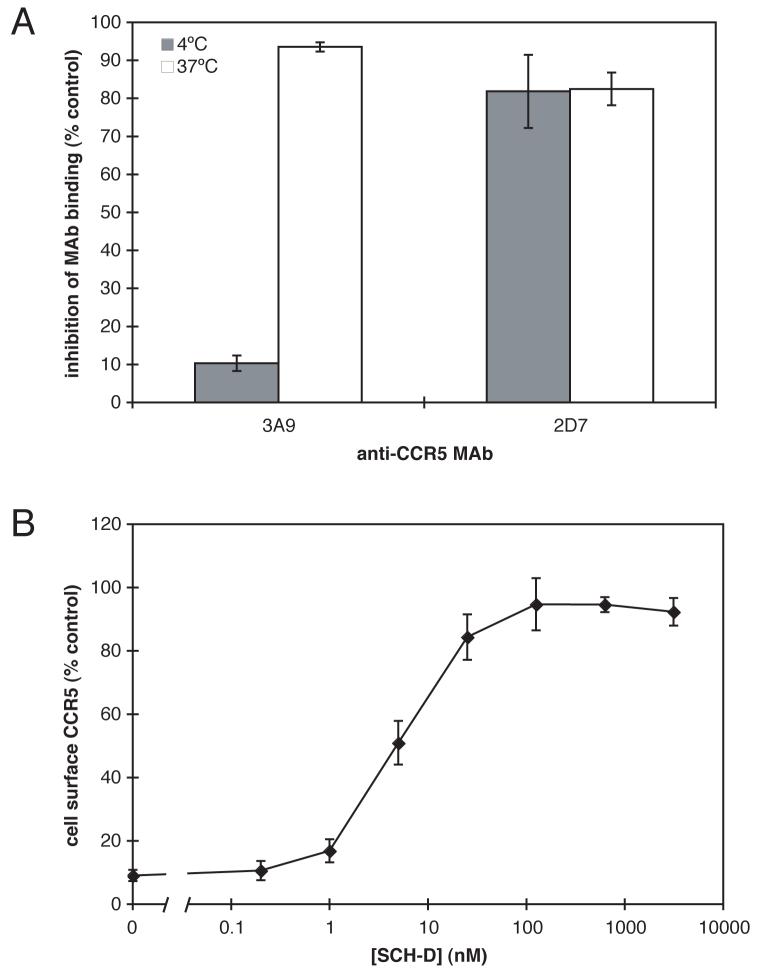
PSC-RANTES is a chemokine derivative that has been reported to rapidly down-regulate CCR5 from the cell surface (Hartley et al., 2004). SCH-D is an antagonist of RANTES-induced Ca2+ flux, guanine nucleotide exchange and chemotaxis mediated by CCR5 (Strizki et al., 2005). We wished to examine how SCH-D and PSC-RANTES interacted with CCR5 when both agents were present. For example, we wanted to determine whether SCH-D could antagonize the action of PSC-RANTES. To carry out this study, it was first necessary to investigate how the treatment of cells with PSC-RANTES affected staining of the cell-surface with anti-CCR5 MAbs. Incubation of HeLa-CD4/CCR5 (clone JC.48) cells with PSC-RANTES (300 nM) for 1 h at 4°C, a temperature at which CCR5 internalization should not occur, reduced the extent of 3A9 binding by only ∼10%, compared to control cells that did not receive PSC-RANTES (Fig.1A). This suggests that the binding of PSC-RANTES to CCR5 does not appreciably occlude the 3A9 epitope by a steric mechanism. In contrast, when the incubation with PSC-RANTES was performed at 37°C to allow CCR5 internalization, 3A9 staining was reduced by 94%, which is consistent with the removal of CCR5 from the cell surface at this temperature (Fig.1A). In contrast, the binding of MAb 2D7 was inhibited by 82% when the cells were treated with 300 nM PSC-RANTES at either 4°C or 37°C under the same conditions (Fig.1A). This finding suggests that the 2D7 epitope is physically occluded by PSC-RANTES binding; indeed, the 2D7 epitope is known to overlap with the RANTES binding site (Lee et al., 1999; Olson et al., 1999). Together, these results show that PSC-RANTES does induce internalization of CCR5 at 37°C in the HeLa-CD4/CCR5 cell line, and that this internalization can be detected by a reduction in staining by the anti-CCR5 MAb 3A9, which binds to CCR5 in a manner that is essentially unaffected by the presence of PSC-RANTES. SCH-D did not inhibit the binding of 3A9 or 2D7 to CCR5 (data not shown).
Figure 1.
SCH-D is an antagonist of the PSC-RANTES induced internalization. A. HeLa-CD4/CCR5 cells were incubated for 1 h at 4°C (shaded bars) or 37°C (open bars) with or without 300 nM PSCRANTES. The cells were then stained for 1 h at 4°C with PE-labeled 3A9 or 2D7. The data shown represent the inhibition of MAb binding after incubation with 300 nM PSC-RANTES relative to cells stained after incubation in the absence of PSC-RANTES. The mean values ± the standard error of the mean (SEM), derived from three independent experiments, are shown. B. The effects of varying SCH-D concentrations on PSC-RANTES-induced CCR5 internalization were determined. HeLa-CD4/CCR5 cells were incubated for 1 h at 37°C with SCH-D at the indicated concentration before the addition of 300 nM PSC-RANTES for 1 h at 37°C. The cells were then stained at 4°C for 1 h with PE-labeled 3A9. The CCR5 expression level was calculated as a percent of the specific 3A9 staining in the same experiment under the same conditions but without PSC-RANTES addition. The data points are the means of between 3 and 7 independent experiments at each SCH-D concentration, and are shown ± SEM.
We next incubated HeLa-CD4/CCR5 cells with varying concentrations of SCH-D for 1 h at 37°C prior to the addition of 300 nM PSC-RANTES for 1 h at 37°C. The cells were then stained with 3A9 at 4°C and analyzed by flow cytometry (Fig.1B). The addition of SCH-D restored 3A9 staining to 96% of the level seen in cells incubated with the same SCH-D concentration, but without PSC-RANTES (Fig.1B). This effect of SCH-D was dose-dependent, with an EC50 of 5 nM. The simplest interpretation of the finding is that SCH-D acts to antagonize CCR5 down-regulation by PSC-RANTES, thereby preserving the 3A9 epitope on the cell surface.
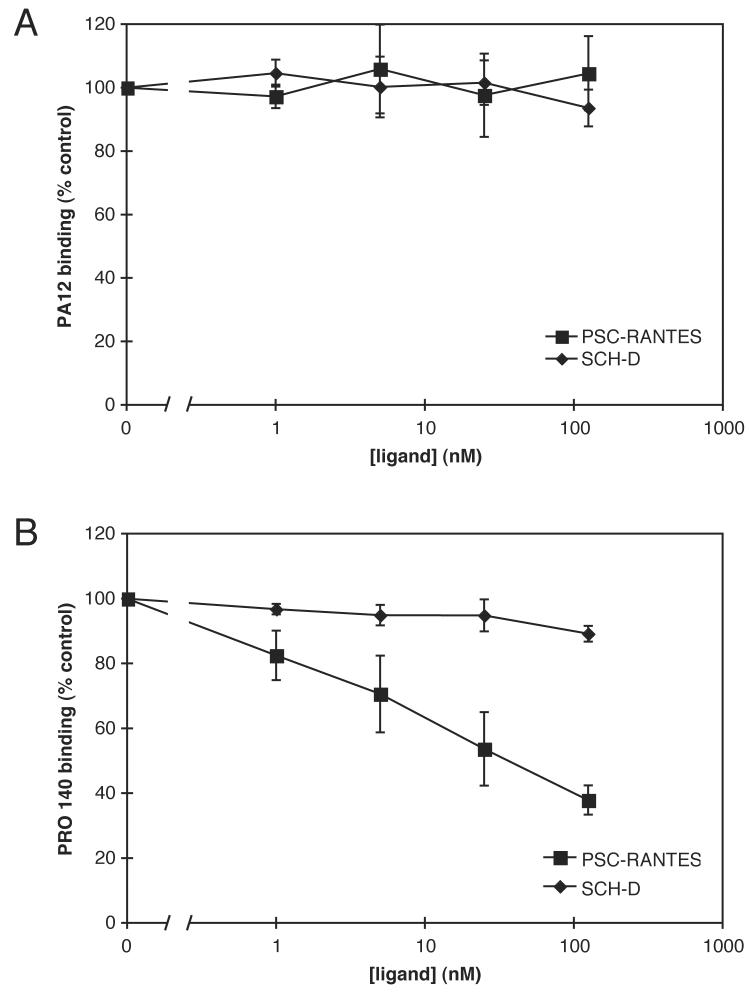
We next sought to investigate how the interactions of SCH-D and PSC-RANTES with CCR5 affect two other CCR5 ligands, the humanized anti-CCR5 MAb PRO 140 and the mouse anti-CCR5 MAb PA12. The HeLa-CD4/CCR5 cells were incubated with varying concentrations of SCH-D or PSC-RANTES for 1 h at 4°C prior to addition of PRO 140 or PA12, also at 4°C. The bound antibodies were detected with an appropriate PE- or FITC-labeled secondary MAb. Neither SCH-D nor PSC-RANTES inhibited PA12 binding to CCR5 in a manner that appeared to be dose-dependent, and the level of PA12 staining at 125 nM SCH-D was not significantly different from that in the absence of SCH-D (P=0.17; paired comparison t-test for 3 independent observations) (Fig.2A). In contrast, PSC-RANTES inhibited PRO 140 binding in a dose-dependent manner with the extent of inhibition reaching 62% at a concentration of 125 nM (Fig.2B). PRO 140 binding was reduced slightly at the highest SCH-D concentration tested (by 11% at 125 nM SCH-D), and the decrease was significant compared to control (i.e., no SCH-D) (P=0.029; paired comparison t-test for 3 independent observations). The partial antagonism of PRO 140 binding by SCH-D is consistent with a recently published report which also shows that SCH-D and PRO 140 act synergistically to inhibit infection by CCR5 inhibitor-sensitive R5 viruses (Murga et al., 2006). Hence other factors must be dominant over the modest interference between these ligands at the level of CCR5 binding.
Figure 2.
How SCH-D and PSC-RANTES interact with PRO 140 and PA12. HeLa-CD4/CCR5 cells were incubated with the indicated concentrations of SCH-D (diamonds) or PSC-RANTES (squares) for 1 h at 4°C, and then with 25 μg/ml PA12 (A) or 30 μg/ml PRO 140 (B) at 4°C for 1 h. Bound MAbs were detected with a PE-labeled anti-mouse IgG1 MAb (A) or a FITC-labeled anti-human IgG4 MAb (B). In both panels, the specific MFI values are expressed as a percentage of those determined under the same conditions, but in the absence of SCH-D or PSC-RANTES, and are the means of three independent experiments ± SEM.
A clonal virus resistant to SCH-D is sensitive to PSC-RANTES
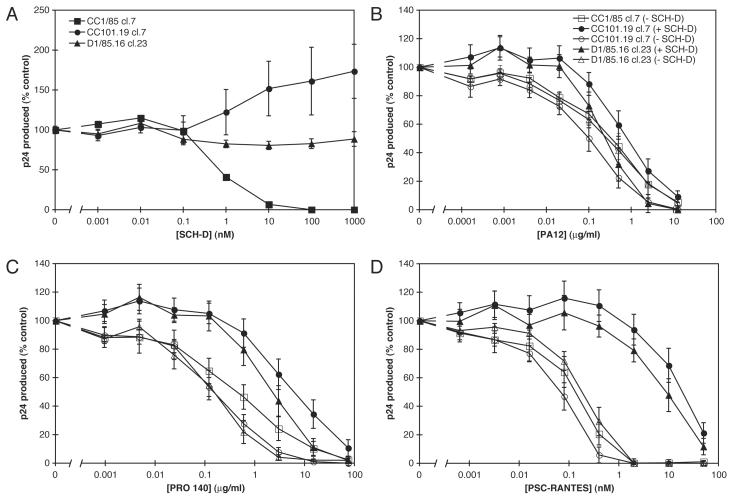
Our investigation of the mechanism of resistance to CCR5 inhibitors was facilitated by the use of clonal viruses produced from plasmids containing HIV-1 env genes in a common proviral background. In the present study, we have used three such clonal proviruses that contain env genes cloned from the CC1/85, CC101.19 and D1/85.16 isolates into the pNL4-3 provirus (Table 1). The Env sequences and properties of the CC1/85 cl.7 (parental) and CC101.19 cl.7 (AD101-resistant) viruses have been described elsewhere (Kuhmann et al., 2004). We showed previously that CC101.19 cl.7, like the isolate from which the env gene was cloned, was resistant to AD101 and SCH-C, but sensitive to inhibition by RANTES and PA14, the mouse MAb from which the humanized PRO 140 MAb was engineered. A partial Env sequence of the D1/85.16 cl.23 virus was published previously, and clonal viruses bearing this Env protein were found to be highly resistant to SCH-D (Marozsan et al., 2005). We now confirm the SCH-D resistance of the D1/85.16 cl.23 virus and show that the AD101-resistant CC101.19 cl.7 clone is cross-resistant to SCH-D (Fig.3A). In fact, at a SCH-D concentration of 1 μM the replication of the CC101.19 cl.7 virus was 73% greater than that of the same virus in the absence of SCH-D. This modest enhancement effect may be specific to the combination of the CC101.19 cl.7 virus and the SCH-D inhibitor, as no such increase in replication was observed with the D1/85.16 cl.23 virus (Fig.3A), or previously when we studied the resistance of the CC101.19 cl.7 virus to AD101 or SCH-C (Kuhmann et al., 2004). However, all three clonal viruses were inhibited by PSC-RANTES with similar IC50 values (Fig.3D, open symbols; Table 3), and each of them was also sensitive to the PA12 and PRO 140 MAbs (Fig.3B,C, open symbols; Table 3). The complete sensitivity of the two SCH-D-resistant clones to three different CCR5 ligands (a chemokine and two MAbs) supports our contention that these Env proteins require the CCR5 coreceptor to enter primary CD4+ T-cells.
Figure 3.
Effects of CCR5 inhibitors alone or in combination with SCH-D on replication of clonal viruses in PBMC. A. PBMC were incubated with the indicated concentration of SCH-D for 1 h at 37°C before the addition of the CC1/85 cl.7 (squares), CC101.19 cl.7 (circles) or D1/85.16 cl.23 (triangles) clonal viruses. After 7 days of culture the production of p24 antigen under each condition was assessed by ELISA. The results show p24 production as a percentage of that derived from cells that were not treated with SCH-D. The values shown are the means ± SEM from 6 independent experiments. B-D. PBMC were incubated with (closed symbols) or without (open symbols) 1 μM SCH-D for 1 h prior to the addition of the indicated concentrations of PA12 (B), PRO 140 (C) or PSC-RANTES (D) for 1 h at 37°C. The CC1/85 cl.7 (squares), CC101.19 cl.7 (circles) or D1/85.16 cl.23 (triangles) clonal viruses were then added. After 7 days of culture p24 production under each condition was assessed by ELISA. The results show p24 production as a percentage of that produced by cells infected in the absence of PA12, PRO 140 or PSC-RANTES, but in the presence (closed symbols) or absence (open symbols) of 1 μM SCH-D. The data for CC1/85 cl.7 in the presence of 1 μM SCH-D could, therefore, not be plotted (no p24 was produced even in the absence of another inhibitor). The addition of PA12, PRO 140 or PSC-RANTES did not reverse the lack of p24 production by CC1/85 cl.7 in the presence of 1 μM SCHD. The values shown are the means ± SEM from 6 or 7 independent experiments.
TABLE 3.
Effect of CCR5 inhibitors alone or in combination.
| ligand titrated | IC50 values for clonal viruses in PBMC |
||
|---|---|---|---|
| CC1/85 cl.7 | CC101.19 cl.7 | D1/85.16 cl.23-21 | |
| SCH-D | 0.82 nM | > 1 μM | >1 μM |
| PA12 | 0.30 μg/ml | 0.088 μg/ml | 0.24 μg/ml |
| PA12 (+ 1 μM SCH-D) | N/Aa | 0.84 μg/ml | 0.25 μg/ml |
| PRO140 | 0.41 μg/ml | 0.15 μg/ml | 0.13 μg/ml |
| PRO140 (+ 1 μM SCH-D) | N/Aa | 6.5 μg/ml | 2.4 μg/ml |
| PSC-RANTES | 0.11 nM | 0.056 nM | 0.17 nM |
| PSC-RANTES (+ 1 μM SCH-D) | N/Aa | 19 nM | 8.4 nM |
N/A = not applicable. Replication of the CC1/85 cl.7 virus was completely inhibited by 1 μM SCH-D, and this inhibition was not affected by the addition of the other CCR5 ligands.
The D1/85.16 cl.23 clonal virus recapitulated the phenotype of the corresponding isolate in respect of its sensitivity to the PRO 140 MAb. We had, however, previously reported that the SCH-D-resistant isolate D1/85.16 was cross-resistant to PSC-RANTES in PBMC cultures (Marozsan et al., 2005), yet we now show that the clonal virus is not. We therefore investigated why the clone and the isolate differ in this regard.
Viruses resistant to small molecule CCR5 inhibitors use CCR5-inhibitor complexes for infection
To further clarify how the clonal viruses interacted with the different CCR5 ligands, we investigated what happened when the ligands were used in combination, bearing in mind the interactions between them that we established earlier (Table 1). It is clear that SCH-D acts as an antagonist of both PSC-RANTES-induced internalization of CCR5 (Fig.1B) and of RANTES signaling activity (Strizki et al., 2005). Furthermore, the SCH-C small molecule ligand inhibits 125I-RANTES binding to CCR5 (Strizki et al., 2001), and this is also true of SCH-D (J. Strizki, personal communication). We therefore considered it likely that SCH-D inhibits the binding of PSC-RANTES to CCR5. Accordingly, we hypothesized that a virus that could use the CCR5-SCH-D complex as a coreceptor would be less affected by the addition of PSC-RANTES when SCH-D was also present in sufficient quantities. We used the clonal CC1/85 cl.7, CC101.19 cl.7 and D1/85.16 cl.23 viruses to explore this scenario. In these experiments, PBMC were incubated with or without 1 μM SCH-D for 1 h prior to the addition of PA12, PRO 140 or PSC-RANTES at various concentrations. Infection by the wild type CC1/85 cl.7 or the SCH-D-resistant CC101.19 cl.7 or D1/85.16 cl.23 clonal viruses was then initiated after an additional 1 h during which the second ligand was also present, or not (Fig.3B-D).
In the absence of SCH-D, the CC1/85 cl.7, CC101.19 cl.7 and D1/85.16 cl.23 viruses were all similarly sensitive to both PA12 (Fig.3B, Table 3) and PRO 140 (Fig.3C, Table 3). The CCR5 MAbs are therefore effective inhibitors of the viruses that have become resistant to the small molecule CCR5 inhibitor, which is consistent with our earlier reports using these and other viruses and CCR5 ligands (Kuhmann et al., 2004; Marozsan et al., 2005; Trkola et al., 2002). All the clones were also comparably sensitive to inhibition by agents not directed at CCR5, such as T1249, CD4-IgG2 and the 2F5 monoclonal antibody (data not shown). SCH-D (1 μM), of course, completely inhibited replication of CC1/85 cl.7 (Fig.3A) and the addition of PA12 or PRO 140 at any concentration did not reverse this inhibition (data not shown). There was no significant effect of 1 μM SCH-D on the inhibition of D1/85.16 cl.23 by PA12, and SCH-D caused only a slight increase (10-fold) in the IC50 for PA12 against CC101.19 cl.7 (Fig.3B, Table 3). These observations are consistent with the earlier observation that SCH-D did not significantly affect PA12 binding to CCR5 (Fig.2A, Table 2). The addition of 1 μM SCH-D caused a rightward shift in the dose-response curve for PRO 140 against the SCH-D-resistant viruses CC101.19 cl.7 and D1/85.16 cl.23, leading to 43- and 18-fold increases in the PRO 140 IC50 values, respectively (Fig.3C, Table 3). In other words, when SCH-D was also present, PRO 140 was a less efficient inhibitor of the SCH-D-resistant viruses than it was in the absence of SCH-D. This effect may arise from the modest inhibition of PRO 140 binding to CCR5 caused by SCH-D at high concentrations (Fig.2B, Table 2).
A similar, but more dramatic, example of the same phenomenon was provided by experiments involving SCH-D and PSC-RANTES. As before, SCH-D by itself was sufficient to inhibit the CC1/85 cl.7 wild type virus at all PSC-RANTES concentrations (data not shown). And, as noted above, all three of the clonal viruses were comparably sensitive to PSC-RANTES in the absence of SCH-D (Fig.3D, Table 3). However, in the presence of 1 μM SCH-D, the inhibitory actions of PSC-RANTES against the SCH-D-resistant viruses CC101.19 cl.7 and D1/85.16 cl.23 were substantially reversed, with rightward shifts in the PSC-RANTES dose-response curves of 340- and 49-fold, respectively (Fig.3D, Table 3). SCH-D therefore significantly antagonizes the actions of PSC-RANTES against the SCH-D-resistant viruses, but not against the wild type virus.
The sensitivities of both SCH-D-resistant viruses to other CCR5 ligands change in a similar manner when SCH-D is also present. The IC50 values for inhibition of CC101.19 cl.7 by PA12, PRO 140 and PSC-RANTES in the presence of 1 μM SCH-D are increased by 10-, 43- and 340-fold, respectively, relative to inhibition in the absence of SCH-D (Table 3). The corresponding values for the D1/85.16 cl.23 virus are 1-, 18- and 49-fold (Table 3). Thus, the magnitude of the change in sensitivity for both viruses correlates with the ability of SCH-D to antagonize the binding or subsequent activity of the other CCR5 ligands (Table 2). The SCH-D-mediated reversal of PSC-RANTES-mediated inhibition of both SCH-D-resistant viruses is substantial. As SCH-D is a chemokine antagonist, the simplest explanation is that by binding to CCR5, SCH-D prevents PSC-RANTES from doing so while creating an alternative, functional binding-site for the SCH-D-resistant viruses to use. The SCH-D-resistant viruses must therefore be able to use CCR5-SCH-D complexes to enter and infect PBMC, whereas wild type viruses can only bind to the ligand-free form of CCR5 (see Discussion).
The PSC-RANTES resistance of the D1/85.16 isolate may be partially due to the presence of residual SCH-D in the virus stock
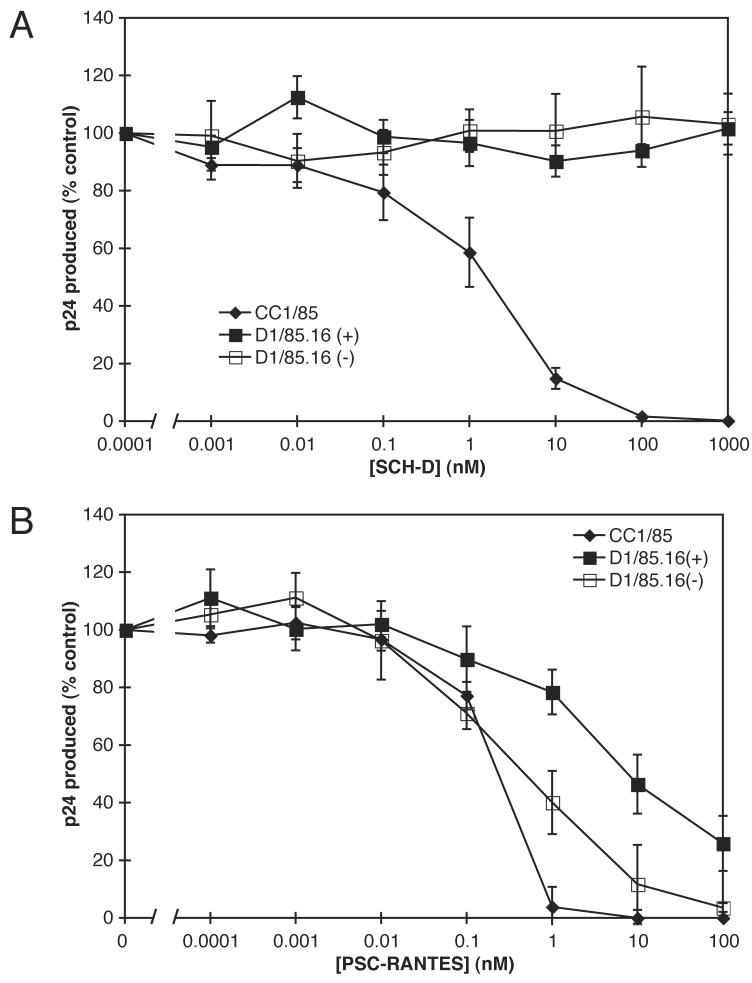
The conditions under which the SCH-D-resistant isolate D1/85.16 was generated involved the gradual escalation of the SCH-D concentration in the cultures up to a level as high as 25 μM, which is ∼25,000-fold greater than the IC50 of the original, SCH-D-sensitive isolate, CC1/85 (Marozsan et al., 2005). The escape mutant isolate was routinely propagated in the presence of 25 μM SCH-D and stocks were frozen down from the cultures for later use, for example as the virus inoculum in studies of cross-resistance. We showed above that SCH-D is able to abrogate the inhibitory effects of PSC-RANTES on two SCH-D-resistant clonal viruses. Hence, we hypothesized that sufficient SCH-D might have been carried over into the frozen virus stocks to make these stocks appear to be cross-resistant to PSC-RANTES.
Too little of the original, apparently PSC-RANTES-resistant stock of the D1/85.16 isolate remained for us to investigate this hypothesis adequately. We therefore determined the maximum effect that residual SCH-D could have in this experimental system. To do this, we propagated the D1/85.16 isolate in PBMC in the presence of 25 μM SCH-D. One day prior to harvesting and freezing the virus stock, the SCH-D was either thoroughly washed out or was left in the culture. The stock containing residual SCH-D is referred to as D1/85.16(+) and that from which the SCH-D was washed out is designated D1/85.16(−). We then assessed the sensitivity of these stocks to SCH-D and PSC-RANTES in PBMC-based virus replication assays under typical conditions (Fig.4). In these replication cultures, the virus stock was diluted by 56-fold. Thus, if all the added 25 μM SCH-D were carried through the propagation and freeze-thaw of the virus stocks, the final SCH-D concentration in the replication cultures would be ∼450 nM, sufficient to significantly inhibit the action of PSC-RANTES (Fig.1B).
Figure 4.
The effect of residual SCH-D on the PSC-RANTES resistance of SCH-D-resistant isolates. PBMC were incubated with the indicated concentration of SCH-D (A) or PSC-RANTES (B) for 1 h at 37°C before the addition of virus isolates CC1/85 (diamonds) or D1/85.16 (squares). Open symbols represent D1/85.16 isolate from which residual SCH-D had been thoroughly removed, whereas filled symbols represent D1/85.16 isolate that was harvested in the presence of 25 μM SCH-D. After 7 days of culture, the amount of p24 antigen produced under each condition was assessed by ELISA. The results show p24 production as a percentage of that produced by cells infected in the absence of any CCR5 inhibitor. The values shown are the means ± SEM from 4 or 5 independent experiments.
We observed that the SCH-D resistance of the D1/85.16 isolate was essentially complete whereas the parental CC1/85 isolate was fully inhibited by SCH-D with an IC50 of 1.3 nM (Fig.4A), a value consistent with the one of 0.95 nM that we had previously reported (Marozsan et al., 2005). The CC1/85 isolate was also fully sensitive to PSC-RANTES with an IC50 of 0.21 nM (Fig.4B,C), again consistent with our previous estimate (0.20 nM) (Marozsan et al., 2005). The D1/85.16(−) isolate was also sensitive to inhibition by PSC-RANTES, with >90% inhibition at a PSC-RANTES concentration of 100 nM (Fig.4B). However, compared to the parental CC1/85 isolate, this virus had a slightly (2.4-fold) reduced sensitivity to PSC-RANTES, with an IC50 value of 0.51 nM (compared to 0.21 nM). This degree of difference in IC50 values may not be significant in this assay system, as the difference in replication levels only reaches significance at 1 nM PSC-RANTES (P=0.03; paired comparison t-test for 4 independent observations), and not at any other PSC-RANTES concentration. It could arise from any SCH-D that was still carried over from the virus-producing cultures despite the use of a washing procedure prior to harvesting.
The presence of 25 μM SCH-D in the virus-producing cultures at the time the isolates were harvested had an additional effect on inhibition by PSC-RANTES. Thus, the IC50 value for the D1/85.16(+) isolate (8.9 nM) was shifted by 17-fold relative to D1/85.16(−), and D1/85.16(+) was able to replicate to 30% of the control level (no inhibitor) when PSC-RANTES was present at 100 nM (Fig.4B). Similar results were obtained for another SCH-D-resistant isolate, D101.12, that we had previously reported to be resistant to PSC-RANTES (data not shown) (Marozsan et al., 2005). Overall, we conclude that residual SCH-D is substantially, but perhaps not entirely, responsible for the high-level resistance to PSC-RANTES we previously reported (Marozsan et al., 2005).
Assay-dependent manifestation of resistance to CCR5 inhibitors
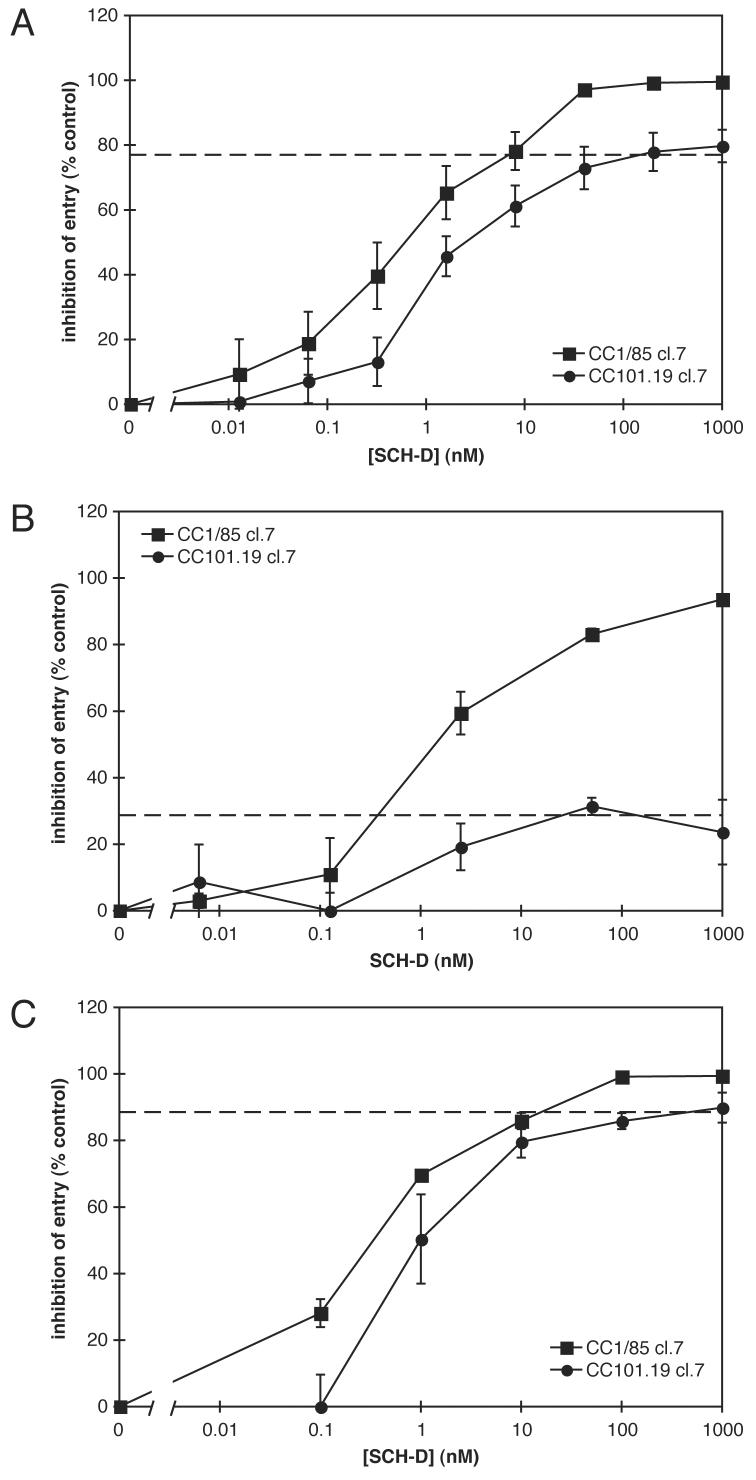
To gain further insights into how the resistant viruses use CCR5 for entry, we used a single cycle infection assay to test one of the same env clones we had evaluated previously in a multi-cycle, PBMC-based replication assay (Fig.5). In the PBMC assay, the env-chimeric virus bearing the Env protein derived from the parental isolate, CC1/85 cl.7, was fully sensitive to SCH-D (IC50 = 0.82 nM) as expected, whereas the CC101.19 cl.7 virus was, again as anticipated, highly resistant to SCH-D (Fig.3A, Table 3). In marked contrast, luciferase-transducing env-defective pseudotype viruses bearing the CC1/85 cl.7 or the CC101.19 cl.7 Env proteins were all inhibited by SCH-D in a single cycle infection assay using U87-CD4/CCR5 target cells (Fig.5A, Table 4). The EC50 values were similar for both Env proteins (0.63 nM and 1.3 nM, respectively). However, the CC101.19 cl.7 Env-pseudotyped viruses were only partially affected by SCH-D, in that the extent of inhibition was incomplete; at high SCH-D concentrations, the inhibition level appears to “plateau” at 77%.
Figure 5.
Single-round infection assays of SCH-D sensitivity of the CC1.85 cl.7 and CC101.19 cl.7 Envs. A. U87-CD4/CCR5 cells were incubated with the indicated concentration of SCH-D for 1 h at 37°C before the addition of luciferase-transducing viruses pseudotyped with the CC1/85 cl.7 (squares) or CC101.19 cl.7 (circles) Env proteins. After three days of incubation, the cells were assayed for luciferase activity. The results are shown as percent inhibition where 0% inhibition is defined as the luciferase activity in cells infected in the absence of SCH-D and 100% inhibition is defined as the luciferase activity measured in cells that were not infected with the pseudoviruses. The values shown are the means ± SEM from 5 independent experiments. The dashed line indicates the apparent plateau in the CC101.19 cl.7 results. B. and C. PBMC (B) or U87-CD4/CCR5 cells (C) were incubated with the indicated concentration of SCH-D for 1 h at 37°C before the addition of hrGFPII-transducing viruses pseudotyped with the CC1/85 cl.7 (squares) or CC101.19 cl.7 (circles) Env proteins. After four days of incubation, the cells were assayed by cytometry for hrGFPII expression. The results are shown as percent inhibition where 0% inhibition is defined as the fraction of hrGFPII+ cells in the absence of SCH-D and 100% inhibition is defined as no hrGFPII+ cells. The values shown are the means ± SEM from 3 independent experiments. The dashed lines indicate the apparent plateau in the CC101.19 cl.7 results.
TABLE 4.
SCH-D inhibition of pseudotype entry in PBMC and U87-CD4/CCR5 cells.
| env clone | reporter gene | |||||
|---|---|---|---|---|---|---|
| luciferase |
hrGFPII |
|||||
| U87-CD4/CCR5 |
cell type PBMC |
U87-CD4/CCR5 |
||||
| EC50a (nM) |
plateaub (%) |
EC50a (nM) |
plateaub (%) |
EC50a (nM) |
plateaub (%) |
|
| CC1/85 cl.7 |
0.63 | 100 | 1.8 | 100 | 0.36 | 100 |
| CC101.19 cl.7 | 1.3 | 77 | 1.4 | 28 | 0.93 | 88 |
The estimated concentration at which the half-maximal inhibition (half-plateau) was reached. When the plateau is estimated to be 100%, this value is also the IC50.
The estimated maximum percent inhibition.
The resistance to SCH-D of the same, clonal Env protein (CC101.19 cl.7) can therefore be manifested in two different ways that depend upon the assay being used. The number of variables (multi-cycle vs. single-cycle, Env-chimeric virus vs. Env-pseudotype, and PBMC vs. U87-CD4/CCR5) does, however, complicate a direct comparison. To overcome this problem, we used a novel pseudotyping backbone that is env-defective and transduces the humanized Renilla green fluorescent protein II (hrGFPII) gene under the control of the CMV promoter, a method that allowed us to now infect PBMC with an Env-pseudotyped virus. The backbone vector was complimented with the CC1/85 cl.7 and CC101.19 cl.7 Env expression vectors, and the resulting pseudotype stocks were used to infect both PBMC and U87-CD4/CCR5 cells in separate cultures under similar conditions. Infection, and its inhibition by SCH-D, was monitored four days post-infection by flow cytometric detection of hrGFPII expression. In PBMC cultures, entry of the pseudotypes bearing the CC1/85 cl.7 Env was, as expected, inhibited in a dose-dependent manner, with an IC50 of 1.8 nM (Fig.5B, Table 4). In contrast to the slight enhancement of replication of the CC101.19 cl.7 Env-chimeric viruses by SCH-D in PBMC (Fig.3A), entry of the CC101.19 cl.7 hrGFPII pseudotypes was partially inhibited, reaching an apparent plateau at 28% inhibition. The half-maximally inhibitory SCH-D concentration (i.e., causing 14% inhibition; EC50) was 1.4 nM, essentially the same as the IC50 value for the CC1/85 cl.7 viruses (Fig.5B, Table 4). The CC1/85 cl.7 pseudotypes were also efficiently inhibited by SCH-D in the U87-CD4/CCR5 cells, with an IC50 of 0.36 nM (Fig.5C, Table 4). Entry of the CC101.19 cl.7 Env-pseudotypes into U87-CD4/CCR5 cells was inhibited by SCH-D, but a plateau was again reached at <100% inhibition, with half-maximal inhibition at 0.93 nM (Fig.5C, Table 4). The plateau value of 88% inhibition was similar to that shown in Fig.5A.
These studies show that, in single-cycle assays, SCH-D-resistance is characterized by the “plateau effect” in dose-response curves, irrespective of whether the target cell is PBMC or U87-CD4/CCR5 cells. However, the nature of the target cell dramatically affects the level of the plateau. In addition, the conditions of the assay (multi- vs single-cycle) also play a role in how resistance is manifested. Thus, SCH-D modestly enhances infection of PBMC by this particular Env (CC101.19 cl.7) in a multi-cycle assay (Fig.3A), but instead causes a plateau at a low level of inhibition in the same cells when a single cycle assay is used (Fig.5B).
PRO 140 and SCH-D antagonism of CCR5 leads to accumulation of CC-chemokines in PBMC cultures
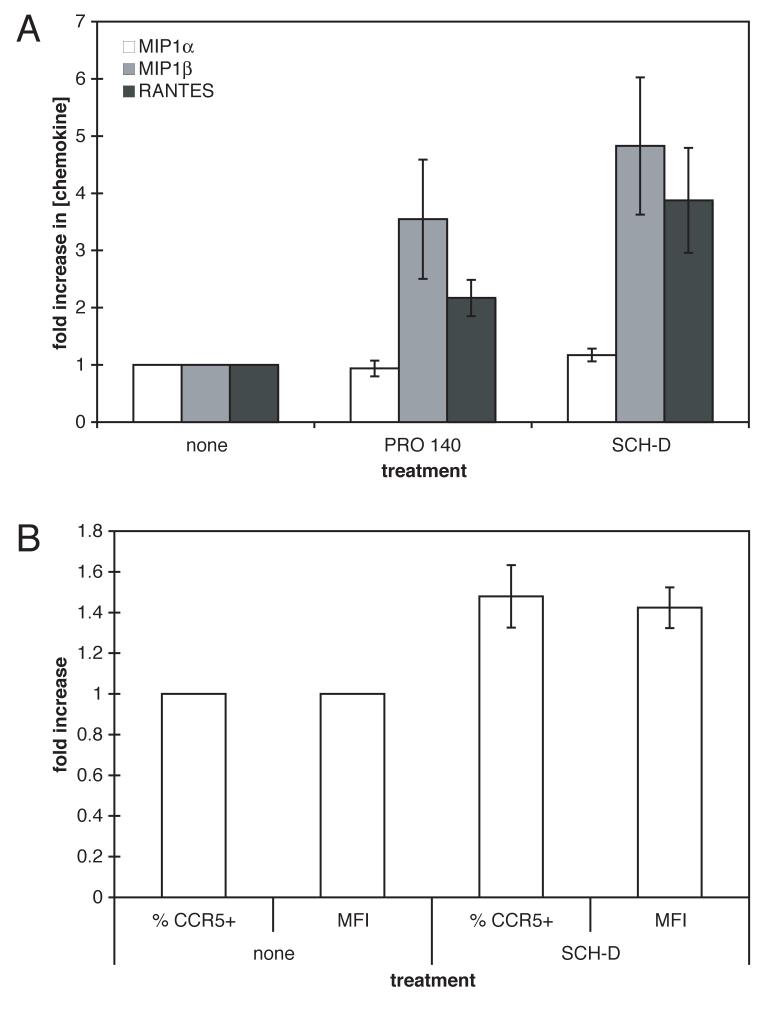
SCH-D antagonizes RANTES signaling mediated by CCR5 (Strizki et al., 2005). PA14, the parent antibody for PRO 140, is also an antagonist of CC-chemokine signaling through CCR5, albeit only at high concentrations (IC50 values for MIP-1α, MIP-1β and RANTES of ∼45 μg/ml) (Olson et al., 1999). We hypothesized that disruption of CCR5-mediated chemokine-signaling pathways in human PBMC cultures could affect the secretion of CC-chemokines by these cells and hence the surface expression of CCR5. To test this, PBMC from seven anonymous donors were mitogen-stimulated for three days prior to treatment for seven days with saturating concentrations of SCH-D (2 μM) or PRO 140 (50 μg/ml). CC-chemokine levels in the culture supernatants were then measured (Fig.6A). In addition, the SCH-D-treated cells were stained with the 2D7 MAb to gauge CCR5 expression levels (Fig.6B). The mean MIP-1α levels were not significantly different for the inhibitor-treated and control cells during the seven-day period (Fig.6A; differences of 0.9- and 1.2-fold for PRO 140 and SCH-D; P=0.30 for both compared to no treatment). However, MIP-1β levels were significantly elevated by both PRO 140 and SCH-D (increases of 3.6- and 4.8-fold for PRO 140 and SCH-D; P=0.05 for both compared to no treatment), as were RANTES levels (increases of 2.2- and 3.9-fold for PRO 140 and SCH-D; P=0.03 and P=0.04, respectively).
Figure 6.
Changes in CC-chemokine production from, and CCR5 expression on, PBMC treated with CCR5 antagonists. A. Stimulated PBMC from seven individuals were incubated for seven days with or without PRO 140 (50 μg/ml) or SCH-D (2 μM) as indicated. The culture supernatants were assayed for MIP-1α (white bars), MIP-1β (grey bars) or RANTES (black bars) content. The data shown are the mean values ± SEM for the fold-increases, compared to no treatment, from the seven donors. B. In the same experiment as in (A), stimulated PBMC from the same seven individuals were incubated for seven days with or without SCH-D as indicated. CD4 expression was assessed with PerCP-labeled anti-CD4, CCR5 expression using the PE-labeled anti-CCR5 MAb 2D7; both the percentage of CD4+ T-cells that were also CCR5+ and the mean fluorescence intensity (MFI) for the CD4+CCR5+ T-cells were determined, as indicated. The data shown are the mean values ± SEM for the fold-increases, compared to no treatment, from the seven donors.
CCR5 expression on CD4+ lymphocytes from the same donors was also significantly increased in the presence of SCH-D (Fig.6B). The percentage of CD4+ T-cells that were CCR5+ after seven days of SCH-D treatment were elevated by an average of 1.5-fold compared to cells from the same donor incubated in the absence of SCH-D (P=0.0003). The average value for the mean fluorescence intensity of 2D7 staining on SCH-D-treated CD4+ T-cells was also increased by 1.4-fold, relative to untreated cells (P=0.004).
Discussion
Here, we have explored the nature of HIV-1 resistance to small molecule CCR5 inhibitors, using clonal infectious viruses or Env-pseudotyped viruses and a variety of analytical techniques. We previously hypothesized that there are two general mechanisms by which HIV-1 might become resistant to small-molecule CCR5 inhibitors while still using CCR5 for entry, and we suggested that both mechanisms were involved when HIV-1 CC1/85 became resistant to AD101 (Trkola et al., 2002). Our new results are entirely consistent with this hypothesis and help us better explain the resistance process, as outlined below. The distinction between the two mechanisms could be important for interpreting the evolution of resistance when CCR5 inhibitors are used clinically to treat HIV-1 infection (Landovitz et al., 2006).
The first mechanism, which we term “competitive resistance”, is manifested by a change in the potency (IC50 value) of a CCR5 inhibitor (Fig.7). In this scenario a gp120 protein would change conformation in such a way as to increase its affinity for inhibitor-free (i.e., wild-type) CCR5. The competitive resistance mechanism would enable a virus to more efficiently use the lower levels of inhibitor-free CCR5 available in the presence of a non-saturating concentration of an inhibitor. Increasing the inhibitor concentration would be sufficient to occupy more CCR5 receptors and thereby overcome competitive resistance. Hence in the face of competitive resistance, the potency of a CCR5 inhibitor will decrease, but complete inhibition could still be achieved if a sufficiently high concentration of inhibitor were present, in vitro or in vivo.
Figure 7.
Schematic depiction of possible mechanisms of resistance to an allosteric small-molecule CCR5 inhibitor. A. CCR5 is depicted with two distinct HIV-1 interaction sites (peaks) and a separate SCH-D (or other allosteric CCR5 inhibitor) binding site (valley). The gp120 protein is depicted as having two interaction sites that are compatible with binding to CCR5, thereby mediating infection. For convenience in drawing the figure, we depict the gp120-CCR5 interaction via one of these sites as being weaker than the other, leaving room for a stronger interaction in (D). This need not necessarily be the case, as the strengthened interaction in (D) could involve both interaction sites. B. In the presence of a high SCH-D concentration, the conformation of one of the interaction regions on CCR5 is altered, prohibiting interaction with gp120 and preventing infection. C. Noncompetitive resistance is depicted as a change in the conformation of gp120 to accommodate the altered CCR5 conformation. In the case of the noncompetitive resistance described in this paper, infection through the SCH-D-free form of CCR5 is also possible for gp120 from the SCH-D-resistant viruses we have studied here. How this form of resistance would be manifested in an entry assay at varying efficiencies of entry through the SCH-DCCR5 complex (relative to free CCR5) is shown below the diagram. D. Competitive resistance is depicted as a change in the conformation of gp120 to increase the affinity of gp120 for CCR5 (here shown by a better fit between the two). In this scenario, gp120 better competes with SCH-D for binding to CCR5. How this form of resistance would be manifested in an entry assay with various degrees of improvement in the gp120-CCR5 interaction (relative to the wild type gp120) is shown below the diagram.
In the second mechanism, which we term “noncompetitive resistance”, a resistant virus continues to enter target cells regardless of the concentration of the inhibitor. Noncompetitive resistance arises when the virus acquires the ability to use the inhibitor-CCR5 complex for entry. When a noncompetitively resistant virus uses the inhibitor-bound form of CCR5 less efficiently than the inhibitor-free form, the outcome is what is often termed the “plateau effect” (i.e., the extent of inhibition does not increase any further once a certain inhibitor concentration is reached, and never reaches 100%, see Fig.5B). Under this scenario, the EC50 (the concentration at which the inhibitor achieves 50% of its maximal effect; i.e., half the plateau height) should be the same as the IC50 for the action of the inhibitor against the wild-type virus (Fig.7). The appearance of a plateau in dose-response curves therefore reflects the difference in the efficiency with which the resistant virus uses the inhibitor-free and inhibitor-bound forms of CCR5. The height of the plateau is a measure of the magnitude of the efficiency difference; the less efficiently the inhibitor-bound of CCR5 is used, the higher the plateau. It therefore follows that, if entry via the inhibitor-free and inhibitor-bound forms of CCR5 occurs with equal efficiency, any switch in entry mediated by one receptor form to the other would not be evident in drug titration experiments. In other words, the plateau would be at 0% inhibition and the virus would appear to be completely resistant to the inhibitor. It follows that a combination of the two mechanisms would result in changes in both the EC50 value and the plateau height.
We have previously demonstrated that the parental CC1/85 isolate became completely resistant to AD101 via a step-wise accumulation of mutations in the V3 region of gp120 (Kuhmann et al., 2004; Trkola et al., 2002). The pathway to resistance seems likely to have involved both the competitive and noncompetitive mechanisms. Thus, an isolate emerged after 4-6 passages in the presence of AD101 in which a single amino acid polymorphism in V3 had undergone purifying selection to dominance, having been present initially at a low frequency (Kuhmann et al., 2004). The isolate from this time point, CC101.6, was modestly resistant to AD101, with an ∼3-fold increase in its IC50 compared to CC1/85 (Trkola et al., 2002). The CC101.6 isolate had a correspondingly increased ability to use low levels of CCR5 for entry into HeLa-CD4/CCR5 cell lines (Trkola et al., 2002). Thus we suggest that this modest, but perhaps clinically significant, degree of resistance is an example of competitive resistance, arising from an increase in the affinity of gp120 for the inhibitor-free configuration of CCR5. The process may be similar to how R5 variants with an apparently improved ability to “scavenge” low levels of CCR5 are thought to emerge during the later stages of HIV-1 infection (Gorry et al., 2002; Koning et al., 2003).
When the AD101-resistance selection experiment was continued, the further accumulation of three additional amino acid substitutions, apparently by de novo mutation, in the V3 region led to an isolate, CC101.19, with complete resistance to AD101 (Trkola et al., 2002). The inability of AD101 to inhibit replication of this isolate, or clones derived from it, at even very high concentrations (up to 20 μM; cf. IC50 of AD101 against CC1/85 of ∼1 nM), led us to hypothesize that the resistance mechanism was noncompetitive, and that its molecular basis would be the use of the inhibitor-CCR5 complex for entry (Kuhmann et al., 2004; Trkola et al., 2002).
In the present study, we have used clonal viruses and a variety of CCR5 ligands to conclude that the fully resistant viruses do indeed use the inhibitor-bound form of CCR5 for entry. We studied two different clonal viruses: CC101.19 cl.7 derived from the CC101.19 isolate, and D1/85.16 cl.23, derived from an isolate that was selected for resistance to SCH-D. We previously reported that the CC101.19 cl.7 virus was completely resistant to AD101 and SCH-C in PBMC replication assays and that D1/85.16 cl.23 was similarly resistant to SCH-D. Here we confirm the SCH-D resistance of the D1/85.16 cl.23 clone and show that the replication of CC101.19 cl.7 in PBMC is not only completely resistant to SCH-D, but is modestly enhanced by it (Fig.3A). Resistance of this magnitude is strongly suggestive of a noncompetitive mechanism, with these viruses using the SCH-D-CCR5 complex as a functional coreceptor. The most compelling evidence that this is the case comes from experiments in which the SCH-D-resistant clones infect PBMC in the presence of both PSC-RANTES and SCH-D. The opposing effect of these two CCR5 ligands, when added together, is most logically explained if the resistant viruses can use the CCR5-SCH-D complex.
Because our results were obtained using clonal viruses bearing Env proteins derived from two different SCH-D-resistant isolates generated under the selection pressures of two different small molecule CCR5 inhibitors (AD101 and SCH-D), we suspect they may have general relevance. Hence the use of the CCR5-inhibitor complex might be the predominant mechanism for the entry of viruses fully resistant to small molecule CCR5 inhibitors. Our general experience has been that viruses resistant to one small molecule CCR5 inhibitor are also cross-resistant to other members of the same inhibitor class (TJK et al., unpublished). However, a virus resistant to maraviroc has been reported to be sensitive to other small molecule CCR5 inhibitors, so cross-resistance may not be a universal phenomenon (Westby et al., 2005).
We do not yet know how the resistant viruses recognize the inhibitor-bound configuration of CCR5. A generally accepted model for how gp120 interacts with CCR5 is that two separate, but topologically adjacent elements of gp120 (the V3 region and the bridging sheet) bind to two domains on CCR5 (respectively, the second extracellular loop and the Tyr-sulfated region of the N-terminus) (Cormier and Dragic, 2002; Cormier et al., 2001; Hartley et al., 2005; Huang et al., 2005; Safarian et al., 2006; Tsamis et al., 2003). Presumably, the binding of the small molecule inhibitor alters the geometry of CCR5 such that the two elements of its gp120 binding site are now positioned in a configuration that cannot be efficiently recognized by gp120 (the allosteric mechanism of inhibition at work). In the resistant virus, however, the accumulated gp120 mutations have altered the geometry of its own binding site such that the new configuration of CCR5 can now be accommodated. It may be that the sequence changes in gp120 enable the resistant virus to now dock stably with only one element on CCR5, and not the two required by the wild-type virus. Whether the interaction between V3 and the second extracellular loop or the one between the bridging sheet and the N-terminus now dominates the gp120-CCR5 interaction remains to be determined. By extension, it is possible that natural isolates with relative resistance to small molecule CCR5 inhibitors (Strizki et al., 2001) are more dependent upon a single domain of CCR5 for entry, so are less affected by the change in receptor conformation created by the binding of the inhibitor. Studies of clonal viruses in single-cycle entry assays may be informative. Do such viruses generate a plateau effect, or is the pattern of inhibition more reflective of the competitive mechanism?
Further evidence of a noncompetitive resistance mechanism for the CC101.19 cl.7 and D1/85.16 cl.23 viruses comes from the use of entry assays in engineered cell lines. In both PBMC and U87-CD4/CCR5 cells the entry of CC101.19 cl.7 Env-pseudotyped viruses is inhibited by SCH-D in a manner that is suggestive of noncompetitive resistance (Fig.5). Thus, the inhibition curves for CC101.19 cl.7 are similar to those for the parental virus CC1/85 cl.7, except that the maximum level of inhibition plateaus at less than 100%. As noted above, this is the hallmark of noncompetitive resistance if the inhibitor-free and inhibitor-bound forms of CCR5 differ in their efficiencies as coreceptors (see Fig.7). We conclude, therefore, that the SCH-D resistance displayed by CC101.19 cl.7 in this system is noncompetitive in nature, and that the efficiency with which the SCH-D-CCR5 complex mediates the entry of this pseudovirus is significantly lower than the efficiency with which the inhibitor-free form of CCR5 does so.
An important conclusion of the results shown in Fig.5 and Table 4 is that the plateau level for the CC101.19 cl.7 Env-pseudotypes varies by cell type. Thus, the plateau level of 28% inhibition for the hrGFPII pseudotypes in PBMC is considerably different from what occurs in U87-CD4/CCR5 cells (77% and 88% for the luciferase and hrGFPII pseudotypes, respectively). That these values differ implies that the efficiency with which the CCR5-SCH-D complex mediates infection varies between the two cell types. This, in turn, suggests that the CCR5 coreceptor itself differs between the two cell types. There are a variety of ways in which CCR5 might vary in a cell type-dependent manner: expression level, post-translational modification and subcellular localization being among them. For example, it is possible that over-expressing CCR5 in a cell line could affect how it is processed and post-translational modified, creating a variety of antigenic forms on the cell surface that are recognized with different efficiencies by wild type and resistant viruses, and by inhibitory ligands. Multiple antigenic forms of CCR5 are also known to be present on PBMC (Lee et al., 1999; Olson et al., 1999). How the presence of different amounts of different CCR5 configurations on different cell types affects entry and its inhibition defies rational analysis at present, for want of hard data on what factors are relevant and to what extent. Nonetheless, the difference between PBMC and U87-CD4/CCR5 cells is a very real one, and one that may have clinical significance: A single-cycle Env-pseudotype assay based on U87-CD4/coreceptor cells (PhenoSense™, Monogram Inc) is now very commonly used to diagnose coreceptor usage and the development of resistance to CCR5 inhibitors (Coakley, Petropoulos, Whitcomb, 2005). This assay is very similar to the Env-pseudotype, luciferase-readout assay used in Fig.5A. Whether an assay of this nature always generates physiologically meaningful information is something to consider.
Whether an assay is conducted in a single-cycle or a multi-cycle format is also relevant to how CCR5 inhibitor resistance is manifested. Noncompetitive resistance of the CC101.19 cl.7 Env protein to SCH-D leads to a plateau effect in a single-cycle entry assay in PBMC (Fig.5B), but appears as modest enhancement in a multi-cycle replication assay in the same cells (Fig.3A). The basis for this difference is not clear at this time, but it is now under investigation. It is possible that the rates and/or extents of CCR5 recycling over time influence how single- or multi-cycle assays provide different information on the magnitude of CCR5 inhibitor resistance. Another variable between single- and multi-cycle assays is the long-term upregulation of chemokine secretion and of CCR5 expression that is triggered by small molecule CCR5 inhibitors in the PBMC cultures used in multi-cycle assays (Fig.6). How an additional CCR5 ligand such as a naturally produced chemokine would influence the overall system cannot easily be determined, but it is a factor that would not apply to a single-cycle assay.
Overall, the interactions between SCH-D (or other CCR5 antagonists) and PSC-RANTES (or other chemokines) and CCR5 are rather complex. SCH-D prevents PSC-RANTES from binding to CCR5, but also causes an increase in RANTES and MIP-1β concentrations in the cultures (at least of lymphoid cells) and elevates the amount of CCR5 present on the cell surface, although almost of that receptor will be in its SCH-D bound form. PSC-RANTES binding to CCR5 internalizes the receptor, but this is antagonized by SCH-D. When both SCH-D and PSC-RANTES are present, the latter has no effect on HIV-1 entry as it is prevented from binding to CCR5 by the former. The same scenario will, presumably, apply to the natural RANTES and MIP-1β chemokines that accumulate over time in PBMC cultures treated with SCH-D or other chemokine antagonists. The increased accumulation of CC-chemokines in the presence of a CCR5 antagonist may occur by a mechanism similar to how PBMC from CCR5-Δ32 homozygotes secrete higher levels of these chemokines (Dragic et al., 1996; Paxton et al., 1996a; Paxton et al., 1996b). Thus, the failure of CCR5 to signal in response to these chemokines may remove a feedback mechanism that controls their constitutive secretion. The same feedback mechanism may also influence the constitutive expression of CCR5 in these cells, explaining the observed upregulation. It is not yet known if changes in circulating chemokine levels or CCR5 expression levels occur in individuals receiving CCR5 inhibitors for prolonged periods. It will be of interest to address these questions, because fully understanding how the mutual interactions between the various CCR5 ligands affect HIV-1 entry and its inhibition requires taking all the possible variables into account.
Finally, we want to revisit an issue that arose in one of our earlier reports (Marozsan et al., 2005). We previously showed that the SCH-D-resistant isolates D101.12 and D1/85.16 were cross-resistant to PSC-RANTES, a finding we confirm here. We now believe, however, that this cross-resistance was probably substantially, although perhaps not completely, an unforeseen and unappreciated consequence of the experimental conditions we used in the original resistance-selection experiments. Specifically, the carry-over of SCH-D from the selection cultures (in which the SCH-D concentration eventually reached 25 μM) into the frozen virus stock could have been sufficient to influence the subsequent assays of PSC-RANTES resistance. Thus, even after dilution of the viral inoculum into the test cultures, the SCH-D concentration may still have been sufficient to nearly fully occupy CCR5 and prevent PSC-RANTES from binding. In consequence, the SCH-D-resistant isolates appeared to be substantially cross-resistant to PSC-RANTES when they were actually entering cells via the SCH-D-bound conformation of CCR5 to which PSC-RANTES cannot bind. Because these viruses can also use the inhibitor-free form of the receptor, the presence of any carried-over SCH-D was not suspected when we wrote our initial report (Marozsan et al., 2005). We have now, however, shown that the SCH-D-resistant isolates D1/85.16(−) (Fig.4) and D101.12(−) (data not shown) appear to be partially cross-resistant to PSC-RANTES. Thus, in the absence of SCH-D, their IC50 values for PSC-RANTES were increased by 2.4- and 11-fold respectively (Fig.4 and data not shown). The SCH-D-resistant clone D1/85.16 cl.23 is fully sensitive to PSC-RANTES. Similarly, we have previously reported that the CC101.19 isolate acquired 10-fold resistance to RANTES, while the clones derived from it did not (Kuhmann et al., 2004; Trkola et al., 2002). It is possible that this level of resistance is due to some low level of SCH-D or AD101 that was carried through in the virus stocks, even when due care was taken to wash the CCR5 inhibitors out of the cultures that produced them. If this modest degree of resistance is, in fact, a property of the viruses in the isolate, it is feasible that the underlying resistance mechanism is competitive in nature: i.e., an increase in the affinity of gp120 for CCR5 enables the virus to compete with PSC-RANTES more efficiently. We have not yet studied this possibility in detail.
Formal studies of viral fitness and comparative studies will be required to place the various escape mutants described by ourselves and others into a suitable quantitative framework. However, one implication of the presently available data is that any CCR5 inhibitor-resistant viruses that emerge in vivo during therapy might be less replication competent, and perhaps even less pathogenic, in vivo than wild type viruses, similar to what has been described for Enfuvirtide-resistant viruses (Lu et al., 2004). If escape mutants to CCR5 inhibitors are also less replication competent than wild type viruses in the absence of the selecting compound, reversion to sensitivity might occur over time after therapy is discontinued. Whether this will occur in clinical practice remains to be determined.
Materials and methods
Cells and cell culture
HeLa-CD4/CCR5 (clone JC.48) cells were a gift of Dr. David Kabat (Oregon Health and Science University, Portland, OR) (Platt et al., 1998). U87-CD4/CCR5 cells (contributed by Dr. HongKui Deng and Dr. Dan Littman) were obtained from the NIH AIDS Research and Reference Reagent Program (ARRRP) (Bjorndal et al., 1997). 293T cells were from the American Type Culture Collection (ATCC; Manassas, VA). HeLa-CD4/CCR5 and 293T cells were maintained in Dulbecco's modified Eagle medium (DMEM; Invitrogen Inc., Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; Invitrogen) and 100 U/ml penicillin + 100 μg/ml streptomycin (1X PenStrep; HyClone, Logan, UT). U87-CD4/CCR5 cells were maintained in DMEM+10% FBS+1X PenStrep supplemented with 0.3 mg/ml G418 and 0.5 μg/ml puromycin (both from Sigma-Aldrich, St. Louis, MO). PBMC were purified from leukopacks obtained from the New York Blood Center (New York, NY) and stimulated as previously described (Kuhmann et al., 2004). Briefly, the leukopacks were depleted of CD8+ cells using the RosetteSep reagent (StemCell Technologies, Vancouver, BC, Canada) and then purified on a Ficoll density gradient (Kuhmann et al., 2004). Cells from each blood donor were split into two cultures, one of which was stimulated for three days with surface-immobilized anti-CD3 MAb (clone OKT3) and the other of which was stimulated for three days with 5 μg/ml of phytohemagglutinin (PHA; Sigma). All PBMC cultures were maintained in PBMC culture medium [Rockwell Park Marine Institute 1640 medium (RPMI 1640; Invitrogen) with 10% FBS, 1X PenStrep and 100U/ml interleukin-2 (IL-2; ARRRP, donated by Hoffmann-La Roche, Inc.)]. All cells were incubated at 37°C in a 5% CO2 atmosphere.
Antibodies and reagents
PSC-RANTES was a gift from Dr. Oliver Hartley (Centre Medicale Universitaire, Geneva). The small molecule CCR5 inhibitor SCH-D (Tagat et al., 2004), the humanized MAb PRO 140 and the mouse MAb PA12 (Olson et al., 1999) were donated by Dr. William Olson (Progenics Pharmaceuticals Inc, Tarrytown, NY). PE-labeled anti-CCR5 MAbs 2D7 and 3A9 were purchased from BD-Biosciences (San Jose, CA), as were PE-labeled anti-mouse IgG2a, PerCP-labeled anti-human CD4 and appropriate isotype controls. FITC-labeled anti-human IgG4 MAb was purchased from Sigma (St. Louis, MO). All restriction enzymes were from New England Biolabs (Ipswich, MA).
Immunocytometry
HeLa-CD4/CCR5 cells were detached with versene (Invitrogen), transferred into Eppendorf tubes, pelleted and washed once with culture medium prior to incubation with inhibitors or antibodies. A total of 1 × 106 cells in a final volume of 1 ml were used for each experimental condition. The cells were incubated with CCR5 inhibitors in cytometry buffer (phosphate buffered saline with 10% FBS) prior to staining (the inhibitor concentrations, temperatures and the order of addition are stated in the text and figure legends). Staining with the appropriate labeled antibodies, diluted according to the manufacturer's recommendations, was carried out in cytometry buffer at 4°C. All cytometry was carried out using an LSRII digital cytometer (BD Bioscience). Because the HeLa-CD4/CCR5 cells were cloned by limiting dilution after transduction of CCR5 expression with a retroviral vector, they express a stable level of CCR5 on 100% of the cells (Platt et al., 1998). Thus, live cells were gated by forward- and side- scatter properties, but no further gating was necessary; the mean fluorescence intensity (MFI) of the entire live cell population was used for further analysis. In all experiments the MFI of cells stained with an appropriate isotype control antibody was used to determine a non-specific fluorescence value that was subtracted from experimentally derived MFI values.
Staining of PBMC was performed in round-bottomed 96-well plates. Approximately 1 × 106 PBMC were seeded per well after three days of stimulation, and then treated for seven days with 2 μM SCH-D in culture medium, or with culture medium only. The cells were then pelleted in the plate and resuspended in 50 μl of PBMC FACS buffer (RPMI 1640 without phenol red + 10% FBS + 0.1% NaN3). A 10 μl aliquot of each of the PE-labeled 2D7 and PerCP-labeled anti-CD4 mAbs, or the appropriate PE-and PerCP-labeled isotype controls, was added to each well. The cells were incubated for 1 h at 4°C, then washed three times with 200 μl of PBMC FACS buffer, resuspended in 200 μl of 2% w/v paraformaldehyde + 2% w/v sucrose in phosphate buffered saline, then further incubated for 15 min at room temperature. A 200 μl aliquot of PBMC FACS buffer was then added prior to flow cytometric analysis.
Plasmid construction
The pCI-env expression plasmids were constructed by insertion of the CC1/85 cl.7 and CC101.19 cl.7 env genes into the multiple cloning site of pCI (Promega, Madison, WI). The env genes were amplified from the previously described proviruses (Kuhmann et al., 2004) by PCR using PfuTurbo polymerase (Stratagene, La Jolla, CA), according to the manufacturer's instructions. The primers used were EcoEnv (5'-GCGGCGGAATTCGACAGTGGCAATGAGAGTGAAGG-3') which is specific for the 5' end of the env gene and added a unique EcoRI restriction site and XhoNef (5'-GCCGCCCTCGAGATACTGCTCCCACCC-3') which overlaps the naturally occuring XhoI restricition site in the nef gene of these viruses. The PCR products were then cut with EcoRI and XhoI and cloned directly into a pCI plasmid that had been treated with the same restriction enzymes.
The pNLluc-AM vector consists of the pNL4-3 proviral plasmid with a portion of the env gene deleted and replaced with an SV40 promoter/firefly luciferase cassette. The construction of the pNLluc-AM vector was accomplished by using a yeast recombination system (Marozsan and Arts, 2003). Briefly, the LEU2 gene was PCR-amplified using the NLleu-S (5'-GGTGGAAATGG GGCACCATGCTCCTTGGGATATTGATGATCTGTAGTCCGCGGAGATTGTACTGAGAGTGCAC-3') and NLleu-AS (5'-CTTTTTTCTCTCTGCACCACTCTTCTCTTTGCCTTGGTGGGTGCTACCTGTGCGGTATTTCACACCG-3') primers. The product was then recombined into the pRecEnv vector as described previously (Marozsan and Arts, 2003), to form the pRecEnvLEU2 vector. Next, the SV40 promoter and luciferase gene were PCR-amplified from pGL3-Control vector (Promega) using the NLluc-S (5'-GGTGGAAATGGGGCACCATGCTCCTTGGGATATTGATGATCTGTAGTTAATGCATCTCAATTAGTCAGCAACCA-3') and NLluc-AS (5'-CTTTTTTCTCTCTGCACCACTCTTCTCTTTGCCTTGGTGGGTGCTACTTACACGGCGATCTTTCCGCCCTTCT-3') primers. The product was then recombined into the pRecEnvLEU2 vector to form pRecEnvLuc. The HIV-Luciferase chimeric sequence from pRecEnvLuc was inserted into pNL4-3 using the EcoRI and XhoI restriction sites. The final pNLluc-AM construct contains the SV40 promoter and luciferase gene recombined into pNL4-3 at positions 6308 to 7692. It has an in-frame stop codon in the env gene prior to the SV40 promoter, to prevent translation through the inserted cassette. This construct leaves all the HIV-1 genes from the NL4-3 provirus intact, with the exception of env.
Similarly, the pNLhrGFPII plasmid consists of the pNL4-3 proviral plasmid with a portion of the env gene replaced with a CMV promoter/hrGFPII cassette. To make this plasmid, the env-containing EcoRI to XhoI fragment of pNL4-3 was inserted into pBluescript II (KS+) (Stratagene) that had been cut with the same enzymes. The resulting plasmid, pKS-NL4-3, was digested with the restriction enzymes NdeI and BglII, as was the plasmid phrGFPII-N (Stratagene). The fragment from phrGFPII-N containing the CMV promoter and the hrGFPII gene was then ligated into pKS-NL4-3 from which a fragment of the env gene between the 3' end of the vpu gene and the 5' end of the Rev-response element had been removed. The EcoRI to XhoI fragment from the resulting plasmid, pKS-NLhrGFPII, was then subcloned back into the pNL4-3 plasmid to form pNLhrGFPII. Similar to pNLluc-AM, this plasmid leaves all of the HIV-1 genes intact with the exception of env. Both the pNLluc-AM and pNLhrGFPII plasmids can be complemented with a pCI-env expression plasmid to form pseudoviruses capable of a single round of infection. These two pseudoviruses transduce expression of firefly luciferase and hrGFPII reporters, respectively.
Virus and pseudovirus stocks
The CC1/85 and D1/85.16 virus isolates were propagated in PBMC as previously described (Trkola et al., 2002). The D1/85.16 virus was propagated in the presence of 25 μM SCH-D. The D1/85.16(−) virus was obtained by pelleting PBMC from the culture medium 24 h prior to harvesting the virus stocks, washing the cells twice in culture medium, and returning them to culture in medium lacking SCH-D. The D1/85.16(+) isolate was propagated in the same way, except that the final culture medium contained 25 μM SCH-D. Infectious, clonal virus stocks were made by transfection of 293T cells with pNL4-3/env plasmids using Lipofectamine 2000 (Invitrogen) as previously described (Kuhmann et al., 2004). All infectious virus stocks were filtered through a 0.45 μm filter, aliquoted and stored at −80°C before use. The infectious titer, measured in 50% tissue culture infective doses (TCID50), was determined in PBMC prior to use by standard methods (Japour et al., 1993).
Pseudotyped viruses were made by cotransfection of 293T cells with a 3:1 ratio of the plasmids pCI-env and pNLluc-AM, or pCI-env and pNLhrGFPII, using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. One day after transfection, the cells were washed twice with culture medium then incubated for an additional day. The pseudovirus-containing supernatants were filtered with a 0.45 μm filter and used immediately afterwards.
HIV-1 replication in primary cells
The assessment of HIV-1 sensitivity to entry inhibitors was performed as described previously (Kuhmann et al., 2004; Pugach et al., 2004). Briefly, 2 × 105 PBMC were seeded into each well of a 96-well culture plate. The PBMC used in this assay consisted of equal numbers of cells from each of the two stimulation conditions and were derived from two or three individuals; the cells were pelleted and resuspended in PBMC culture medium after three days of stimulation as described above. They were then treated with inhibitors as described in the text and figure legends, the recorded concentrations of the inhibitors taking into account the added volume of inoculum virus. Each well of the 96-well plate was then inoculated with 100 TCID50 of the appropriate virus. The three replication-competent clonal viruses were produced by transfection under comparable conditions, and the titers were similar for all three. Hence, these viruses were all used at the same dilution, and similar quantities of p24 were therefore added to each well. For example, the average input quantities of p24 per well in the experiment shown in Fig.3B were 1.7 ± 0.3 ng/ml, 1.4 ± 0.3 ng/ml and 1.8 ± 0.4 ng/ml for the CC1/85 cl.7, CC101.19 cl.7 and D1/85 cl.23 viruses, respectively. The ensuing production of p24 antigen was also similar; thus, in the above experiment, the amounts of p24 produced (in the absence of PA12) were 14.1 ± 3.5 ng/ml, 11.1 ± 3.7 ng/ml and 10.0 ± 3.1 ng/ml for the same three viruses, respectively.
For the inhibitor-combination assays, the cells were incubated with SCH-D for 1 h at 37°C prior to addition of a second inhibitor for 1 h at 37°C, and then the replication-competent virus. Production of the viral p24 antigen after 7 days of culture was quantified using an in-house ELISA (Trkola et al., 1995). In each assay, each data point was derived from triplicate wells. The amount of p24 produced was corrected by subtracting the residual p24 remaining from the added virus. The data were used only if replication in the absence of any inhibitor produced at least 5 ng/ml of p24 (typically, the cultures produced 10 to 20 ng/ml of p24). The results obtained using inhibitors are expressed as a percent of control. For the single inhibitor experiments, the control was replication in the absence of any inhibitor; for combination experiments, the control was replication in the presence of 1 μM SCH-D, but the absence of any additional inhibitor.
HIV-1 entry assays
U87-CD4/CCR5 cells were seeded in white 96-well plates at a density of 3000 cells per well in a volume of 100 μl. The following day, 50 μl of SCH-D diluted in culture medium to four times the desired final concentration was added to each well for a 1 h incubation at 37°C. Freshly harvested supernatants from a pCI-env/pNLluc-AM co-transfection culture (50 μl) which contained the appropriate Env-pseudotyped viruses were then added, and the plates were incubated for 3 days at 37°C. At that time, 100 μl of supernatant was removed from each well plate and was replaced with 100 μl of Bright-Glo Luciferase Substrate (Promega). After 5 min, the plates were analyzed in a Victor3 1420 plate-reading luminometer (Perkin Elmer, Wellesley, MA). No background luminescence was evident in uninfected cells. Each data point in each experiment was derived from the average of triplicate wells. The percent inhibition of entry was calculated by taking 100×[1−(lumtest/lumcon)], where lumcon is the luminescence (in arbitrary RLU) of the control infection (no inhibitor), and lumtest is the luminescence obtained from the test condition (e.g., an inhibitor present).
For the hrGFPII entry assays, 3 × 105 U87-CD4/CCR5 cells were seeded into a six well plate 24 h prior to infection. On the day of infection 2× 106 PBMC consisting of equal number of cells derived from both stimulation conditions and from two or three donors were seeded in a six well plate. Both cell types were incubated with twice the desired final SCH-D concentration in 1 ml of the appropriate culture medium for 1 h at 37°C. A 1 ml aliquot of freshly harvested supernatant from the appropriate pCI-env/pNLhrGFPII transfection culture was then added, and the cells were incubated for 4 days at 37°C. The U87-CD4/CCR5 cells were then lifted from the wells using trypsin (Invitrogen), and pelleted. The PBMC were removed from the plates and also pelleted. The cell pellets were resuspended in 1 ml of ICFix cell fixation buffer (Invitrogen) and incubated for 10 min at 25°C, at which time 1 ml of cytometry buffer was added. The cells were then assayed for hrGFPII expression by cytometry. Gating of the U87-CD4/CCR5 cells or the lymphocyte population from the PBMC cultures was based on forward- and side-scatter parameters. Approximately 2 × 105 U87-CD4/CCR5 cells and 2 × 106 lymphocytes were analyzed for each data point in each experiment. The gate for hrGFPII+ cells was set by exclusion of all events when running an uninfected sample of the same cell type. The percent inhibition of entry was calculated as 100×[1−(rattest/ratcon)], where ratcon is the ratio of hrGFPII+ cells to the total cells in the control infection (no inhibitor), and rattest is the ratio of hrGFPII+ cells to the total cells used in the test culture. For both entry assays, the CC1/85 cl.7 and CC101.19 cl.7 pseudotypes were always produced in comparable transfection cultures at the same time, and the PBMC and U87-CD4/CCR5 cells were always tested at the same time under comparable conditions in the hrGFPII entry assay.
The Env-pseudotype, luciferase-readout system is a reporter assay in which enzyme activity can be assumed to be proportional to viral entry. In contrast, the hrGFPII pseudotypes create a focal-infectivity assay; the cells are scored as either being infected or not infected, based on hrGFPII expression, but we did not assume that the intensity of hrGFPII fluorescence in the infected cells was related to the efficiency of entry. The readout in each assay, luciferase activity or the fraction of hrGFPII+ cells, was proportional to the amount of input virus when serial dilutions were applied to U87-CD4/CCR5 cells in the absence of an inhibitor (data not shown). This finding validates the use of each assay to measure viral entry.
Analysis of replication and entry data
The IC50 data reported in the text and in Table 3 are based on the data in Fig.3. They are derived from a non-linear sigmoidal dose-response curve fit performed in Prism (GraphPad Software, San Diego, CA) that assumes the maximal infection level to be 100% and the minimum to be 0%. The plateau levels reported in the text and in Table 4 were estimated from the data in Fig.5, and the plateau was considered to be 100% if the extent of inhibition at 1 μM SCH-D was not significantly different from 100% (P≥0.05 in a paired comparison t-test). In that scenario, the IC50 was determined as described in Table 3, assuming 100% maximum inhibition. If, however, the extent of inhibition at 1 μM SCH-D was significantly different from 100% (P<0.05 in a paired comparision t-test), both the plateau level and the EC50 at which the half-plateau was reached were determined from a non-linear sigmoidal dose-response curve fit of the data.
Chemokine ELISA
PBMC isolated from individual donors were stimulated for three days, then seeded at 106 cells in 1 ml volume in 24 well plates. The cells were incubated without treatment, with 50 μg/ml PRO 140, or with 2 μM SCH-D for seven days before the supernatants were collected for analysis. MIP-1α, MIP-1β and RANTES levels were assayed using Quantikine ELISA kits (R&D Systems, Minneapolis, MN) according to the manufacturer's instructions.
Acknowledgements
We thank William Olson, Julie Strizki and David Kabat for providing several reagents used in this study. We are grateful to Oliver Hartley for providing PSC-RANTES and technical advice on its use. We appreciate critical reading of the manuscript and helpful comments from Per Johan Klasse, Julie Strizki and Christoph Seibert. This work was funded by NIH awards R01 AI 41420 and U19 AI 66329, by the NIH Immunology Training Grant T32 AI 07621 (PP) and by the NIH NRSA Fellowship F32 AI 062664 (SEK). The Department of Microbiology and Immunology gratefully acknowledges the support of the William Randolph Hearst Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Billick E, Seibert C, Pugach P, Ketas T, Trkola A, Endres MJ, Murgolo NJ, Coates E, Reyes GR, Baroudy BM, Sakmar TP, Moore JP, Kuhmann SE. The differential sensitivity of human and rhesus macaque CCR5 to small-molecule inhibitors of human immunodeficiency virus type 1 entry is explained by a single amino acid difference and suggests a mechanism of action for these inhibitors. J. Virol. 2004;78(8):4134–4144. doi: 10.1128/JVI.78.8.4134-4144.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorndal A, Deng H, Jansson M, Fiore JR, Colognesi C, Karlsson A, Albert J, Scarlatti G, Littman DR, Fenyo EM. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J. Virol. 1997;71(10):7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coakley E, Petropoulos CJ, Whitcomb JM. Assessing chemokine co-receptor usage in HIV. Curr. Opin. Infect. Dis. 2005;18(1):9–15. doi: 10.1097/00001432-200502000-00003. [DOI] [PubMed] [Google Scholar]
- Cormier EG, Dragic T. The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J. Virol. 2002;76(17):8953–8957. doi: 10.1128/JVI.76.17.8953-8957.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier EG, Tran DN, Yukhayeva L, Olson WC, Dragic T. Mapping the determinants of the CCR5 amino-terminal sulfopeptide interaction with soluble human immunodeficiency virus type 1 gp120-CD4 complexes. J. Virol. 2001;75(12):5541–5549. doi: 10.1128/JVI.75.12.5541-5549.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, Cayanan C, Maddon PJ, Koup RA, Moore JP, Paxton WA. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381(6584):667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- Dragic T, Trkola A, Thompson DA, Cormier EG, Kajumo FA, Maxwell E, Lin SW, Ying W, Smith SO, Sakmar TP, Moore JP. A binding pocket for a small molecule inhibitor of HIV-1 entry within the transmembrane helices of CCR5. Proc. Natl. Acad. Sci. USA. 2000;97(10):5639–5644. doi: 10.1073/pnas.090576697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatkenheuer G, Pozniak AL, Johnson MA, Plettenberg A, Staszewski S, Hoepelman AI, Saag MS, Goebel FD, Rockstroh JK, Dezube BJ, Jenkins TM, Medhurst C, Sullivan JF, Ridgway C, Abel S, James IT, Youle M, van der Ryst E. Efficacy of short-term monotherapy with maraviroc, a new CCR5 antagonist, in patients infected with HIV-1. Nat. Med. 2005;11(11):1170–1172. doi: 10.1038/nm1319. [DOI] [PubMed] [Google Scholar]
- Gorry PR, Taylor J, Holm GH, Mehle A, Morgan T, Cayabyab M, Farzan M, Wang H, Bell JE, Kunstman K, Moore JP, Wolinsky SM, Gabuzda D. Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate. J. Virol. 2002;76(12):6277–6292. doi: 10.1128/JVI.76.12.6277-6292.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley O, Gaertner H, Wilken J, Thompson D, Fish R, Ramos A, Pastore C, Dufour B, Cerini F, Melotti A, Heveker N, Picard L, Alizon M, Mosier D, Kent S, Offord R. Medicinal chemistry applied to a synthetic protein: development of highly potent HIV entry inhibitors. Proc. Natl. Acad. Sci. USA. 2004;101(47):16460–16465. doi: 10.1073/pnas.0404802101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley O, Klasse PJ, Sattentau QJ, Moore JP. V3: HIV's switch-hitter. AIDS Res. Hum. Retroviruses. 2005;21(2):171–189. doi: 10.1089/aid.2005.21.171. [DOI] [PubMed] [Google Scholar]
- Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310(5750):1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Japour AJ, Mayers DL, Johnson VA, Kuritzkes DR, Beckett LA, Arduino JM, Lane J, Black RJ, Reichelderfer PS, D'Aquila RT, Crumpacker CS. Standardized peripheral blood mononuclear cell culture assay for determination of drug susceptibilities of clinical human immunodeficiency virus type 1 isolates. The RV-43 Study Group, the AIDS Clinical Trials Group Virology Committee Resistance Working Group. Antimicrob. Agents Chemother. 1993;37(5):1095–1101. doi: 10.1128/aac.37.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmierski W, Bifulco N, Yang H, Boone L, DeAnda F, Watson C, Kenakin T. Recent progress in discovery of small-molecule CCR5 chemokine receptor ligands as HIV-1 inhibitors. Bioorg. Med. Chem. 2003;11(13):2663–2676. doi: 10.1016/s0968-0896(03)00161-5. [DOI] [PubMed] [Google Scholar]
- Kenakin T. G-protein coupled receptors as allosteric machines. Receptors Channels. 2004;10(2):51–60. doi: 10.1080/10606820490464316. [DOI] [PubMed] [Google Scholar]
- Koning FA, Kwa D, Boeser-Nunnink B, Dekker J, Vingerhoed J, Hiemstra H, Schuitemaker H. Decreasing sensitivity to RANTES (regulated on activation, normally T cell-expressed and -secreted) neutralization of CC chemokine receptor 5-using, non-syncytium-inducing virus variants in the course of human immunodeficiency virus type 1 infection. J. Infect. Dis. 2003;188(6):864–872. doi: 10.1086/377105. [DOI] [PubMed] [Google Scholar]
- Kuhmann SE, Pugach P, Kunstman KJ, Taylor J, Stanfield RL, Snyder A, Strizki JM, Riley J, Baroudy BM, Wilson IA, Korber BT, Wolinsky SM, Moore JP. Genetic and phenotypic analyses of human immunodeficiency virus type 1 escape from a small-molecule CCR5 inhibitor. J. Virol. 2004;78(6):2790–2807. doi: 10.1128/JVI.78.6.2790-2807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landovitz R, Faetkenhauer G, Hoffmann C, Horst H, Strizki J, Whitcomb J, Gheyas F, Knepp D, Greaves W. Abstracts of the XV International Drug Resistance Workshop; Stiges, Spain. 2006. [Google Scholar]
- Lee B, Sharron M, Blanpain C, Doranz BJ, Vakili J, Setoh P, Berg E, Liu G, Guy HR, Durell SR, Parmentier M, Chang CN, Price K, Tsang M, Doms RW. Epitope mapping of CCR5 reveals multiple conformational states and distinct but overlapping structures involved in chemokine and coreceptor function. J. Biol. Chem. 1999;274(14):9617–9626. doi: 10.1074/jbc.274.14.9617. [DOI] [PubMed] [Google Scholar]
- Lu J, Sista P, Giguel F, Greenberg M, Kuritzkes DR. Relative replicative fitness of human immunodeficiency virus type 1 mutants resistant to enfuvirtide (T-20) J. Virol. 2004;78(9):4628–4637. doi: 10.1128/JVI.78.9.4628-4637.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marozsan AJ, Arts EJ. Development of a yeast-based recombination cloning/system for the analysis of gene products from diverse human immunodeficiency virus type 1 isolates. J. Virol. Methods. 2003;111(2):111–120. doi: 10.1016/s0166-0934(03)00166-6. [DOI] [PubMed] [Google Scholar]
- Marozsan AJ, Kuhmann SE, Morgan T, Herrera C, Rivera-Troche E, Xu S, Baroudy BM, Strizki J, Moore JP. Generation and properties of a human immunodeficiency virus type 1 isolate resistant to the small molecule CCR5 inhibitor, SCH-417690 (SCH-D) Virology. 2005;338(1):182–199. doi: 10.1016/j.virol.2005.04.035. [DOI] [PubMed] [Google Scholar]
- Murga JD, Franti M, Pevear DC, Maddon PJ, Olson WC. Potent Antiviral Synergy between Monoclonal Antibody and Small-Molecule CCR5 Inhibitors of Human Immunodeficiency Virus Type 1. J. Virol. 2006;50(10):3289–3296. doi: 10.1128/AAC.00699-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson WC, Rabut GE, Nagashima KA, Tran DN, Anselma DJ, Monard SP, Segal JP, Thompson DA, Kajumo F, Guo Y, Moore JP, Maddon PJ, Dragic T. Differential inhibition of human immunodeficiency virus type 1 fusion, gp120 binding, and CCchemokine activity by monoclonal antibodies to CCR5. J. Virol. 1999;73(5):4145–4155. doi: 10.1128/jvi.73.5.4145-4155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxton WA, Dragic T, Koup RA, Moore JP. The beta-chemokines, HIV type 1 second receptors, and exposed uninfected persons. AIDS Res. Hum. Retroviruses. 1996a;12(13):1203–1207. doi: 10.1089/aid.1996.12.1203. [DOI] [PubMed] [Google Scholar]
- Paxton WA, Martin SR, Tse D, O'Brien TR, Skurnick J, VanDevanter NL, Padian N, Braun JF, Kotler DP, Wolinsky SM, Koup RA. Relative resistance to HIV-1 infection of CD4 lymphocytes from persons who remain uninfected despite multiple high-risk sexual exposure. Nat. Med. 1996b;2(4):412–417. doi: 10.1038/nm0496-412. [DOI] [PubMed] [Google Scholar]
- Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 1998;72(4):2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugach P, Kuhmann SE, Taylor J, Marozsan AJ, Snyder A, Ketas T, Wolinsky SM, Korber BT, Moore JP. The prolonged culture of human immunodeficiency virus type 1 in primary lymphocytes increases its sensitivity to neutralization by soluble CD4. Virology. 2004;321(1):8–22. doi: 10.1016/j.virol.2003.12.012. [DOI] [PubMed] [Google Scholar]
- Safarian D, Carnec X, Tsamis F, Kajumo F, Dragic T. An anti-CCR5 monoclonal antibody and small molecule CCR5 antagonists synergize by inhibiting different stages of human immunodeficiency virus type 1 entry. Virology. 2006 doi: 10.1016/j.virol.2006.05.016. [DOI] [PubMed] [Google Scholar]
- Schuermann D, Pechardscheck C, Rouzier R, Nougarede R, Faetkenheuer G, Ochlast I, Raffi F, Hoffman C, Greaves W, Sansone A. The 3rd International AIDS Society Conference on HIV Pathogenesis and Treatment; Rio De Janeiro, Brazil. 2005. [Google Scholar]
- Seibert C, Sakmar TP. Small-molecule antagonists of CCR5 and CXCR4: a promising new class of anti-HIV-1 drugs. Curr. Pharm. Des. 2004;10(17):2041–2062. doi: 10.2174/1381612043384312. [DOI] [PubMed] [Google Scholar]
- Seibert C, Ying W, Gavrilov S, Tsamis F, Kuhmann SE, Palani A, Tagat JR, Clader JW, McCombie SW, Baroudy BM, Smith SO, Dragic T, Moore JP, Sakmar TP. Interaction of small molecule inhibitors of HIV-1 entry with CCR5. Virology. 2006;349(1):41–54. doi: 10.1016/j.virol.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Strizki JM, Tremblay C, Xu S, Wojcik L, Wagner N, Gonsiorek W, Hipkin RW, Chou CC, Pugliese-Sivo C, Xiao Y, Tagat JR, Cox K, Priestley T, Sorota S, Huang W, Hirsch M, Reyes GR, Baroudy BM. Discovery and characterization of vicriviroc (SCH 417690), a CCR5 antagonist with potent activity against human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 2005;49(12):4911–4919. doi: 10.1128/AAC.49.12.4911-4919.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strizki JM, Xu S, Wagner NE, Wojcik L, Liu J, Hou Y, Endres M, Palani A, Shapiro S, Clader JW, Greenlee WJ, Tagat JR, McCombie S, Cox K, Fawzi AB, Chou CC, Pugliese-Sivo C, Davies L, Moreno ME, Ho DD, Trkola A, Stoddart CA, Moore JP, Reyes GR, Baroudy BM. SCH-C (SCH 351125), an orally bioavailable, small molecule antagonist of the chemokine receptor CCR5, is a potent inhibitor of HIV-1 infection in vitro and in vivo. Proc. Natl. Acad. Sci. USA. 2001;98(22):12718–12723. doi: 10.1073/pnas.221375398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagat JR, McCombie SW, Nazareno D, Labroli MA, Xiao Y, Steensma RW, Strizki JM, Baroudy BM, Cox K, Lachowicz J, Varty G, Watkins R. Piperazine-based CCR5 antagonists as HIV-1 inhibitors. IV. Discovery of 1-[(4,6-dimethyl-5-pyrimidinyl)carbonyl]- 4-[4-[2-methoxy-1(R)-4-(trifluoromethyl)phenyl]ethyl-3(S)-methyl-1-piperaz inyl]- 4-methylpiperidine (Sch-417690/Sch-D), a potent, highly selective, and orally bioavailable CCR5 antagonist. J. Med. Chem. 2004;47(10):2405–2408. doi: 10.1021/jm0304515. [DOI] [PubMed] [Google Scholar]
- Trkola A, Kuhmann SE, Strizki JM, Maxwell E, Ketas T, Morgan T, Pugach P, Xu S, Wojcik L, Tagat J, Palani A, Shapiro S, Clader JW, McCombie S, Reyes GR, Baroudy BM, Moore JP. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proc. Natl. Acad. Sci. USA. 2002;99(1):395–400. doi: 10.1073/pnas.012519099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Pomales AB, Yuan H, Korber B, Maddon PJ, Allaway GP, Katinger H, Barbas CF, 3rd, Burton DR, Ho DD, et al. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J. Virol. 1995;69(11):6609–6617. doi: 10.1128/jvi.69.11.6609-6617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsamis F, Gavrilov S, Kajumo F, Seibert C, Kuhmann S, Ketas T, Trkola A, Palani A, Clader JW, Tagat JR, McCombie S, Baroudy B, Moore JP, Sakmar TP, Dragic T. Analysis of the mechanism by which the small-molecule CCR5 antagonists SCH-351125 and SCH-350581 inhibit human immunodeficiency virus type 1 entry. J. Virol. 2003;77(9):5201–5208. doi: 10.1128/JVI.77.9.5201-5208.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson C, Jenkinson S, Kazmierski W, Kenakin T. The CCR5 receptor-based mechanism of action of 873140, a potent allosteric noncompetitive HIV entry inhibitor. Mol. Pharmacol. 2005;67(4):1268–1282. doi: 10.1124/mol.104.008565. [DOI] [PubMed] [Google Scholar]
- Westby M, Mori J, Smith-Burchnell C, Lewis M, Mosley M, Perruccio F, Mansfield R, Dorr P, Perros M. Abstracts of the XIV International Drug Resistance Workshop; Quebec, Canada. 2005. [Google Scholar]
- Westby M, van der Ryst E. CCR5 antagonists: host-targeted antivirals for the treatment of HIV infection. Antivir. Chem. Chemother. 2005;16(6):339–354. doi: 10.1177/095632020501600601. [DOI] [PubMed] [Google Scholar]