Abstract
Purpose:
The purpose of this study is to demonstrate that the expression of rhodopsin can be down regulated in vivo by AAV-delivered siRNA. This is the first step in an RNA replacement strategy for the allele-independent treatment of Autosomal Dominant Retinitis Pigmentosa (ADRP).
Methods:
HEK 293 cells were co-transfected with a plasmid carrying mouse RHO cDNA driven by the CMV promoter and a chemically synthesized siRNA duplex of 21 nucleotides. Reduction of RHO mRNA was confirmed by RT-PCR. One active siRNA and a control siRNA were embedded in a small hairpin RNA (shRNA) and cloned in Adeno-associated virus (AAV) vector under regulation of the H1 promoter and containing a GFP reporter. AAV5 expressing either active siRNA or an irrelevant siRNA were subretinaly injected into the right eyes of wild-type or RHO+/− heterozygote mice at postnatal day 16. At 1 and 2 months post injection, animals were analyzed by electroretinography (ERG). Animals were then sacrificed, and retinas were examined by western blot, RT-PCR, histology and immunohistochemistry.
Results:
All of the siRNAs tested in HEK 293 cells caused degradation of RHO mRNA, although the efficiency varied from 25% to 80%. In vivo siRNA delivery to the retina led to more than 40% reduction of scotopic a-and b-wave amplitudes in RHO+/− heterozygotes. Although the reduction of RHO mRNA was estimated at 30% compared to control animals, western blots revealed 60% decrease in rhodopsin content. Histological analysis showed significant reduction in the thickness of the ONL, ranging between 53% and 86%.
Conclusions:
AAV-siRNA delivery into the subretinal space resulted in the reduction of retinal function caused by diminished RHO mRNA and protein content. This level of reduction may permit the replacement of endogenous mRNA with siRNA-resistant mRNA encoding wild-type RHO.
Keywords: retina, retinitis pigmentosa, gene therapy, rhodopsin, RNA interference, Adeno-associated virus
1. Introduction
Retinitis Pigmentosa (RP) is an heterogeneous group of diseases clinically characterized by loss of the night vision followed by progressive loss of peripheral vision (Krauss and Heckenlively, 1982 ). The disease is caused by heritable defects in rod photoreceptor cells or the RPE (retinal pigment epithelium) cells and may be transmitted in an autosomal dominant (ADRP), autosomal recessive (ARRP) or X-linked (XLRP) fashion. Over 120 mutations in the RHO gene, which encodes rhodopsin, have been identified. The majority cause ADRP.
Gene therapy for ADRP can adopt a direct or an indirect strategy. Indirect approaches support the survival of rod cells without affecting expression of the mutated protein. For example, neurotrophic factors like GDNF (McGee Sanftner et al., 2001 ) and antiapoptotic proteins such as XIAP (Petrin et al., 2003 ) may preserve vision in ADRP by blocking apoptotic death of photoreceptors. The direct approach involves modulating relative levels of mutant and wild-type protein. The study of dominant negative forms of RP, often associated with toxicity of mutated RHO, suggests that the mutant gene must be repaired or silenced. Two strategies have been proposed for RHO mRNA silencing. The first includes the use of allele-specific inhibitors, which block the expression of only the defective mRNA and allow expression of the normal allele. Expression of only the wild-type allele should be sufficient to maintain the function of surviving rod cells (Liang et al., 2004 ). To be widely applicable, however, the considerable heterogeneity among RHO mutations would likely require the development of a large number of mutation-specific inhibitors. The second strategy takes an allele-independent approach: antisense agents are designed to suppress all RHO alleles, mutant and wild-type (Farrar et al., 2002 ). Therefore, allele independent RNA inhibitors are more useful, since a single reagent can, in theory, be used against different alterations in the RHO gene. For optimal therapy, they should be used in combination with wild-type cDNA containing silent mutations that block base pairing with the antisense inhibitor.
A variety of such antisense inhibitors are available. These include small catalytic RNA (ribozymes), antisense oligonucleotides, small interfering RNA (siRNA) and antisense transcripts that regulate alternative mRNA splicing (Alfano et al., 2005 ). It has already been established that ribozymes can limit gene expression by cleavage of targeted mRNA in the retina (Drenser et al., 1998 ; Gorbatyuk et al., 2005 ; Lewin et al., 1998 ). Despite this successful application of ribozymes in vivo, many researchers consider siRNA a more potent and more durable approach. It has been estimated that the half-maximal inhibition levels of siRNA are some 100-to 1,000-fold lower than an optimal antisense oligonucleotide directed against the same target (Miyagishi et al., 2003 ). Several experiments suggest that RNA interference using siRNA is also more effective than ribozymes or DNAzymes (Akashi et al., 2005 ; Yokota et al., 2004 ). Small interfering RNAs can be used in an allele independent approach for ADRP gene therapy. For example, Kiang et al. have designed siRNAs targeting the RHO gene and created a resistant RHO gene with 7 mismatches to replace the native one (Kiang et al., 2005 ). They demonstrated in tissue culture experiments that the modified RHO mRNA was resistant to siRNA-mediated attack, even at high concentrations of the siRNA. In vivo RNA interference has been used to reduce the expression of growth factors in the retina, suggesting that this approach should be applicable if the correct siRNA and delivery system are employed (Kwak et al., 2000 ; Nakamura et al., 2004 ; Reich et al., 2003 ; Saishin et al., 2003 ). Small interfering RNA can be processed from small hairpin RNA (shRNA) driven by an RNA pol III promoter like U6 or by a pol II promoter like CMV (Xia et al., 2002 ).
Only just recently has RHO directed gene therapy been attempted in the retina using siRNA (Tessitore et al., 2006 ). In their study, Tessitore et al. used AAV5 containing the U6 promoter to deliver an shRNA preferentially targeting the mouse P23H RHO transgene. Expression of this allele-specific siRNA reduced the mutant RHO mRNA in P23H line 3 rats and, presumably, should have decreased the level of P23H rhodopsin. However, suppression of the P23H RHO allele did not lead to the rescue of vision in these transgenic animals. The authors concluded that more robust shRNA expression in the retina may be required to achieve therapeutic efficacy in vivo.
In the current study, instead of targeting a specific mutant allele we designed shRNA molecules that would cause degradation of both wild-type and mutant alleles of RHO. We used the H1 promoter system and AAV serotype 5 to transfer genes for short hairpin RNAs acting as a precursor for RHO-specific siRNAs. Our goal was to demonstrate that the expression of RHO, the most abundant retinal protein, can be down-regulated by AAV-delivered siRNA to the retina. This is the first step in an RNA replacement strategy for the allele-independent treatment of ADRP, requiring, as a second step, supplementation with an siRNA-resistant RHO to retinas treated with siRNA.
2. Materials and methods
2.1. Design and screening of siRNAs in cultured cells
We designed siRNAs targeting the mouse RHO coding sequence based on recommendations described by Jagla et al. (Jagla et al., 2005 ). The targets of the siRNAs comprised of 19 nucleotides of the mouse RHO gene (Table 1). Some siRNA targets were conserved between mouse and human or mouse and dog RHO mRNA. These target sequences were used to screen GenBank using BLAST to avoid possible off-target effects in the mouse. Duplexes of the designed siRNAs were purchased from Dhramacon, Inc. (Lafayette, CO). The numeration of all siRNAs came from the location of the targeting segment on RHO mRNA sequences.
Table I.
RHO siRNA target sequences
| siRNA | targeted region in RHO sequence | specificity |
|---|---|---|
| siRNA301 | CCTCTTCACGCTCTACGTC | mouse, human, dog |
| siRNA589 | GAGGTCAACAACGAATCCT | mouse |
| siRNA750 | CCCGCATGGTTATCATCAT | mouse |
| siRNA855 | AGAGCTCTTCCATCTATAA | mouse |
To test siRNAs in vitro, HEK 293 cells were co-transfected with synthetic siRNAs and with plasmids expressing mouse RHO driven by the CMV promoter. An irrelevant siRNA (targeting a cardiac-specific mRNA) was used as a control. Transfection of HEK 293 cells, RNA extraction and quantitation of RHO mRNA by RT-PCR was performed as described in an earlier publication (Gorbatyuk et al., 2005 ).
2.2 rAAV-siRNA constructs
For delivery and prolonged expression in animal tissues, siRNA genes were inserted into pSilencer plasmid (Ambion) under control of the H1 RNA polymerase III promoter and an oligoT terminator sequence. The expression cassette was then cloned into the Sal I sites of AAV2 vectors (pTR-UF11) that also contained the GFP gene driven by the CMV enhancer/chicken β-actin (CBA) chimeric promoter (Fig. 2). This vector was packaged in AAV5 capsids, which have been shown to lead to more rapid and robust transduction of photoreceptor cells than AAV2 (Auricchio and Rolling, 2005 ; Rabinowitz et al., 2002 ). Infected cells expressing both the siRNA and GFP could be monitored by GFP fluorescence.
Fig.2.
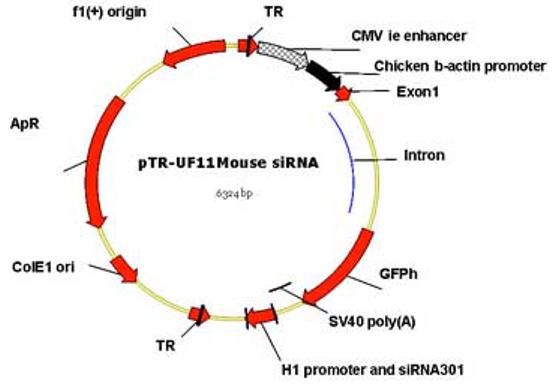
Map of the rAAV vector. Expression of siRNA was driven by an H1 promoter. Green fluorescent protein (GFP) was expressed by the hybrid CMV enhancer – chicken β-actin (CBA) promoter. Localization of siRNA301 was monitored by detection of humanized GFP (GFPh). The region packaged in AAV was flanked by the inverted terminal repeats of AAV2 (TR). Viruses were psuedotyped by packaging into AAV-5 capsids.
2.3. Subretinal vector injection
Mice were treated in accordance with the policies of the University of Florida IACUC and in compliance with the ARVO Statement for the Use of Animals for Ophthalmic and Vision Research. The siRNA vector was tested in heterozygous RHO knockout mice (RHO+/−) and C57BL/6J (RHO+/+). Heterozygous mice were created by crossing C57BL/6 (RHO+/+) mice with RHO knockout mice (RHO−/−). The RHO knockout mice were generated by disruption of both opsin alleles (Lem et al., 1999 ) and were a generous gift of Dr. J. Lem.
Mice were injected subretinally in their right eyes at post natal day 16 (P16) with AAV5 siRNA301 or AAV5 expressing an irrelevant siRNA (specific for cardiac phospholamban mRNA, GenBank gi:257745). Both siRNAs were cloned and packaged identically. The subretinal injection method was described by Timmers et al. (Timmers et al., 2001 ). 0.5 μl of AAV5-ribozyme (1.3×1012 vector genomes) was injected subretinally into the right eyes of RHO+/− mice. Left eyes served as an untreated control. A single cohort of animals of each genotype was used for ERG analysis (see below) at 1 and 2 months and for analysis of rhodopsin mRNA and protein content at the end of the experiment.
2.4. Visual function tests
For ERG analysis, mice were dark-adapted overnight, and scotopic ERGs, which primarily measure rod function, were elicited with 10 μsec flashes of white light at intensities of 0.02, 0.018 and 2.68 cd-s/m2. Five flashes were averaged at each light intensity. Details of analysis are described in Gorbatyuk et al. (Gorbatyuk et al., 2005 ).
2.5. Isolation of total RNA for quantitative analysis of RHO mRNA
Total RNA was isolated from each retina with an RNAqueous isolation kit (Ambion) following the manufacturer's procedure. Isolated RNA was treated with DNAse I (Ambion) to remove genomic DNA contamination. Quantitative RNA analysis was performed by comparison of RHO and β-actin amplification products from right and left eyes of individual mice (Gorbatyuk et al., 2005 ). The amount of RHO mRNA in a total RNA sample was normalized to the level of β-actin PCR product in each sample using Sybr-Green staining.
2.6. Protein analysis
Individual retinal protein extracts were obtained from dissected retinas by sonication in Laemmli loading buffer. 10 μg of total retinal protein was used in each lane. Protein samples were separated by electrophoresis on 12% SDS polyacrylamide gels. Then proteins were transferred to a nitrocellulose membrane, which was incubated overnight with a primary antibodies. As a primary antibody against opsin, we used the B6-30 antibody (a gift from Dr. Paul Hargrave). As an internal control, β-actin was detected by anti-β-actin antibody (Sigma-Aldrich). Detection of protein-primary antibody complexes was done with alkaline phosphatase-conjugated secondary antibodies. The complexes were visualized using the color reagent, 5-bromo-4-chloro-3-indolyl phosphate (Zymed Inc.) and band intensities were analyzed using the BioRad Gel Analysis Software.
2.7. Histological and immunohistochemical analysis
Eyes were fixed in 4% paraformaldehyde overnight at 4°C. Eye cups were then transferred into phosphate buffered saline and submerged sequentially in solutions of 10%, 20% and 30% sucrose. Eye cups were then embedded in OCT medium (Sakura Finetek, Inc.) in order to produce 12 μm frozen sections. The frozen sections were used to measure the outer nuclear layer (ONL) thickness as an indicator of treatment-induced retinal degeneration. Sections of mouse retina fixed as described above were stained with propidium iodide. Images were taken on a Zeiss fluorescent microscope (Axiovert 200) for full retina mapping retina, and 400 μm radial sections, starting from optic nerve (ON), were used to count the ONL lengths by using Axiovision 4.4 Software. Immunostaining with B6-30 antibody was conducted to detect RHO protein. Detection of RHO was done by secondary antibody conjugated with CY5 (Jackson Lab Immunoresearch Co).
2.8. Statistical analysis of data
Student's t-test for paired data was employed to test significance of ERG, protein and RNA measurements. Results were expressed as means with standard error of the mean (SEM). Data from ERG and RNA analysis are presented as the average of the ratios of the injected eye versus uninjected eye in each animal injected with AAV5 expressing siRNA301 or AAV5 expressing an irrelevant siRNA. Protein analysis results were expressed in the direct measurement of right and left eye samples. For comparison of the ONL thickness, results were examined using the one-way ANOVA test.
3. Results
3.1. Screening of siRNA in cultured cells
All siRNAs (301, 589, 750 and 855) were designed to degrade mouse RHO mRNA, with the exception of siRNA301 which also targets dog and human RHO (Table 1). Co-transfection experiments in HEK 293 cells showed that each of the siRNAs could cause degradation of RHO mRNA, although the level of efficiency varied (Fig. 1). SiRNA549 showed about 25% reduction of RHO mRNA, siRNA301 and siRNA750 resulted in 50% of mRNA reduction, while siRNA855 demonstrated 80% knock down of RHO mRNA compared to an irrelevant siRNA. This siRNA efficiently suppresses the synthesis of cardiac specific phospholamban (>50%) in rat primary cardiomyocytes (Andino et al, manuscript in preparation). Despite the fact that siRNA301 reduced mouse RHO mRNA by only 50%, we proceeded to test this inhibitor in mice, since it could also be tested in a large animal model of ADRP as a prelude to clinical testing.
Fig.1.
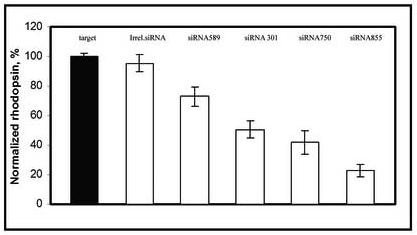
siRNA leads to RHO mRNA degradation in vitro. HEK 293 cells were transfected with a plasmid expressing the mouse RHO gene using the CMV immediate early promoter and duplexes of an irrelevant siRNA (against phospholamban[PLN]), siRNA301, siRNA549, siRNA750 or siRNA855 targeted against RHO mRNA. The high efficiency of the irrelevant siRNA was shown in vitro in neonatal rat cardiomyocytes by Andino et al. (manuscript in preparation). Levels of RHO transcript were measured in triplicate using reverse transcription polymerase chain reaction (RT-PCR) and were normalized to levels of β-actin mRNA amplified in the same reactions. Products were analyzed as described previously (Gorbatyuk et al., 2005 ).
3.2. Delivery of siRNA to the subretinal space
Delivery of siRNA into subretinal space was accomplished by cloning the siRNA sequence in AAV as a small hairpin RNA (shRNA) under the control of the H1 promoter (Fig. 2). Delivery was verified by detection of GFP, which was co-expressed in this vector using a chimeric CMV enhancer - CBA promoter (Fig. 2 ). Transduction extended over 80% of the retina, and expression was restricted to photoreceptor cells. However, not all photoreceptors were infected and, in many animals, the inferior hemisphere was preferentially transduced (data not shown). Mice heterozygous for a disruption of the RHO gene (RHO+/−) and wild-type CB57BL/6 mice (RHO+/+) were injected subretinally in their right eyes at P16 with AAV5 siRNA301, AAV5 expressing an irrelevant siRNA.
3.3. Attenuation of a- and b- wave amplitudes of ERG in response to AAV5 siRNA301
Animals were analyzed by full-field scotopic electroretinography (ERG) at 1 and 2 months after injection. AAV expressing siRNA301 a modest but statistically insignificant reduction in the ERG response of treated eyes in RHO+/+ mice at 1 and 2 months after injection (for a- and b- waves amplitudes p values < 0.08 and 0.2 and 0.08 and < 0.12, respectively) (Fig. 3A). This observation was similar to that previously obtained using a RHO-specific ribozyme in wild-type mice (Gorbatyuk et al., 2005 ). In contrast, in RHO +/− mice AAV delivery of siRNA301 led to a significant reduction in both a- and b-wave maximum amplitudes (Fig. 3B and C). At 1 month after injection, the reduction of a-wave amplitude was 40% ( p<0.012) relative to control eyes injected with an AAV vector expressing irrelevant phospholamban siRNA. At 2 months, this reduction was over 50 %, (p<0.027). ERG b-wave amplitudes were diminished as well. One month after injection, the b-wave amplitude was 25% lower in experimental eyes compared to controls (p<0.027). After the second month, this reduction increased to over 50% (p<0.005).
Fig. 3.
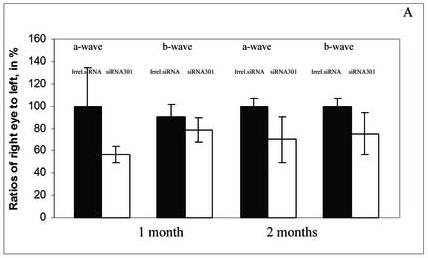
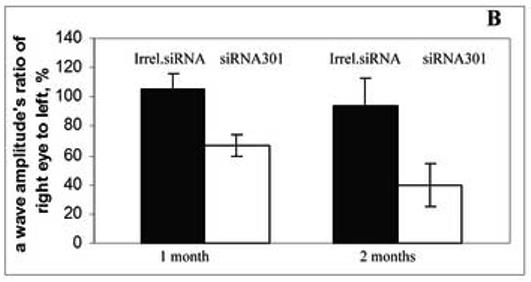
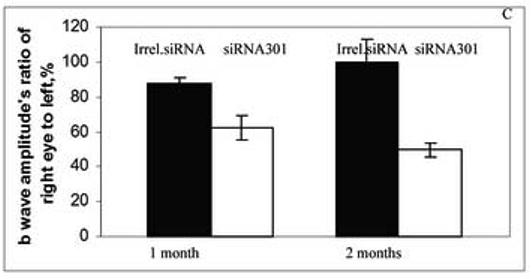
Expression of siRNA leads to reduction of a- and b-wave amplitudes of the scotopic ERG. Panel A: a- and b-wave amplitudes of C57BL/6 RHO+/+ mice at 1 and 2 months after injection with an irrelevant siRNA or with siRNA 301 into their right eyes (N=9 in each group). Although diminution in a- and b-wave amplitudes was observed, these differences did not achieve statistical significance. P values for a- and b waves were < 0.08 and <0.2 at 1 month and <0.08 and < 0.12 at 2 months, respectively. Panel B: a-wave amplitudes in eyes of RHO+/− mice injected with siRNA301 was about 40% lower compared to control eyes at 1 month after injection, p < 0.012 (N=15 in each group). At 2 months after injection, the amplitude was diminished more significantly (57%) compared to eyes injected with control siRNA (p value < 0.027). Panel C: Reduction of b-wave amplitudes in eyes of RHO+/− mice injected with siRNA301 was 25% at 1 month after injection compared to eyes injected with control siRNA (p < 0.027). At 2 months after injection, this reduction was more pronounced (over 50%) in experimental eyes.
3.4. AAV5 siRNA301 reduction of RHO mRNA
RHO+/+ mice with no obvious visual phenotype and RHO+/− mice were sacrificed for mRNA analysis 2 months after injection. Reverse transcription PCR was used to detect levels of RHO transcripts, which were normalized by dividing the band intensity of the RHO product by the band intensity of β-actin PCR product from the same sample. Results were expressed as a ratio of the normalized RHO level of the AAV-treated eyes to that of the untreated eyes. Despite of the fact that we did not see a significant ERG difference between injected and uninjected eyes of RHO+/+ mice, the level of RHO mRNA was diminished by 49% (p<0.04) in siRNA301 treated eyes (Fig. 4A). Delivery of AAV5 siRNA301 to RHO+/− retinas led to a reduction of the RHO mRNA level by over 30% (p < 0.012) compared to mRNA extracted from eyes injected with irrelevant siRNA (Fig. 4B). Therefore, in wild-type mice, residual rhodopsin synthesis following siRNA treatment supports an ERG response that is only slightly diminished relative to control eyes, while in RHO+/− mice, further reduction of rhodopsin expression led to a decrease in ERG amplitudes.
Fig. 4.
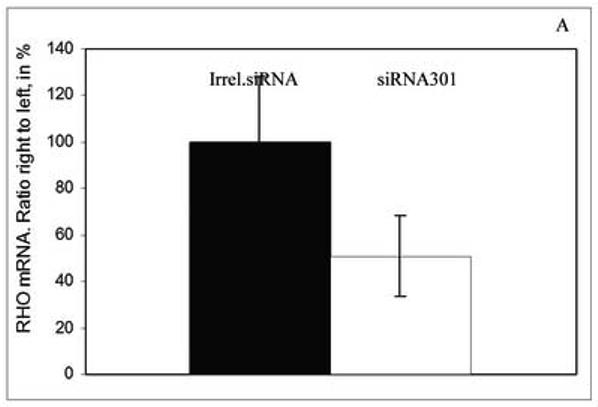
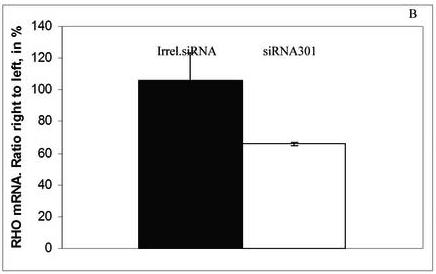
Expression of siRNA301 in retinas of mice injected at P16 leads to reduction of RHO mRNA. At 2 months after injection, the reduction of RHO mRNA (normalized to β-actin mRNA) was about 50% in RHO+/+ mice, p <0.04 (N=6 in each group) ( panel A) and over 30% in RHO+/− mice, p <0.0076 (N=5 in each group) (panel B). Bars represent the ratio of the normalized rhodopsin mRNA level in the siRNA301-treated eyes to the normalized rhodopsin mRNA values in the control eyes and are given in percent.
3.5. AAV5 siRNA301 reduced the steady state level of rhodopsin protein in mouse retinas
Expression of siRNA301 in the retina led to a decrease in the level of rhodopsin protein as detected by western blot. (Table II). We compared the level of rhodopsin protein in eyes of RHO+/− mice treated with siRNA301 with eyes treated with an irrelevant siRNA. Densities of the opsin bands in western blots were normalized to the level of β-actin from the same samples. In this experiment, opsin reduction was 60%, p < 0.013, compared to the control. We infer that the remaining 30-40% of RHO protein was sufficient to support 40-50% of the ERG response, consistent with the fraction of remaining rods (see below).
Table II.
Rhodopsin Protein Reduction Following siRNA or Control Injection
| Vector | Right eyes, Rhodopsin protein | Left eyes, Rhodopsin protein | P value |
|---|---|---|---|
| siRNA301 | 40.4 ± 10.6 * | 100 ± 17.6 | <0.0127 |
| Irrelevant. siRNA | 125.7 ± 31.8 | 100 ± 10.7 | <0.445 |
Values (ratios of the intensity of the RHO band to the β-actin band in a Western blot) represent the content of rhodopsin protein in %. Experiment was done in triplicate. Right eyes were injected with either siRNA301 or irrelevant (phospholamban) siRNA. Left eyes were uninjected.
3.6. AAV-siRNA301 treatment causes thinning of the ONL in RHO+/− mice
In mice treated with siRNA301, we measured a reduction in the ONL thickness at 2 months post injection (Fig. 5). Expression of this RHO-specific siRNA resulted in the reduction of the ONL thickness in the range of 53% to 86% compared to eyes injected with AAV-expressing an irrelevant siRNA (p <0.004). The region located within 1200 μm of optic nerve head (ONH) was most affected by siRNA301 treatment. ONL thinning in the inferior hemisphere region between 800 and 2000 μm from ONH and in the superior hemisphere up to 1200 μm from the ONH was statistically significant. Analysis of ONL thickness and RHO protein levels determined by western blot, suggests that loss of rhodopsin from surviving cells, in addition to the loss of photoreceptors, contributed to the 60% reduction of opsin protein that we observed (Table II).
Fig. 5.
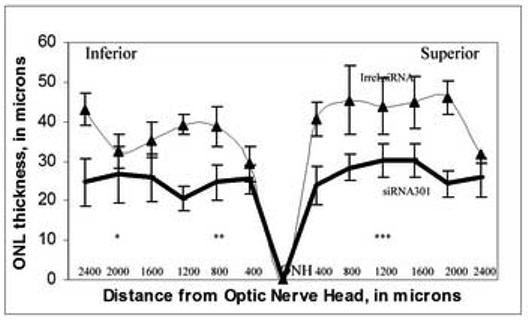
Expression of siRNA301 in the mouse retina causes reduction in ONL thickness. Retinal nuclei were detected by propidium iodide staining. Retinas of six mice in each group were divided into 400 μM sectors starting from optic nerve head (ONH), and the thickness of ONL was quantitated using the Zeiss Axiovision 4.4 software. Reduction in the ONL thickness of mice injected with siRNA301 (heavy line) ranged from 53 to 86 % across the full retina compared to retinas injected with the control siRNA (thin line). The most affected regions siRNA301 treated retinas were in the inferior hemisphere, in the sector between 800 and 2000 μm from the ONH and in the superior hemisphere 1200 μm from the ONH.(*- p value < 0.04, **- p value< 0.0097, ***- p value < 0.002.)
4. Discussion
Allelic heterogeneity poses a serious problem for efforts to treat ADRP by sequence specific reagents. Additionally, there are technical limitations to the designing ribozymes or siRNAs specific for each of the RHO mutation. In the case of ribozymes, there may be no cleavage sites near the sites of mutations, while in the case of siRNA, a single nucleotide change may not provide enough specificity to distinguish mutant from wild-type mRNA. To overcome these difficulties, we and other groups are trying to design RNA inhibitors that do not discriminate between mutant and wild-type transcripts, but reduce the expression of both. To be useful for therapy, these allele non-specific inhibitors will need to be delivered along with a wild-type cDNA of RHO that has been rendered resistant to the siRNA or ribozyme through silent mutations. Our previous studies have been based on the application of ribozymes to control expression of the RHO gene by means of direct RNA cleavage. In this paper, we extend this approach to include RISC-mediated RNA interference as a first step in the development of a general strategy to replace RHO transcripts in living animals.
RNA interference has been used by others to target the expression of the RHO gene (Cashman et al., 2005 ; Kiang et al., 2005 ). Experiments conducted in vitro and in the liver in vivo following hydrodynamic tail vein injection of siRNA and a RHO-expressing plasmid demonstrated that an siRNA can effectively block RHO expression in non-retinal cells or in retinal organ culture. Tessitore et al. (Tessitore et al., 2006 ), showed that mouse RHO mRNA could be knocked down in the retina of a living rat. We have now shown that endogenous rhodopsin protein (and RNA) can be suppressed in the retina in vivo. SiRNA301 was created for use in a canine model of ADRP containing T4R rhodopsin (Kijas et al., 2002 ). For in vivo validation of this siRNA, we choose heterozygous RHO+/− mice as a working model, because they already contain a 40% reduction in the level of rhodopsin protein. AAV delivered gene expression was distributed across the full retina, although the inferior hemisphere was transduced to a higher extent than the superior retina. This region-specific transduction was reflected in a greater decrease in the ONL thickness in the inferior retina, suggesting that the regional delivery of vector may be a critical factor for siRNA potency in vivo.
Expression of siRNA in vivo led to a reduction in the visual response as reflected in diminished scotopic ERG a- and b- wave amplitudes. However, over the two months of analysis, the rate of decline of the a- and b-waves was not identical. A-wave amplitudes declined faster in response to the expression siRNA301 in photoreceptors than did the b-wave (Fig. 3C). The difference in the rate of the decline of a- and b- wave amplitudes could be due to the compensatory effects occurring at the synaptic level between rods and rod bipolar cells or due to the stabilization of the remaining photoreceptors cells. An analogous and much more detailed observation was reported by Richards et al. (Richards et al., 2006 ) in response to light damage to the mouse retina. We would expect siRNA delivered by AAV5 to be expressed in photoreceptor cells by the 1 month time point (Auricchio et al., 2001 ). The fact that the ERG response continues to decline suggests that photoreceptors continue to degenerate during the second month.
The ERG data correlated with the reduction in the ONL thickness of retinas injected with AAV-siRNA301 measured at 2 months after injection. ONL thinning was significant overall (p value < 0.04) but was not uniform along the full retina, and ranged from 53% to 86% compared to the control eye. The most dramatic reduction of the ONL thickness was observed in the inferior hemisphere in a region approximately 1200 μm away from ONH, where a greater percentage of photoreceptors were transduced by AAV. SiRNA301 led to the loss of more than 50% of the photoreceptors from these retinas (Fig. 5). At this stage, reduction of rhodopsin protein was 60-70%, and since this level was normalized to β-actin, this probably reflects reduced rhodopsin production in remaining cells. It is hard to explain the difference between the protein decrease and the 30% reduction measured in mRNA, but the assays are completely independent and have their own sources of variation. However, it is clear from these experiments that RNA interference with siRNA can down-regulate expression of the RHO gene in vivo.
As the second step toward a generalized ADRP gene therapy, the supplementation with wild type RHO will be required. Designing an siRNA-resistant RHO gene should not be a major impediment. Kiang et al. (Kiang et al., 2005 ) and Cashman et al. (Cashman et al., 2005 ) have demonstrated how this might be done for RHO. The mouse-specificity of the RHO siRNAs designed by Tessitore et al. was due to 4 mismatches with the endogenous rat mRNA (Tessitore et al, 2006). A more important issue is balancing the expression of the siRNA and the replacement rhodopsin so that rhodopsin levels remain sufficiently high to sustain photoreceptors but not so high that they become toxic (Tan et al., 2001 ). In addition, over-expression of shRNA has been shown to lead to competition between shRNA and miRNAs for limiting cellular factors required for the processing of various small RNAs (Grimm et al., 2006 ). Consequently, over-expression of shRNAs must also be avoided.
Another issue we need to explore is the time-frame for ADRP therapy. Working with RHO+/− mice, we simply validated the use of siRNAs delivered by AAV. The treatment of a real ADRP mouse model, however, requires the delivery of therapeutic agents early enough to prevent the massive loss of photoreceptor cells. Because of the rapid course of retinal degeneration in many rodent models, the intervention using RNA inhibitors (siRNAs or ribozymes) may have to be initiated before substantial of retinal degeneration has begun. The appropriate timing may vary with the model and the treatment.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akashi H, Matsumoto S, Taira K. Gene discovery by ribozyme and siRNA libraries. Nat.Rev.Mol.Cell Biol. 2005;6:413–422. doi: 10.1038/nrm1646. [DOI] [PubMed] [Google Scholar]
- Alfano G, Vitiello C, Caccioppoli C, Caramico T, Carola A, Szego MJ, McInnes RR, Auricchio A, Banfi S. Natural antisense transcripts associated with genes involved in eye development. Hum.Mol.Genet. 2005;14:913–923. doi: 10.1093/hmg/ddi084. [DOI] [PubMed] [Google Scholar]
- Auricchio A, Kobinger G, Anand V, Hildinger M, O'Connor E, Maguire AM, Wilson JM, Bennett J. Exchange of surface proteins impacts on viral vector cellular specificity and transduction characteristics: the retina as a model. Hum.Mol.Genet. 2001;10:3075–3081. doi: 10.1093/hmg/10.26.3075. [DOI] [PubMed] [Google Scholar]
- Auricchio A, Rolling F. Adeno-associated viral vectors for retinal gene transfer and treatment of retinal diseases. Curr.Gene Ther. 2005;5:339–348. doi: 10.2174/1566523054065020. [DOI] [PubMed] [Google Scholar]
- Cashman SM, Binkley EA, Kumar-Singh R. Towards mutation-independent silencing of genes involved in retinal degeneration by RNA interference. Gene Ther. 2005;12:1223–1228. doi: 10.1038/sj.gt.3302512. [DOI] [PubMed] [Google Scholar]
- Drenser KA, Timmers AM, Hauswirth WW, Lewin AS. Ribozyme-targeted destruction of RNA associated with autosomal-dominant retinitis pigmentosa. Invest Ophthalmol.Vis.Sci. 1998;39:681–689. [PubMed] [Google Scholar]
- Farrar GJ, Kenna PF, Humphries P. On the genetics of retinitis pigmentosa and on mutation-independent approaches to therapeutic intervention. EMBO J. 2002;21:857–864. doi: 10.1093/emboj/21.5.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbatyuk MS, Pang JJ, Thomas J, Jr., Hauswirth WW, Lewin AS. Knockdown of wild-type mouse rhodopsin using an AAV vectored ribozyme as part of an RNA replacement approach. Mol.Vis. 2005;11:648–656. [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Jagla B, Aulner N, Kelly PD, Song D, Volchuk A, Zatorski A, Shum D, Mayer T, De Angelis DA, Ouerfelli O, Rutishauser U, Rothman JE. Sequence characteristics of functional siRNAs. RNA. 2005;11:864–872. doi: 10.1261/rna.7275905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiang AS, Palfi A, Ader M, Kenna PF, Millington-Ward S, Clark G, Kennan A, O'Reilly M, Tam LC, Aherne A, McNally N, Humphries P, Farrar GJ. Toward a gene therapy for dominant disease: validation of an RNA interference-based mutation-independent approach. Mol.Ther. 2005;12:555–561. doi: 10.1016/j.ymthe.2005.03.028. [DOI] [PubMed] [Google Scholar]
- Kijas JW, Cideciyan AV, Aleman TS, Pianta MJ, Pearce-Kelling SE, Miller BJ, Jacobson SG, Aguirre GD, Acland GM. Naturally occurring rhodopsin mutation in the dog causes retinal dysfunction and degeneration mimicking human dominant retinitis pigmentosa. Proc.Natl.Acad.Sci.U.S.A. 2002;99:6328–6333. doi: 10.1073/pnas.082714499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss HR, Heckenlively JR. Visual field changes in cone-rod degenerations. Arch.Ophthalmol. 1982;100:1784–1790. doi: 10.1001/archopht.1982.01030040764011. [DOI] [PubMed] [Google Scholar]
- Kwak N, Okamoto N, Wood JM, Campochiaro PA. VEGF is major stimulator in model of choroidal neovascularization. Invest Ophthalmol.Vis.Sci. 2000;41:3158–3164. [PubMed] [Google Scholar]
- Lem J, Krasnoperova NV, Calvert PD, Kosaras B, Cameron DA, Nicolo M, Makino CL, Sidman RL. Morphological, physiological, and biochemical changes in rhodopsin knockout mice. Proc.Natl.Acad.Sci.U.S.A. 1999;96:736–741. doi: 10.1073/pnas.96.2.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin AS, Drenser KA, Hauswirth WW, Nishikawa S, Yasumura D, Flannery JG, LaVail MM. Ribozyme rescue of photoreceptor cells in a transgenic rat model of autosomal dominant retinitis pigmentosa. Nat.Med. 1998;4:967–971. doi: 10.1038/nm0898-967. [DOI] [PubMed] [Google Scholar]
- Liang Y, Fotiadis D, Maeda T, Maeda A, Modzelewska A, Filipek S, Saperstein DA, Engel A, Palczewski K. Rhodopsin signaling and organization in heterozygote rhodopsin knockout mice. J.Biol.Chem. 2004;279:48189–48196. doi: 10.1074/jbc.M408362200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee Sanftner LH, Abel H, Hauswirth WW, Flannery JG. Glial cell line derived neurotrophic factor delays photoreceptor degeneration in a transgenic rat model of retinitis pigmentosa. Mol.Ther. 2001;4:622–629. doi: 10.1006/mthe.2001.0498. [DOI] [PubMed] [Google Scholar]
- Miyagishi M, Hayashi M, Taira K. Comparison of the suppressive effects of antisense oligonucleotides and siRNAs directed against the same targets in mammalian cells. Antisense Nucleic Acid Drug Dev. 2003;13:1–7. doi: 10.1089/108729003764097296. [DOI] [PubMed] [Google Scholar]
- Nakamura H, Siddiqui SS, Shen X, Malik AB, Pulido JS, Kumar NM, Yue BY. RNA interference targeting transforming growth factor-beta type II receptor suppresses ocular inflammation and fibrosis. Mol.Vis. 2004;10:703–711. [PubMed] [Google Scholar]
- Petrin D, Baker A, Coupland SG, Liston P, Narang M, Damji K, Leonard B, Chiodo VA, Timmers A, Hauswirth W, Korneluk RG, Tsilfidis C. Structural and functional protection of photoreceptors from MNU-induced retinal degeneration by the X-linked inhibitor of apoptosis. Invest Ophthalmol.Vis.Sci. 2003;44:2757–2763. doi: 10.1167/iovs.02-0729. [DOI] [PubMed] [Google Scholar]
- Rabinowitz JE, Rolling F, Li C, Conrath H, Xiao W, Xiao X, Samulski RJ. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J.Virol. 2002;76:791–801. doi: 10.1128/JVI.76.2.791-801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich SJ, Fosnot J, Kuroki A, Tang W, Yang X, Maguire AM, Bennett J, Tolentino MJ. Small interfering RNA (siRNA) targeting VEGF effectively inhibits ocular neovascularization in a mouse model. Mol.Vis. 2003;9:210–216. [PubMed] [Google Scholar]
- Richards A, Emondi AA, Rohrer B. Long-term ERG analysis in the partially light-damaged mouse retina reveals regressive and compensatory changes. Vis.Neurosci. 2006;23:91–97. doi: 10.1017/S0952523806231080. [DOI] [PubMed] [Google Scholar]
- Saishin Y, Saishin Y, Takahashi K, Lima e Silva, Hylton D, Rudge JS, Wiegand SJ, Campochiaro PA. VEGF-TRAP(R1R2) suppresses choroidal neovascularization and VEGF-induced breakdown of the blood-retinal barrier. J.Cell Physiol. 2003;195:241–248. doi: 10.1002/jcp.10246. [DOI] [PubMed] [Google Scholar]
- Tan E, Wang Q, Quiambao AB, Xu X, Qtaishat NM, Peachey NS, Lem J, Fliesler SJ, Pepperberg DR, Naash MI, Al Ubaidi MR. The relationship between opsin overexpression and photoreceptor degeneration. Invest Ophthalmol.Vis.Sci. 2001;42:589–600. [PubMed] [Google Scholar]
- Tessitore A, Parisi F, Denti MA, Allocca M, Di Vicino U, Domenici L, Bozzoni I, Auricchio A. Preferential silencing of a common dominant rhodopsin mutation does not inhibit retinal degeneration in a transgenic model. Mol.Ther. 2006;14:692–699. doi: 10.1016/j.ymthe.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Timmers AM, Zhang H, Squitieri A, Gonzalez-Pola C. Subretinal injections in rodent eyes: effects on electrophysiology and histology of rat retina. Mol.Vis. 2001;7:131–137. [PubMed] [Google Scholar]
- Xia H, Mao Q, Paulson HL, Davidson BL. siRNA-mediated gene silencing in vitro and in vivo. Nat.Biotechnol. 2002;20:1006–1010. doi: 10.1038/nbt739. [DOI] [PubMed] [Google Scholar]
- Yokota T, Miyagishi M, Hino T, Matsumura R, Tasinato A, Urushitani M, Rao RV, Takahashi R, Bredesen DE, Taira K, Mizusawa H. siRNA-based inhibition specific for mutant SOD1 with single nucleotide alternation in familial ALS, compared with ribozyme and DNA enzyme. Biochem.Biophys.Res.Commun. 2004;314:283–291. doi: 10.1016/j.bbrc.2003.12.098. [DOI] [PubMed] [Google Scholar]


