Abstract
Combined confocal microscopy and freeze-fracture replica immunogold labeling (FRIL) were used to examine the connexin identity at electrical synapses in goldfish brain and rat retina, and to test for “co-localization” vs. “close proximity” of connexins to other functionally interacting proteins in synapses of goldfish and mouse brain and rat retina. In goldfish brain, confocal microscopy revealed immunofluorescence for connexin35 (Cx35) and NMDA-R1 (NR1) glutamate receptor protein in Mauthner Cell/Club Ending synapses. By FRIL double labeling, NR1 glutamate receptors were found in clusters of intramembrane particles in the postsynaptic membrane extraplasmic leaflets, and these distinctive postsynaptic densities were in close proximity (0.1–0.3 μ m) to neuronal gap junctions labeled for Cx35, which is the fish ortholog of connexin36 (Cx36) found at neuronal gap junctions in mammals. Immunogold labeling for Cx36 in adult rat retina revealed abundant gap junctions, including several previously unrecognized morphological types. As in goldfish hindbrain, immunogold double labeling revealed NR1-containing postsynaptic densities localized near Cx36-labeled gap junction in rat inferior olive. Confocal immunofluorescence microscopy revealed widespread co-localization of Cx36 and ZO-1, particularly in the reticular thalamic nucleus and amygdala of mouse brain. By FRIL, ZO-1 immunoreactivity was co-localized with Cx36 at individual gap junction plaques in rat retinal neurons. As cytoplasmic accessory proteins, ZO-1 and possibly related members of the membrane-associated guanylate kinase (MAGUK) family represent scaffolding proteins that may bind to and regulate the activity of many neuronal gap junctions. These data document the power of combining immunofluorescence confocal microscopy with FRIL ultrastructural imaging and immunogold labeling to determine the relative proximities of proteins that are involved in short- vs. intermediate-range molecular interactions in the complex membrane appositions at synapses between neurons.
Introduction
The nearly complete decoding of the human genome (Venter et al., 2001; Internat. Human Genome Seq. Cons., 2001) has spawned the field of “proteomics”, which is focused on protein sequence identification, elucidation of protein modifications, and delineation of protein-protein interactions (Giot et al., 2003), with the ultimate goal of understanding complete functional sets of proteins and the precise nature of their macromolecular arrangements within individual cells and defined subcellular domains. A central concept emerging is that functionally related biochemical reactions, cell signaling cascades, and subcellular structural and regulatory components often involve multiple interacting proteins that are either in direct molecular contact or are compartmentalized in sufficiently close proximity that diffusion of substrate or product between interacting proteins occurs in nanoseconds to microseconds. Coincident with these evolving concepts, it now is appropriate to consider new strategies and technologies for the systematic identification and mapping of the myriad protein constituents of mammalian cells. One set of strategies in the arsenal of proteomics is based on high-resolution imaging and simultaneous mapping of interacting proteins.
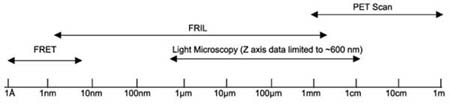
Biological imaging methods cover a wide range of formats and resolutions–from whole body scans for protein localization at the tissue and organ levels, to x-ray crystallography for atomic-scale analysis of protein structure (see Table 1 for overlapping resolution and imaging ranges of anatomical imaging techniques). Between these extremes are imaging approaches that provide essential information at many different levels of resolution. Light microscopy (including laser scanning confocal microscopy), combined with immunohistochemistry, allows assignment of proteins to cells and subcellular structures, giving a general view of protein distribution, abundance, localization and co-association, and provides the basis for more detailed ultrastructural and molecular analyses. At the other end of the spectrum, fluorescence resonance energy transfer (FRET) allows identification of protein molecules that are in direct molecular contact or that are separated by < 5 – 10 nm (Verveer et al., 2003). However, due to diffraction-limited phenomenon for focusing photons of any wavelength, FRET is not capable of visualizing proteins or mapping them within cells to any greater resolution than conventional light microscopy. Consequently, there remains the problem of identifying and mapping proteins or protein arrays that interact functionally over “intermediate” distances (i.e., 10 nm to 300 nm, which is below the limit of resolution of light microscopy [LM]) and mapping them to nanometer resolutions. This gap in separately resolving multiple, closely-spaced proteins is a particularly vexing problem in studies of the vertebrate CNS, where cellular heterogeneity and morphological complexity often give rise to plasma membranes of three, four, or more neuronal and/or glial cell processes within the smallest volume of tissue resolvable by LM (Rash et al., 2001a). To decipher the two- and three-dimensional distribution of multiple proteins within this convoluted entanglement of neuronal and glial processes, ultrastructural approaches are required.
Table 1.
| Terms | ||
|---|---|---|
| Physiology and biochemistry | Anatomy | Functional significance |
| Short-range molecular interaction | Co-localization | Molecules in direct contact; diffusion in nanoseconds |
| Intermediate-range interaction | Close proximity/co-association | Subcellular; diffusion distances < 0.3 μ m; diffusion in microseconds to < 1 millisecond |
| Long-range interactions | Histological proximity | Diffusion in milliseconds to minutes; cellular level to histological level; transport may involve circulatory system |
FRET (fluorescence resonance energy transfer) identifies molecules that are in molecular contact or that are separated by < 10 nm (i.e., “short-range interactions’’). Spatial localization in X- and Y-coordinates is limited to > 300 nm by optical diffraction phenomena. Laser scanning confocal microscopy provides biochemical, immunocytochemical, and structural data in the X- and Y-axes from 300 nm to 30 millimeters, and from 0.6 μ m to ~600 μ m in the Z-axis. FRIL provides structural and immunocytochemical data from about 2 nm to 2 mm (six orders of magnitude). PET scans (positron emission tomography) have about 1 mm resolution and collect data over ~0.5 m2. Thus, FRIL spans the gap from FRET to PET scans.
Conventional thin-section electron microscopy (TEM), combined with pre-embedding or post-embedding immunogold labeling methods, has provided invaluable information regarding simultaneous high-resolution mapping of two or more membrane proteins (Ottersen & Landsend, 2003), while at the same time revealing the ultrastructural correlates for at least some of the membrane proteins that are visualized immunohistochemically (e.g. gap junctions, tight junctions). However, when no recognizable ultrastructural correlate is discernible, extrapolation of the location or ultrastructural identification of labeled proteins from one sample to another is difficult if not impossible. Equally important, detergents and solvents are often used in pre-embedding immunolabeling to permeabilize cell membranes to allow penetration of antibodies and immunogold beads. Those reagents significantly reduce tissue contrast and definition, making it problematic to identify many small cell processes as either neuronal or glial.
Recently, combined confocal microscopy and freeze-fracture replica immunogold labeling techniques [collectively called FRIL1 (Rash et al., 1995; Rash & Yasumura, 1999)] have been used to visualize membrane proteins, to identify replicated proteins within ultrastructurally defined cellular structures, and to measure distances between labeled protein complexes. For example, Fujimoto2 1995, 1997 used simultaneous double immunogold labeling to show that connexin32 (Cx32) is present in hepatocyte gap junctions, whereas the zonula occludens-1 (ZO-1) protein is expressed separately in nearby tight junctions. Similarly, FRIL double labeling was used for simultaneous identification of aquaporin-4 (AQP4) in “square arrays” and Cx43 in nearby gap junctions of astrocyte and ependymocyte plasma membranes (Rash et al., 1998b; Rash & Yasumura, 1999).
Here, we demonstrate combined confocal microscopy and FRIL for discriminating between “co-localized” proteins vs. proteins in “close-proximity” in rodent and goldfish brain. We determined the relative proximity and subcellular location of two proteins that physiological measurements (Cx35/NR-1) and immunological and biochemical analyses (Cx36/ZO-1) suggest to be functionally associated. In “mixed” (chemical plus electrical) synapses in goldfish and rat brain, we show that Cx35 (a principal protein forming the intercellular channels of neuronal gap junctions in goldfish) is closely associated with glutamate receptor subunit NR1, which is present in distinctive clusters of intramembrane particles that represent the freeze-fracture correlate of the postsynaptic densities (PSDs) seen in conventional thin section electron micrographs. By FRIL, we confirmed that similar glutamatergic mixed synapses occur in rat inferior olive, with Cx36-containing gap junctions occurring within 0.1 μ m of NR1-containing PSDs. Using FRIL, we identified several new morphological classes of gap junctions that, nevertheless, are abundant between neurons in adult rat retina. By confocal immunofluorescence microscopy, we mapped the overall distribution of Cx36 in amygdala and reticular thalamic nucleus in mice and showed co-localization of Cx36 with ZO-1 protein, a primary scaffolding protein of apical plasma membranes. After FRIL confirmation of the efficacy of ZO-1 antibody labeling on tight junctions of adult rat liver, we confirmed co-localization of Cx36 and ZO-1 in individual gap junction plaques in rat retina. Thus, we have demonstrated that the combination of confocal microscopy and FRIL provides for unprecedented visualization and nanometer-scale mapping of interacting integral and peripheral membrane proteins—from their subcellular domains to their gross anatomical locations within the functional vertebrate “proteome”.
Materials and methods
IMMUNOHISTOCHEMISTRY OF GOLDFISH NEURONS
Goldfish were perfused intracardially with 4% formaldehyde in phosphate buffered saline (PBS, 0.12 M at pH 7.4) for 15 minutes, kept overnight in phosphate-buffered saline, and sectioned (20–50 μ m) with a TPI Vibratome (Technical Products International, St. Louis, MO). Sections were rinsed several times with PBS, incubated overnight with either anti-Cx36 (Ab298; 1:1000/5000; from JI Nagy), anti-NR1 (Apteronotus aptNR1, 1:500/1000 [courtesy of R. Dunn, McGill University; Berman et al., 2001]) or anti-Cx35 (MAB3045, 1:200/2000; Chemicon International, Temecula, CA). Primary antibodies and source data are listed in Table 2. Samples were rinsed in PBS, incubated for 2 hours with Texas Red-conjugated secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA), and rinsed with PBS. Sections were mounted on slides, dehydrated, cleared, covered, and examined under transmitted light or using fluorescence and confocal microscopes (Leitz Aristoplan and BioRad Radiance 2000 Laser Scanning Confocal Microscope). Control sections were incubated with secondary antibodies in the absence of primary antibodies. To allow anatomical identification of Mauthner cells during confocal grid mapping, electrodes were filled with a 5% solution of Lucifer yellow (CH, lithium salt; Molecular Probes, Eugene, OR) in distilled water, and this solution was iontophoretically-injected into the Mauthner cell somata (400 ms pulses of 50 nA for 20 minutes). For FRIL, goldfish hindbrains were sectioned to uniform 150-μ m thickness and frozen according to procedures detailed below.
Table 2.
Primary antibodies used in this study, the generating species, protein target, target amino acid sequence (if available) and species reactivities, and private vs. commercial sources.
| Primary Antibody | Type | Target | Specificity | Source |
|---|---|---|---|---|
| Ab298 | rabbit poly | Cx35/Cx36 | C-terminus aa298-318 Human (fish, rodent) | JI Nagy |
| MAB3045 | mouse mono | Cx35/Cx36 | intracellular loop; seq. NA Fish (monkey/mouse) | Chemicon |
| Ab36-4600 | rabbit poly | Cx36 | C-terminus; seq. NA Mouse | Zymed |
| Ab51-6300 | rabbit poly | Cx36 | C-terminus; seq. NA Mouse | Zymed |
| Ab37-4600 | mouse mono | Cx36 | C-terminus; seq. NA Mouse | Zymed |
| AptNR1 | rabbit poly | NR1 subunit | extraplasmic loop aa844-901, Fish | R. Dunn |
| Ab55-6308 | mouse mono | NR1 subunit | extraplasmic loop Human (rodent) | B-D Biosciences/PharMingen |
| Ab33-9100 | mouse mono | ZO-1 | aa334-6341 | Zymed |
| Ab61-7300 | rabbit poly | ZO-1 | aa463-11092 | Zymed |
| MAB3069 | mouse mono | Cx32 | cytoplasmic loop aa95-125 | Chemicon |
Monoclonal antibodies to connexins are made against target sequences of about 11–17 amino acids, whereas monoclonal antibodies to ZO-11 were made against a target sequence of 300 amino acids (Zymed Product Information Sheet), and rabbit polyclonal antibody to ZO-12 was made against a cytoplasmic sequence of > 600 amino acids (Zymed Product Information Sheet), possibly yielding additional epitope specificities, not all of which have been completely characterized (Li et al., 2004b). NA, Not available; seq., sequence; mono, monoclonal; poly, polyclonal.
LM IMMUNOHISTOCHEMISTRY OF MOUSE BRAIN
Ten wildtype (WT) and ten Cx36-knockout (KO) C57/BL6-129vEv mice were used for immunohistochemical analysis of Cx36 in brain, and for studies of Cx36 co-localization with ZO-1 in various brain regions. Heterozygous pairs of Cx36+/− mice (Deans et al., 2001) were kindly provided by Dr. David Paul (Harvard University), and animals were bred at the University of Manitoba. Care and treatment of mice was in accordance with approved experimental protocols of the University of Manitoba Central Animal Care Services. Verification of WT and Cx36 KO genotype was performed as described elsewhere (Li et al., 2004a). Sixteen-day-old and adult mice were deeply anesthetized and perfused transcardially with 3 ml of pre-fixative (50 mM phosphate buffer, 0.1% sodium nitrite, and 1 unit/ml heparin), followed by 40 ml of fixative (160 mM sodium phosphate buffer, pH 7.6, 0.2% picric acid and 1% formaldehyde from freshly depolymerized paraformaldehyde), followed by perfusion with a sucrose wash (10% sucrose, 25 mM sodium phosphate buffer, pH 7.4). Brains were removed and stored at 4° C in cryoprotectant (10% sucrose, 25 mM phosphate buffer, pH 7.4, 0.04% sodium azide) for 16 h. Cryostat sections (10 μ m thick) were collected on gelatinized glass slides. Double-immunofluorescence labeling was performed with all primary and secondary antibodies diluted in TBST (50 mM Tris-HCl buffer, pH, 7.4, 1.5% NaCl, 0.3% Triton X-100) containing 10% normal goat serum. Sections were incubated for 16 h at 4° C with either rabbit polyclonal anti-Cx36 antibody (Ab36-4600, Zymed Laboratories Inc., South San Francisco, CA) or anti-Cx36 (Ab51-6300; Zymed) at a concentration of 3 μ g/ml, and simultaneously with mouse monoclonal anti-ZO-1 antibody (Ab33-9100; Zymed) at 4 μ g/ml. Alternatively, sections were incubated with monoclonal anti-Cx36 (Ab37-4600; Zymed) in combination with polyclonal anti-ZO-1 (Ab61-7300; Zymed), followed by incubation with the appropriate combination of secondary antibodies. [The polyclonal anti-ZO1 antibody has an additional cross-reactivity in brain tissue with a protein of 60kDa, the identity of which is uncertain; Li et al. 2004a.] Sections were washed for 1 h in TBST, and incubated for 1.5 h at room temperature with AlexaFluor488-conjugated goat anti-rabbit IgG (Molecular Probes) diluted 1:1000, and simultaneously with Cy3-conjugated goat anti-mouse IgG (Jack-son) diluted 1:200. Sections were then washed in TBST for 20 min, followed by a rinse in 50 mM Tris-HCl buffer (pH 7.4) for 30 min, dried and coverslipped with anti-fade medium. Non-immunostained sections and sections labeled only for Cx36 were counterstained by incubation for 10 min with Neuro-Trace 530/615 (Molecular Probes) Nissl stain diluted 1:200 in PBS (100 mM sodium phosphate buffer, pH 7.4, 0.9% NaCl). Fluorescence was examined using a Zeiss Axioscop2 fluorescence microscope and an Olympus Fluoview IX70 confocal microscope. Images were acquired using Axiovision 3.0 (Carl Zeiss Vision GmbH, Germany) and Fluoview 2.1 software, respectively, and assembled using Photoshop 7.0 (Adobe Systems, San Jose, CA).
PREPARATION OF TISSUES FOR FRIL
Adult Sprague-Dawley rats (120–540 g) were anesthetized with Ketamine/Xylazine (80 mg/kg; 8 mg/kg) and fixed by whole body perfusion with 1% or 2% formaldehyde in Soren-son’s phosphate buffer, pH 7.4. The eyes, brain, and liver were removed, dissected, cut into 100–150 μ m-thick slices using a refrigerated Lancer 1000 Vibratome (Technical Products International), and the slices were prepared for FRIL.
Rat and goldfish tissue slices were infiltrated with 30% glycerol (as a cryoprotectant to minimize damage during freezing) and frozen by contact with a liquid nitrogen-cooled metal mirror (Phillips & Boyne, 1984). Frozen samples were fractured in a JEOL 9010C freeze-fracture device (RMC Products, Tucson, AZ), pre-coated with 1nm of vaporized carbon (Winkler et al., 2002), replicated with 1–1.5 nm of vaporized platinum, and coated with 5–20 nm of vaporized carbon. Each replicated but still frozen sample was bonded to a gold “index” grid (G200-F1-Au; Electron Microscopy Sciences, Fort Washington, PA) using 1.5–2% Lexan (GE Plastics, Pittsfield, MA; available from local plastic suppliers) dissolved in ethylene dichloride. The ethylene dichloride solvent was evaporated by slowly raising the sample temperature from −95° C to −35° C. The resulting Lexan-replica-tissue-grid “sandwich” was thawed and mapped by confocal microscopy prior to immunogold labeling.
IMMUNOGOLD LABELING
Bulk tissue remnants were removed from the Lexan-stabilized replicas by washing in 2.5% SDS detergent in TRIS-HCl buffer (pH 8.9) for 29 hr at 48.5° C. (For mammalian tissues, this solution was replaced for 3 hr with 2% collagenase D (Boehringer-Mannheim, Mannheim, Germany) in 150 mM phosphate buffer. This washing procedure leaves a thin film of lipid and protein molecules adhering to the highly absorptive platinum/carbon replica, plus additional molecules that are bound to those molecules that are directly adsorbed to the replica (Fujimoto, 1995; Rash & Yasumura, 1999). SDS-washed replicas were rinsed in “blocking buffer” [10% heat-inactivated goat serum, 1.5% fish gelatin in Sorenson’s phosphate buffer, pH 7.4; (Dinchuk et al., 1987)] and labeled for 60-220 minutes using various combinations of mouse monoclonal and rabbit polyclonal antibodies. Antibodies to Cx36 included polyclonal Ab298 (Rash et al., 2000; Pereda et al., 2003) and polyclonal Ab36-4600 (Zymed) and monoclonal Ab51-6300 (Zymed). Monoclonal antibodies to glutamate receptor subunit NR1 (Ab55-6308) were obtained from BD Biosciences PharMingen (San Diego, CA), and antibodies to ZO-1 were obtained from Zymed (monoclonal Ab33-9100 and poly-clonal Ab61-7300). Anti-Cx32 (mouse monoclonal MAB3069; Chemicon) was used to label adult rat liver. Labeled samples were rinsed and counter-labeled using goat anti-rabbit IgG and goat anti-mouse IgG, each coupled to a separate size of uniform-diameter gold beads (6, 12, or 18 nm; Jackson).
After immunogold labeling, each sample was air dried and coated on the labeled side with 10–20 nm of evaporated carbon, to anneal cracks in the replica, as well as to stabilize the immunogold beads. The Lexan support film was removed by immersing the grids in ethylene dichloride solvent for 1–2 h, labeled replicas were air dried, and the grids were examined by TEM.
ELECTRON MICROSCOPY
FRIL samples were examined in JEOL 2000 EX-II and JEOL 1200 EX transmission electron microscopes, and stereoscopic images (8° included angle; tilt range ± 60° and 45° ±) were obtained at TEM magnifications from 10,000X to 100,000X. Negatives were digitized by an ArtixScan 2500f digital scanner (Microtek; Carson, CA), and processed using Photoshop 7.01 (Adobe Systems). Selected regions are presented as stereoscopic “triplets” consisting of a stereo pair (left pair when using a stereopticon-type viewer) and a reverse or “intaglio” pair (right pair). (Stereoscopy is reversed when viewed by the crossed-eyes method.) Stereoscopic images are essential for assessing complex three-dimensional membrane topography, as well as for confirming that each label is on the tissue-side of the replica, whereas relatively rare non-specific binding is primarily on the Lexan-coated side of the replica (Rash & Yasumura, 1999). The reverse stereo (or “intaglio”) images are especially useful for determining the “sidedness” of the electron opaque gold beads with respect to the electron-translucent platinum replicas, as well as for spatial discrimination of the smaller 6 nm gold beads from the equally electron-opaque granularity of the platinum replica (Rash & Yasumura, 1999).
Neurons and glia were identified in freeze-fracture replicas based on 24 inclusive and exclusive criteria described by Rash et al. 1997. For neurons, these included the presence of > 25 uniform diameter synaptic vesicles in nerve terminal cytoplasm, pre- and postsynaptic membrane specializations called active zones and postsynaptic densities, and absence of glial cell markers (GFAP filaments, square arrays, etc.) Gap junctions were identified based on well established criteria (reviewed in Rash et al., 1998a), including: (a) uniform 8–9 nm diameter intramembrane particles in replicated protoplasmic leaflets [“P-faces”; the internationally-recognized freeze-fracture terminology of Branton et al. 1975 is used in this report.] and uniform 8–9 nm diameter pits in the extraplasmic leaflet (E-face); (b) close packing of IMPs/pits, often in hexagonal close packing; (c) narrowing of the extracellular space within the border of the gap junction; (d) maintained alignment of IMPs and pits across the step from E- to P-face. In addition, immunogold labeling restricted to gap junctions within a single cell type was considered confirmatory evidence.
Results
CX35 IN NEURONAL GAP JUNCTIONS OF LARGE MYELINATED CLUB ENDING SYNAPSES ON GOLDFISH MAUTHNER CELLS
In initial immunolabeling experiments to identify connexins in goldfish neuronal gap junctions, we used anti-Cx36 Ab298 because it was generated against an amino acid sequence that is similar in goldfish Cx35 (Pereda et al., 2003), the fish ortholog of Cx36 (O’Brien et al., 1998). (Thus, when used to label goldfish, this antibody is designated anti-Cx35.) Confocal immunofluorescence microscopy revealed abundant Cx35 in Mauthner Cell/Large Myelinated Club Ending (MC/CE) synapses (Fig. 1A). Typically, 50–100 fluorescent puncta and small patches were present at most MC/CE. Thus, by confocal microscopy, MC/CE synapses were positively identified by their uniquely large size and distinctive subcellular location on the distal portion of the Mauthner cell lateral dendrites, as well as by their distinctive distribution of multiple connexin-immunoreactive puncta.
Fig. 1.

Confocal images of Mauthner cells labeled for Cx35 and glutamate receptor NR1, with companion FRIL immunogold labeling of gap junctions in goldfish Mauthner cells. (A) Confocal immunofluorescence image (three Z sections of 2 μ m) showing Cx35 (MAB 3045) at a Club Ending on distal portion of a Mauthner Cell lateral dendrite. These terminals, identified by their large size, exhibit multiple sites of punctate labeling for Cx35. (B and C) Cx35 (Ab298) labeling of conventional “plaque” gap junctions (red overlay) in MC/CE synapses. Both images are from the same goldfish MC/CE synapse, showing primarily postsynaptic P-face (B) and presynaptic E-face (C). In both images, all labeling is on postsynaptic connexins. (D) Laser scanning immunofluorescence image (three Z sections of 2 μ m) of a Club Ending on a Mauthner-cell lateral dendrite after labeling with anti-NR1 antibody. (E–H) Simultaneous co-localization of Cx35 at gap junctions and NR1 glutamate receptors in E-face particles arrays (yellow overlay) in goldfish Mauthner cell. (E) E-face view of the postsynaptic membrane of goldfish Mauthner cell. E-face image of gap junction in postsynaptic membrane, but with unfractured and unreplicated presynaptic plasma membrane beneath. It is these unreplicated presynaptic connexins that are labeled (18 nm gold). In contrast, the two arrays of E-face IMPs represent glutamate receptor clusters, which are immunogold labeled for NR1 (12 nm gold beads in the extracellular space; see Fig. 2B–E for explanatory diagram). Inscribed areas are shown at higher magnification (F–H). (F) Stereoscopic (left pair) and reverse stereoscopic images (right pair) of a neuronal gap junction labeled for Cx35. (G and H) Portions of two PSDs after immunogold labeling for glutamate receptor NR1 by two gold beads (G) and one gold bead (H). The gap junction and the PSD on the right are separated by ~50 nm, or less than the limit of resolution of light microscopy. For confocal images A and D, calibration bars = 5 μ m. In all FRIL replicas, calibration bars = 0.1 μ m, unless otherwise designated.
FRIL ANALYSIS OF Cx35 AT MAUTHNER CELL/CLUB ENDING SYNAPSES
Combining physiological and anatomical techniques facilitated identification of Club Endings by FRIL (Pereda et al., 2003). Mauthner cells, as identified by electrophysiological criteria, were photomapped by confocal microscopy, and the same cells subsequently were examined by FRIL. MC/CE synapses contained 80–150 gap junctions, each labeled by multiple immunogold beads representing Cx35 immunoreacitivity (Fig. 1B and C). [For low magnification overviews of this same Mauthner cell, see Pereda et al. 2003. Additional overviews of mixed synapses are provided in Rash et al. (1996, 1998a, 2000, 2001b).] These mixed synapses had 12 nm gold beads beneath gap junction plaques in the replicated P-face of the Mauthner cell plasma membrane (Fig. 1B), as well as the E-face of the overlying presynaptic club ending plasma membrane, as viewed toward the postsynaptic membrane (Fig. 1C). In both of these views of the same synaptic contact, immunogold labeling is to cytoplasmic epitopes of the connexon hemiplaques of the Mauthner cell (below the replica), and not of the connexons of the overlying Club Ending that had been removed by freeze fracturing (interpretive drawing, Fig. 2B and 2C, left side). As commonly observed (Fujimoto, 1995; Fujimoto, 1997; Rash & Yasumura, 1999), labeling of connexons beneath replicated E-face plaques was somewhat higher than for connexons replicated as P-face plaques. The structural basis for labeling beneath both P-face particles and E-face pits in the paired hemi-plaques at gap junctions is shown diagrammatically in Figure 2B. However, the molecular basis for somewhat greater labeling of connexons beneath replicated E-face pits than beneath platinum-coated P-face particles is not yet known. Possibly, the additional half membrane of the extraplasmic leaflet, superimposed on the unfractured membrane below, may protect the connexons of the underlying cell against heating damage by vaporized platinum and carbon (Zingsheim et al., 1970). Alternatively, because the connexons of the subjacent membrane are not coated with platinum or carbon, which otherwise potentially immobilizes the protein, the antibodies may have better access to epitopes (Pereda et al., 2003; Fujimoto, 1995, 1997; Rash et al., 1998b; Rash & Yasumura, 1999).
Fig. 2.
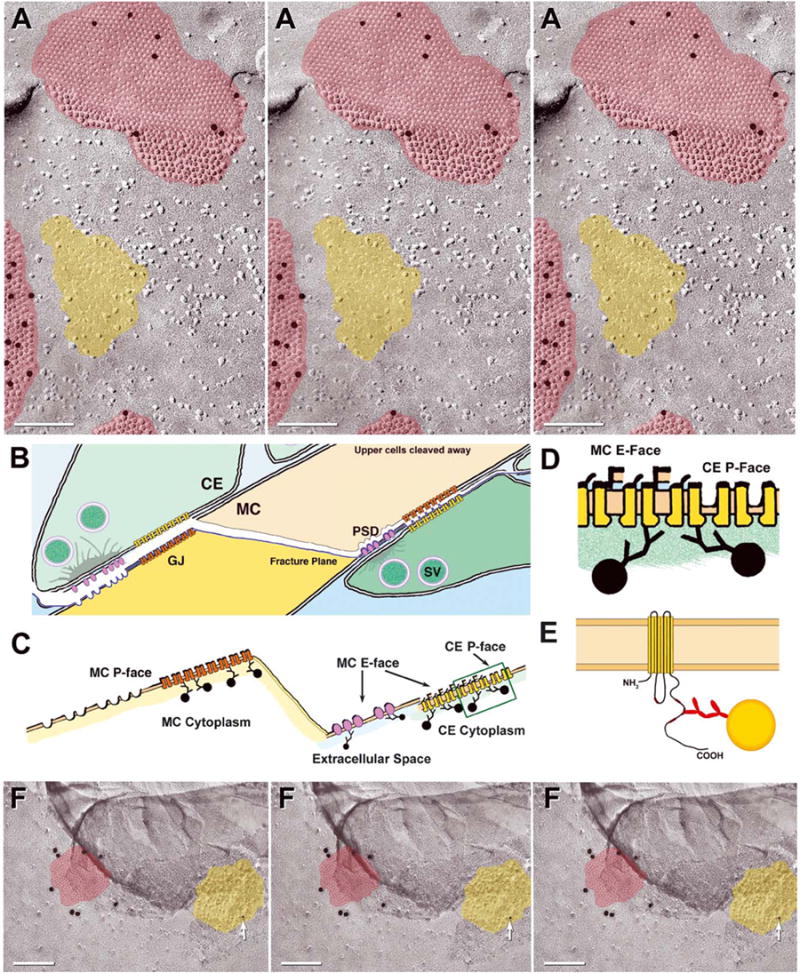
Immunogold labeling of glutamate receptor PSDs and nearby gap junctions. (A) Portions of three Cx35-immunogold-labeled gap junctions (red overlays) surrounding a P-face imprint of a PSD (yellow overlay), corresponding to the arrays of IMPs on E-faces (see Fig. 1G and H). The PSD P-face pits are devoid of NR-1 labeling. (B–E) Drawing of fracture plane through two Club Ending synapses on a Mauthner cell. (B and C) From left to right, the fracture plane (blue line) sequentially fractures within the postsynaptic plasma membrane, through a glutamate receptor PSD (lavender) and through a postsynaptic gap junction (left side of image), across the Mauthner cell cytoplasm (center of image), then to within the Mauthner cell plasma membrane (right side), exposing the postsynaptic membrane E-face particles corresponding to glutamate receptors, then the gap junction E-face pits (postsynaptic membrane) and, finally, gap junction P-face particles (presynaptic membrane of lower club ending). (B and C) PSD P-face pits are not labeled because no glutamate receptor proteins remain with the P-face pits (left side). However, P-face connexon IMPs (orange connexons, left side) in the adjacent gap junction are labeled. Glutamate receptor E-face particles are labeled on their extracellular determinants (right side), whereas connexons of presynaptic gap junctions are labeled on their cytoplasmic epitopes (D and E). In the gap junction on the right (2C, box) and in closer view (D), postsynaptic E-face pits and presynaptic P-face particles are replicated, but in both cases, it is the presynaptic connexons (yellow connexons) of the underlying Club Ending that are labeled. (E) Diagram showing immunogold labeling of epitope in the cytoplasmic carboxy terminus of a single connexin molecule by Ab298 (Pereda et al., 2003). (F) Stereoscopic and reverse stereoscopic image of portion of mixed synapse in adult rat inferior olive labeled for Cx36 (20 nm gold; red overlay) and NR1 glutamate receptors (10 nm gold, arrow; yellow overlay). Reverse stereoscopy (right pair) is helpful for discerning the 10 nm gold bead superimposed on the equally electron-dense platinum replica, as well as for discriminating between the two layers of replica that arose when a portion of replica was displaced (darker area at top).
CO-LOCALIZATION OF Cx35 AND NR1 AT LARGE MYELINATED CLUB ENDINGS
Activity-dependent modification of the strength of electrical transmission at MC/CE “mixed” synapses depends on functional interactions with co-localized or neighboring glutamatergic synapses (Yang et al., 1990; Pereda & Faber, 1996; Smith & Pereda, 2003). Confocal microscopic analysis revealed the presence of glutamate receptor NR1-immunofluorescence labeling within Club Ending synapses (Fig. 1D). The immunofluorescence labeling was largely localized to the periphery of the contacts, consistent with ultrastructural data describing the predominance of PSDs that consisted of distinctive arrays of E-face IMPs in the periphery of freeze-fractured nerve terminals (Tuttle et al., 1986).
SIMULTANEOUS FRIL IMMUNOGOLD LABELING OF Cx35 AND NR1 GLUTAMATE RECEPTORS
In a different, double-labeled replica of an MC/CE synapse (Fig. 1E) viewed from the post-synaptic Mauthner cell E-face toward the presynaptic Club Ending (not fractured, but beneath the MC E-face), Club Ending gap junctions were labeled for Cx35 (in this case, by 18 nm gold beads; Fig. 1F), and nearby distinctive postsynaptic clusters of E-face IMPs (corresponding to PSDs as seen in conventional thin sections) were labeled for NR1 glutamate receptor subunits (12 nm gold beads; Figs. 1G and H). In high magnification stereoscopic and reverse stereoscopic images, gold beads were detected solely on the extracellular side of the E-face IMPs (i.e., labels were in the residual extracellular space after SDS washing). In the case of non-paired structures such as these postsynaptic receptor clusters, sufficient protein remained after SDS washing to be labeled by anti-NR1 antibodies (interpretive drawing, Fig. 2C, right side). On the other hand, gap junctions represent paired or mirror image membrane structures that, because of their unique cleaving properties, were labeled for Cx35 beneath both E-face pits and P-face IMPs (Figs. 1B, C and F and 2A, red overlays). In both P- and E-face images of gap junctions, labeling was on the cytoplasmic side of the connexons in the plasma membrane of the lower cell, whether or not the labeled plasma membrane was visible in that portion of the replica (interpretive drawing, Fig. 2C–E). For example, when a different, double-labeled replica was viewed from the opposite side of the cell (i.e., from Club Ending toward Mauthner cell, Fig. 2A; similar view also shown in Fig. 1B and C) so that the P-face of the postsynaptic membrane and the E-face of the presynaptic membrane were present in the same gap junction (Fig. 2A, top; red overlay), gold beads were beneath both P- and E-faces. However, labeling in both cases was to those connexins in the Mauthner cell plasma membrane—even for those portions beneath the replicated E-face of the Club Ending. In contrast to gap junction labeling, clusters of Mauthner cell P-face pits representing the impressions where glutamate receptors had been removed (Fig. 2A, yellow overlay) were not labeled. As always, NR1 labeling was to extraplasmic epitopes exposed in the extracellular space. Unlike connexons, NR1 receptors remain associated with the E-face, where they are resolved as distinctive clusters of E-face IMPs. Thus, labeling of NR1 receptors occurred only beneath E-face IMPs, and not beneath P-face pits (interpretive drawing, Fig. 2C, right side vs. left side). The absence of labeling of those distinctive arrays of P-face pits in the postsynaptic membrane demonstrates that NR1, as a transmembrane protein, is removed completely from the protoplasmic leaflet by freeze fracturing, leaving no epitopes to label in the Mauthner cell cytoplasm.
Distinctive IMPs in E-face arrays similar to the NR1 arrays of goldfish also occur in mammalian mixed synapses (Rash et al., 1998a, 2000, 2001b). In preliminary FRIL studies of rat inferior olive (Fig. 2F), gap junctions labeled for Cx36 (20 nm gold) were found in close proximity to E-face IMP arrays that were labeled for NR1 (10 nm gold). (These immunogold labels no longer available from Chemicon.) Thus, as in goldfish, neurons in rat hindbrain have “mixed” (electrical plus chemical) synapses containing Cx36 in their gap junctions and NR1-labeled glutamate receptors in nearby clusters of E-face IMPs, confirming an early conjecture that on morphological grounds, similar clusters of E-face IMPs were identified in hippocampal neurons as glutamate receptor PSDs (Harris & Landis, 1986).
Cx36-IMMUNOGOLD LABELING REVEALED MULTIPLE TYPES OF NEURONAL GAP JUNCTIONS IN RAT RETINA
By FRIL, Cx36-immunoreactivity revealed abundant gap junctions (Fig. 3) in outer and inner plexiform layers and in the ganglion cell layer of adult rat retina (n > 500). These included conventional regular and irregular “plaque” gap junctions (Fig. 3A and Fig. 3D, upper left), as well as several previously unrecognized but abundant classes of gap junctions, including single- and multi-strand “string” gap junctions (Fig. 3B), single-and multi-strand “ribbon” gap junctions (Fig. 3C and D, lower right), and relatively rare “meandering” and “reticular” gap junctions (Rash et al., 1998a) [the latter possibly representing “lace” gap junctions (Raviola & Gilula, 1975)] (not shown).
Fig. 3.

FRIL images of “plaque”, “string’’ and “ribbon” gap junctions (red overlays) in inner and outer plexiform layers of adult rat retina after immunogold labeling for Cx36. (Criteria for identifying gap junctions are at the end of Materials and Methods.) (A) Two small, regular plaque gap junctions labeled for Cx36 (18 nm gold beads). (B) Two single-strand “string’’ gap junctions’’, with both P-face particles and E-face pits occurring primarily as single rows of particles or pits. Immunogold beads are associated with most of the connexon strands. (C) Immunogold-labeled multi-strand “ribbon’’ gap junction. Each “ribbon’’ is 2–6 IMPs (or pits) wide, with up to 500 connexons per compound ribbon gap junction. (D) One plaque-type gap junction consisting of ~12 pits (upper left) and labeled by four 12 nm immunogold beads, as well as one simple, single-strand ribbon gap junction (lower right) labeled by three 12 nm gold beads. Due to their small diameter, neither of these two gap junctions would be reliably resolved in a conventional thin section TEM image.
“Plaque” gap junctions consist of continuous areas of densely packed connexons, either in regular hexagonal array (Fig. 3A) or in irregular clusters (this abundant subtype not shown). Plaque gap junctions were present in all layers of the retina except the outer nuclear layer and the photoreceptor outer segments. Plaque gap junctions in retina contained as many as 1000 connexons (not shown) to fewer than a dozen IMPs/pits (Fig. 3D, upper left corner). It is unlikely that plaque gap junctions consisting of fewer than 60 connexons would be detectable by thin-section electron microscopy, whereas gap junctions with fewer than ten connexons were readily detected by FRIL.
Single-strand and multi-strand “string” gap junctions (Fig. 3B) consisted of 1–10 curvilinear rows of connexons, with membrane areas between the strands containing few P-face IMPs or E-face pits. Multi-strand string gap junctions usually consisted of linked strands, tightly or loosely aggregated into a compound string gap junction (Fig. 3B, left side), whereas the rarer, single-strand string gap junctions consisted of a single straight or curved strand (not shown) or a closed loop (Fig. 3B, right side). Each string of connexons was usually one IMP wide (occasionally two IMPs wide) and up to 50 IMPs long, with large IMP-free areas often included within the margin. The component single strands superficially resembled tight junction strands, but instead of continuous IMP ridges or E-face furrows, each string consisted of individual 8-nm IMPs/pits, with a center-to-center spacing of 10 nm, as is characteristic of most other types of gap junctions. Single-strand string gap junctions were equally as well or more highly labeled for Cx36 than nearby “plaque” gap junctions. Labeling efficiencies (defined in Rash & Yasumura, 1999) for single-strand string gap junctions were usually > 1:10, whereas labeling of plaque gap junctions was often 1:30. Other than in an illustrated abstract (Rash et al., 2001a), these images represent the first documentation of Cx36 in string gap junctions, and the first demonstration of multi-strand string gap junctions in mammalian CNS.
“Ribbon” gap junctions (Fig. 3C and D, lower right) consisted of 1–10 curvilinear compound rows of connexons, with each row being 2–6 connexons wide and containing up to 100 connexons per ribbon, and up to 500 connexons per gap junction. Each component ribbon was surrounded by a 30 nm-wide particle-free zone (Fig. 3C). Multi-stranded ribbon gap junctions were observed in configurations resembling the letters “C” (Fig. 3D, lower right) and “O”, and the numeral “8”, as well as in compound gap junctions containing 3–12 ribbons of IMPs/pits (Fig. 3C). In the most abundant sup-type of multi-stranded ribbon gap junctions, connexons in each ribbon were in regular hexagonal arrays (Fig. 3C), while in another subtype, the IMPs/pits were irregularly distributed (not shown). Most multi-stranded ribbon gap junctions were labeled at an efficiency equivalent to plaque gap junctions (i.e., LE = ~1:30). [“Ribbon” gap junctions are not to be confused with “ribbon’’chemical synapses seen in thin section images of retina (Raviola & Gilula, 1975).]
Cx36 EXPRESSION AND CO-LOCALIZATION WITH ZO-1 IN AMYGDALA AND RETICULAR THALAMIC NUCLEUS
Several different connexins are reported to associate with the protein ZO-1 (Nielsen et al., 2001, 2002; Duffy et al., 2002). To investigate ZO-1 in association with neuronal Cx36, double immunofluorescence labeling of Cx36 and ZO-1 was examined in the reticular thalamic nucleus (RTN) of 16-day-old WT mice (Fig. 4). At low magnification at the anterior-posterior level of Bregma-0.9 [according to the mouse atlas of Hof et al. 2000], robust immunofluorescent puncta representing Cx36 labeling were seen, clearly delimiting the boundaries of the RTN in brains of WT mice (Fig. 4A1). Little or no Cx36-labeling was seen in the adjacent ventroposterolateral nucleus of the thalamus (left of RTN) or the internal capsule (area to the right of RTN). A similar distribution and density of Cx36-positive puncta was evident throughout the RTN at more anterior and posterior levels. Labeling for ZO-1 in the same field was even more robust, and appeared as both coarse and fine puncta with only a slightly higher density in the RTN than in surrounding structures (Fig. 4A2). In addition, threadlike labeling for ZO-1 was seen along blood vessels throughout the brain. Overlay of low magnification double-labeled images indicated some Cx36 co-localization with ZO-1 (Fig. 4A3); however, the extent of this apparent co-localization was difficult to discern at low magnification.
Fig. 4.
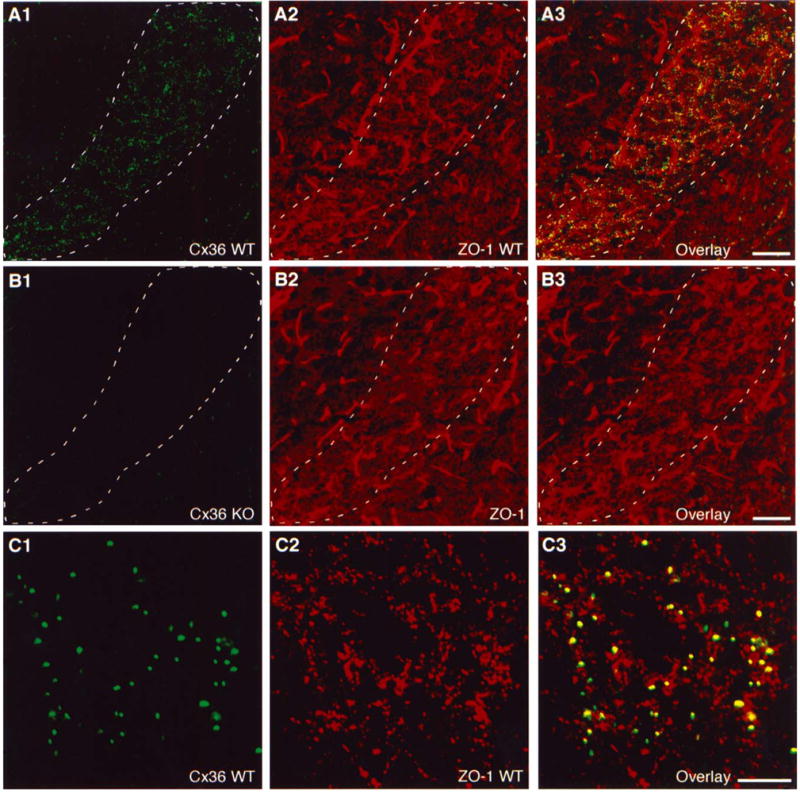
Expression of Cx36 in the reticular thalamic nucleus (RTN) and co-localization with ZO-1 in 16-day-old mouse brains. (A) Low magnification double immunofluorescence showing punctate labeling of Cx36 (A1) and, in the same field, more widely distributed punctate labeling of ZO-1 (A2) in the RTN (outlined by dashed line) of a wildtype (WT) mouse, with substantial Cx36/ZO-1 co-localization shown by yellow in overlay of images (A3). (B) Low magnification double immunofluorescence images of a Cx36 knockout (KO) mouse. A field corresponding to that in A shows an absence of specific labeling for Cx36 (B1) and no discernible loss of specific labeling for ZO-1 (B2). Overlay (B3) shows reduced yellow in the RTN (outlined by dashed line). Residual green and red fluorescence in thalamic areas of Cx36 KO mice represents non-specific attachment of primary and secondary antibodies. Bright, threadlike labeling for ZO-1 is associated with blood vessels. (C) Laser scanning confocal double immunofluorescence showing magnification of Cx36 (C1) and ZO-1 (C2) co-localization (yellow in overlay images, (C3) in a field of RTN in a WT mouse. Scale bars: A and B, 100 μ m; C, 10 μ m.
Immunofluorescence analysis with anti-Cx36 antibody in brains of 16-day-old Cx36 KO mice demonstrated a complete absence of punctate labeling for Cx36 in the RTN (Fig. 4B1), indicating the specificity of the anti-Cx36 antibody employed. Immunofluorescence labeling for ZO-1 in the RTN of Cx36 KO mice was similar to that in WT mice or was slightly reduced (Fig. 4B2), and overlay of images from Cx36 KO mice showed an absence of double labeling (Fig. 4B3) as a consequence of the elimination of Cx36 in these mice. Analysis of double immunofluorescence by laser scanning confocal microscopy revealed punctate labeling for Cx36 (Fig. 4C1) and for ZO-1 (Fig. 4C2) in the RTN of WT mice. Nearly all Cx36-immunopuncta were co-localized with a subset of ZO-1-positive puncta (Fig. 4C3). Conversely, however, only a small percentage of ZO-1-positive puncta in the RTN was co-localized with Cx36.
Immunohistochemical analysis of Cx36 and ZO-1 in the amygdala of WT and Cx36 KO mice is shown in Figure 5. Fluorescence Nissl staining of the amygdala is shown in Figure 5A, illustrating the basomedial (BMA) and basolateral (BLA) nuclei of the amygdala. Sparse punctate immunolabeling of Cx36 was observed throughout the amygdala of WT mice, but dense labeling appeared to be associated specifically with the BMA (Fig. 5B), extending to the cortical regions of the amygdala (not shown). Immunolabeling of Cx36 in the BLA (Fig. 5C), shown for comparison, was far less dense. Immunofluorescence labeling of Cx36 was absent in the BMA (Fig. 5D) as well as the BLA and cortical amygdala regions of Cx36 KO mice (not shown). By low magnification, double immunofluorescence labeling in BMA of WT mice revealed a similar distribution of punctate labeling for Cx36 (Fig. 5E1) and ZO-1 (Fig. 5E2), but with a greater density and variation in intensity of punctate labeling for ZO-1. In addition, ZO-1, but not Cx36, was localized along blood vessels. Overlay of double-labeled images revealed a high degree of Cx36 co-localization with ZO-1 in the BMA (Fig. 5E3), as well as in the cortical amygdala and surrounding regions, which contained much lower levels of Cx36 (not shown). Analysis by confocal microscopy revealed Cx36-positive (Fig. 5F1) and ZO-1-positive (Fig. 5F2) puncta arrayed in either random or linear patterns and, as in RTN, overlay images indicated that nearly all labeling for Cx36 was co-localized with ZO-1, whereas only a small proportion of the more abundant ZO-1 labeling overlapped with Cx36 (Fig. 5F3). In the RTN and amygdala, as well as other brain regions of WT and Cx36 KO mice, results obtained using combinations of polyclonal anti-Cx36 and monoclonal anti-ZO-1 antibodies (data shown) were similar to those obtained with combinations of monoclonal anti-Cx36 and polyclonal anti-ZO-1 antibodies (not shown).
Fig. 5.

Expression of Cx36 in the amygdala and Cx36 co-localization with ZO-1 in adult mouse brains. (A) Fluorescence Nissl-stained section showing the basomedial (BMA) and basolateral (BLA) nuclei of the amygdala (dashed lines). (B–D) Immunofluorescence labeling of Cx36 showing a dense concentration of Cx36-positive puncta in the BMA (B) of wildtype (WT) mouse, much sparser distribution of Cx36-positive puncta in the BLA of WT mouse (C), and an absence of punctate labeling for Cx36 in the BMA of a Cx36 knockout (KO) mouse (D). (E, F) Low magnification (E) and higher magnification confocal (F) double immunofluorescence showing labeling of Cx36 (E1, F1) and, in the same fields, labeling of ZO-1 (E2, F2, respectively) in the BMA of WT mouse, with Cx36/ZO-1 co-localization seen as yellow in overlay of images (E3, F3). Punctate labeling for ZO-1 is much denser than that for Cx36, and some intense ZO-1 labeling is seen in association with blood vessels (E2). Scale bars: A, 200 μ m; B–D, 100 μ m; C, 10 μ m.
FRIL DEMONSTRATION OF SEPARATE DISTRIBUTION OF TWO DISSIMILAR PROTEINS, Cx32 AND ZO-1, IN RAT LIVER GAP JUNCTIONS
To test the efficacy of antibodies to ZO-1 for use in FRIL, as well as to confirm the relative proximity of Cx32 and ZO-1 in rat hepatocyte junctional complexes (Fujimoto, 1995), freeze-fracture replicas of rat liver were double-labeled with polyclonal antibody to ZO-1 and monoclonal antibody to Cx32 (Fig. 6A). Immunogold labeling for Cx32 (12 nm gold) was restricted to gap junctions (Fig. 6A, white arrowheads), whereas immunogold labeling for ZO-1 (18 nm gold) was restricted to the tight junction strands (Fig. 6A, black arrowheads). [Stereoscopic images (Fig. 6A, left pair) and reverse stereoscopic images (right pair) allow confirmation that all of the labels were beneath the replica.] Such images initially led investigators to assume that ZO-1 represented the primary transmembrane protein of tight junctions [reviewed in (Gonzalez-Mariscal et al., 2000, 2003)]. However, subsequent experiments from a variety of disciplines have revealed that the transmembrane proteins of tight junctions, at the minimum, consist of “claudins” and “occludins” (Gonzalez-Mariscal et al., 2000, 2003), whereas ZO-1 is one of several cytoplasmic accessory proteins that bind to transmembrane proteins in the apical plasma membranes of some cells. Thus, in rat liver junctional complexes, Cx32 and ZO-1 have separate locations.
Fig. 6.
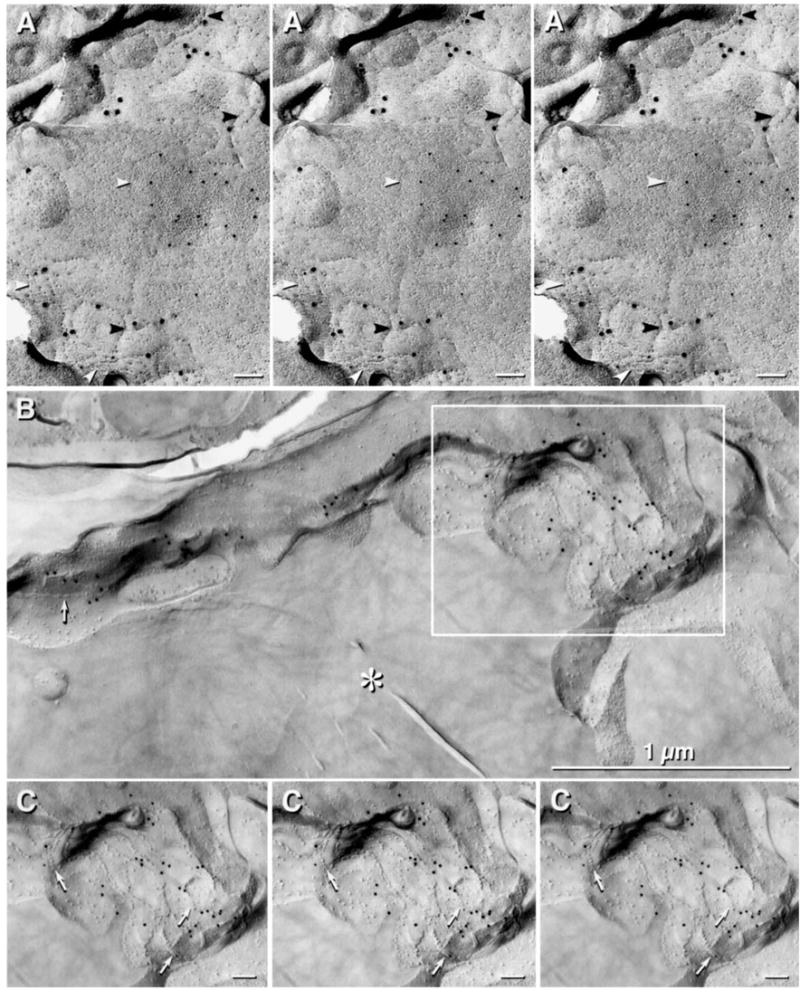
Simultaneous labeling of Cx32 in gap junctions and ZO-1 in tight junctions of rat liver, and comparison with ZO-1 labeling in tight junctions of capillary in adult rat retina. (A) Stereoscopic imaging (left pair) and reverse stereoscopic imaging (right pair) reveals that Cx32 labeling (12 nm gold) is directly associated with gap junctions (white arrowheads), whereas labeling for ZO-1 (18 nm beads) is associated exclusively with tight junctions strands (black arrowheads). (B) Portions of five capillary endothelial cells linked by tight junctions (arrow), each immunogold labeled for ZO-1 (12 nm gold beads). Asterisk (*) denotes capillary lumen. (C) Stereoscopic and reverse stereoscopic images of tight junctions (arrows), with ZO-1 labeling beneath the replica.
ZO-1 IN TIGHT JUNCTIONS IN RETINAL CAPILLARIES
In adult rat retina, polyclonal antibodies to ZO-1 labeled tight junctions in capillary endothelial cells (Fig. 6B and C). This observation established that tight junctions in rat retinal capillary endothelial cells retain ZO-1 immunoreactivity after SDS washing, thereby potentially allowing the use of that antibody to identify other structures in which ZO-1 immunoreactivity was found by LM. Based on confocal and biochemical demonstrations of Cx36 interaction with ZO-1 in neurons and some peripheral cell types that express Cx36 (Li et al., 2004a), we turned our attention to the use of ZO-1 as a possible alternative method for using FRIL to find and identify gap junctions in neurons.
ULTRASTRUCTURAL CO-LOCALIZATION OF Cx36 AND ZO-1 IN GAP JUNCTIONS OF ADULT RAT RETINA
In samples of rat retina that had been double-labeled for Cx36 and ZO-1 (monoclonal and polyclonal antibodies, respectively), immunogold beads for both Cx36 (6 nm and 12 nm) and ZO-1 (18 nm) were co-localized within individual gap junction plaques (Fig. 7A–C) in neuronal processes of both the outer and inner plexiform layers. Both large and small “plaque” gap junctions in neuronal processes were consistently labeled for ZO-1 by the polyclonal antibody, but string gap junctions were labeled less consistently. ZO-1 labeling of most plaque gap junctions in retinal neurons is to be contrasted with the absence of ZO-1 labeling of hepatocyte gap junctions (Fig. 6A, above). The basis for this differential labeling of gap junctions by ZO-1 antibodies in neurons vs. hepatocytes is attributed to the presence of a ZO-1 binding site in Cx36 (see Discussion) vs. the absence of ZO-1 binding sites in Cx32. However, it should be noted that labeling of neuronal gap junctions by ZO-1 monoclonal antibody has yet to be detected, whereas polyclonal anti-ZO-1 antibody consistently labeled neuronal gap junctions in retina.
Fig. 7.

(A–C) Simultaneous co-localization of Cx36 (6 nm gold beads [arrows] and 12 nm gold beads) and ZO-1 (18 nm gold beads) in “plaque” gap junctions linking unidentified neurons in adult rat retina. (C) Reverse stereoscopy facilitates detection of the smallest gold beads (Fig. 7B, right pair of images), as well as the “sidedness” of the labels. (C) Neuronal gap junction with seven 18 nm gold beads (ZO-1) and four 12 nm gold beads (Cx36).
Finally, we note that many small- to medium-diameter plaque gap junctions and a few large gap junction plaques were immunogold labeled for ZO-1 but not for Cx36. Polyclonal antibody ZO-1 labeling of a distinct group of neuronal gap junctions in retina in the absence of labeling for Cx36 suggested that ZO-1 binds to additional neuronal connexins other than Cx36. Those studies to identify additional neuronal connexins are in progress (Rash, Kamasawa, Nagy, et al., unpublished observations).
Discussion
We have used confocal microscopy and FRIL to reveal the chemical identities, structural associations, and relative proximities of Cx35/Cx36, NR1 glutamate receptors and ZO-1 proteins in vertebrate synapses. Using FRIL, we identified several new types of neuronal gap junctions in adult rat retina and showed that most of those contain Cx36. By FRIL double labeling of goldfish “mixed” synapses, we confirmed Cx35 in gap junctions and glutamate NMDA-R1 subunits in PSDs that were within 0.1 μ m of the gap junctions, or sufficiently close for microsecond diffusion of ions and signaling molecules between the two types of structures. Similarly, glutamate receptor PSDs were also observed near gap junctions in mixed synapses of rat inferior olive. Using confocal microscopy, we established the abundance and overall distribution of Cx36 in adult mouse RTN and amygdala, and showed, to the limits of resolution of confocal microscopy, that Cx36 frequently was co-localized with ZO-1 immunofluorescent puncta in those tissues. Finally, we used combined confocal microscopy and FRIL to distinguish between close proximity of Cx32 in gap junctions and ZO-1 in tight junctions of rat liver vs. co-localization of Cx36 and ZO-1 immunoreactivity within individual gap junction plaques in retinal neurons of adult rats.
HIGH-RESOLUTION MAPPING OF PROTEIN ARRAYS INTERACTING AT “INTERMEDIATE” DISTANCES
Auditory afferents on goldfish Mauthner cells, known as Large Myelinated Club Endings, represent one of the largest and most easily accessible mixed synapses in the vertebrate CNS. These synapses represent an ideal experimental model to study the properties of gap junctions between CNS neurons, as well as to explore possible functional interactions of gap junctions with the other major form of inter-neuronal communication—i.e., chemically-mediated transmission (Robertson, 1963; Furshpan, 1964; Yang et al., 1990; Pereda et al., 1998; Smith & Pereda, 2003). In this report, we provided additional evidence (Pereda et al., 2003) that Cx35, the fish ortholog of the mammalian Cx36, is on both sides of gap junctions at these mixed synapses. These gap junctions interact with their co-localized glutamatergic synapses by intermediate-range cytoplasmic signaling that causes gap junction conductance to increase. It has been proposed that NMDA glutamate receptors (of which, NR1 is an essential subunit) acting postsynaptically via local calcium signaling (Yang et al., 1990) cause activation of calcium/calmodulin-dependent kinase II, which is essential for enhancing gap junction conductance (Pereda & Faber, 1996; Pereda et al., 1998). Those electrophysiological findings are supported by additional confirmation of glutamate receptor immunolabeling at Club Ending synapses. The PSD-mediated signaling mentioned here [see also (Pereda et al.,1998, 2003)] may be relevant not only to Mauthner cell synapses, but potentially, to mammalian mixed synapses (Rash et al., 1996). In support of this notion, we provided FRIL images from mammalian CNS neurons revealing that NR1-containing E-face PSDs similar to those in Mauthner Cell/Club Ending synapses (Tuttle et al., 1986) are located at similar close distances from gap junction plaques in mammalian brain. Such arrangements suggest that this form of intermediate-range modulation of gap junctions by nearby chemical synapses constitutes a widespread property of electrical synapses where glutamatergic transmission and gap junctions co-exist.
Cx36 EXPRESSION IN RETICULAR THALAMIC NUCLEUS AND AMYGDALA
In the latter half of the last century, numerous now classical reports [reviewed in Nagy & Dermietzel (2000)] provided ultrastructural and electrophysiological evidence for the existence, if not prevalence, of gap junctions between neurons in the mammalian brain, indicating the potential importance of electrical and mixed synapses in the CNS of higher vertebrates. The abundance and widespread distribution of gap junctions between neurons has now been verified by the cloning of Cx36, the first connexin identified to be expressed in neurons (Condorelli et al., 1998; Willecke et al., 2002; see also O’Brien et al., 1998), by the establishment of the presence of Cx36 in gap junctions between neurons in widespread regions of brain (Rash et al., 2000, 2001b), and by demonstrations of the physiological role of electrical synapses in various brain regions (Deans et al., 2001; Hormuzdi et al., 2001). Our finding of a particularly dense concentration of Cx36 in the RTN of 16-day-old mice, with reduced but still striking levels in this region of adult mice, and very much lower levels in adjacent thalamic nuclei, indicates an especially prominent contribution of electrical synapses to the organization of neuronal circuitry in the RTN. This is consistent with reports demonstrating the presence of electrical synapses in the RTN, and with dense Cx36 mRNA expression and Cx36 promoter-driven β -gal expression in the RTN (Landisman et al., 2002; Condorelli et al., 2003). Although reports of electrical coupling in the amygdala are rare, neuronal gap junctions have recently been observed ultrastructurally in this region (Muller et al., 2002), and the basomedial amygdala contains an abundance of Cx36 mRNA (Belluardo et al., 2000). The high levels of Cx36-immunoreactive puncta in the basomedial amygdala (this report) suggest a high density of neuronal gap junctions in yet another mammalian brain region.
CO-LOCALIZATION OF Cx36 WITH ZO-1 IMMUNOREACTIVITY
Most cellular structural elements (e.g., tight junctions and gap junctions, both of which have the most deceivingly simple organization as viewed by thin-section or freeze-fracture electron microscopy) are in fact composed of many different interlocking proteins forming macromolecular complexes. A major aim of proteomics is to understand the composition of these multi-protein complexes and the rules regulating their assembly and composite function. Deciphering these rules is especially challenging where cellular structures with seemingly identical ultrastructural appearance in different tissues (e.g., gap junctions) are in fact constituted from extended families of proteins with tissue-specific and cell-specific expression patterns (Rash et al., 2001b). Despite these challenges, progress has been made toward identification of connexin interacting proteins (Nielsen et al., 2001, 2002; Duffy et al., 2002). Several connexins, including Cx31.9, Cx43, Cx45, Cx46 and Cx50 (Giepmans & Moolenaar, 1998; Laing et al., 2001; Kausalya et al., 2001; Nielsen et al., 2001, 2002, 2003), are localized to gap junctions and directly interact with ZO-1, a protein that was previously thought to be a component only of tight junctions and adherens junctions (reviewed by Gonzalez-Mariscal et al., 2003). Although the functions of connexin/ZO-1 associations are not yet known, their co-localization in neuronal gap junctions provides precedent to search for other ZO-1 binding partners that serve regulatory or structural roles at gap junctions (Nielsen et al., 2003).
Our demonstration of Cx36/ZO-1 co-localization in mouse brain and rat retina extends the list of connexins with which ZO-1 is associated and, more specifically, indicates targeting of ZO-1 to at least one class of gap junctions that is formed between neurons. Further, we have determined that Cx36 binds directly to the first of the three PDZ domains in ZO-1 (Li et al., 2004a, 2004b). Several points in relation to these findings are noteworthy:
In most brain regions that we have examined by confocal microscopy, nearly all Cx36 puncta were co-localized with ZO-1. However, by FRIL, some large neuronal plaque gap junctions in retina did not show ZO-1 immunogold labeling, whereas many small gap junctions had strong ZO-1 immunoreactivity but no reactivity for Cx36. Thus, in retina, ZO-1-positive neuronal gap junctions that apparently are devoid of Cx36 imply the existence of at least one additional neuronal connexin, and that at least one of those additional connexins binds ZO-1.
Only a small proportion of overall punctate labeling for ZO-1 was associated with Cx36, indicating that ZO-1 is associated with other structures, including blood vessels [see Vorbrodt & Dobrogowski (2003)] and adherens junctions [as observed in peripheral tissues (Gonzalez-Mariscal et al., 2000)]. LM immunolabeling for ZO-1 that we observed in brain was far more extensive than that described in most previous studies of this protein, and their widespread distribution was confirmed by FRIL at tight junctions of blood vessels. It thus bears emphasis that relatively weak tissue fixation conditions (see Methods) were required to visualize the more extensive distribution of ZO-1 in the CNS.
Although neuronal expression of ZO-1 has been described only rarely (Miragall et al., 1994; Inagaki et al., 2003), widespread neuronal expression of Cx36 and substantial Cx36 co-localization with ZO-1 in brain that we observed indicates widespread expression of ZO-1 in subpopulations of neurons that form gap junctions. Thus, ZO-1 may serve an important role in directing connexins and other related proteins to the same target areas of the plasma membrane in specific subpopulations of neurons.
A potential discrepancy noted by FRIL but not by LM was that monoclonal antibody to ZO-1 did not label neuronal gap junctions in retina. The basis for this difference in labeling by monoclonal vs. polyclonal antibodies to ZO-1 may be due to a technical limitation, masking of epitopes that would be detected by the monoclonal antibody, or to an unidentified cross-reactivity of the polyclonal antibody to some other protein localized at neuronal gap junctions.
APPLICATIONS OF FRIL
The high visibility of immunogold beads in FRIL facilitates detection and identification of both rare and common structures, including small or difficult-to-recognize structures. For example, “string” and “ribbon” gap junctions are abundant and easily detected by FRIL, but there are few published examples of “string” gap junctions, and those were of single IMP strands in freeze fracture replicas of retina (Raviola & Gilula, 1975; Gold & Dowling, 1979). Likewise, despite their abundance in FRIL images, there are no previously published examples of “ribbon” gap junctions.
A second advantage is that FRIL allows recognition of many different classes of intramembrane proteins and protein arrays. Thus, when an easily identified morphological class of protein arrays is consistently labeled in a particular cell type, to the exclusion of all other morphological classes of particle arrays in the same cell type, substantial confidence develops regarding the assignment of immunolabeled proteins to that specific ultra-structural feature.
In contrast, FRIL has yet to be of value for identifying individual IMPs interspersed among other IMPs in complex membranes. For this task, it would be useful to visualize the antibody bridges linking immunogold beads to individual IMPs. Indeed, osmium/ferricyanide mixtures have been used to delineate antibody molecules by conventional thin section TEM (Rash et al., 1978), but those differential staining methods have yet to be applied to FRIL. Alternatively, avidin-coated gold beads may permit higher-resolution labeling based on direct avidin-gold binding to biotinylated membrane proteins, thereby reducing the 15–20 nm “radius of uncertainty” that occurs when using double-antibody “sandwich” immunolabeling methods, as used here.
Despite its short history, FRIL in combination with confocal microscopy has become a valuable tool of proteomics. By combining high-resolution imaging, wide-area mapping, structural discrimination between a wide variety of membrane proteins, and immunogold identification and simultaneous mapping of multiple membrane proteins, the combination of confocal microscopy and FRIL provides essential information regarding protein molecules and molecular arrays that are in sufficiently close proximity to participate in short-range vs. intermediate-range biochemical and biophysical interactions.
Acknowledgments
We thank Aaron Magnie for assistance with preparation of stereoscopic images. Funded by NIH grants NS-38121, NS-44010, NS-44395 (JER), MH-59995 (FED), DC03186 (AP) and by the Canadian Institutes of Health Research (JIN).
Footnotes
We distinguish “FRIL” from “SDS-FRL” because Fujimoto’s original technique (Fujimoto, 1995, 1997) did not permit tissue visualization or “grid mapping” by confocal microscopy, both of which are requirements for high-resolution proteomic mapping in the vertebrate CNS.
Professor Kazushi Fujimoto died at age 49 on November 28, 2003. His introduction of SDS-FRL for proteomic mapping is gratefully acknowledged in this report, which extends his pioneering methodology.
References
- BELLUARDO N, MUDO G, TROVATO-SALINARO A, LE GURUN S, CHAROLLAIS A, SERRE-BEINIER V, AMATO G, HAEFLIGER JA, MEDA P, CONDORELLI DF. Expression of connexin36 in the adult and developing rat brain. Brain Research. 2000;19:121–138. doi: 10.1016/s0006-8993(00)02300-3. [DOI] [PubMed] [Google Scholar]
- BERMAN N, DUNN R, MALER L. Function of NMDA receptors and persistent sodium channels in a feedback pathway of the electrosensory system. J Neurophysiol. 2001;86:1612–1621. doi: 10.1152/jn.2001.86.4.1612. [DOI] [PubMed] [Google Scholar]
- BRANTON D, BULLIVANT S, GILULA NB, KARNOVSKY MJ, MOOR H, NORTHCOTE DH, PACKER L, SATIR B, SATIR P, SPETH V, STAEHELIN LA, STEERE RL, WEINSTEIN RS. Freeze-etching nomenclature. Science. 1975;190:54–56. doi: 10.1126/science.1166299. [DOI] [PubMed] [Google Scholar]
- CONDORELLI DF, PARENTI R, SPINELLA F, SALINARO AT, BELLUARDO N, CARDILE V, CICIRATA F. Cloning of a new gap junction gene (Cx36) highly expressed in mammalian brain neurons. Eur J Neurosci. 1998;10:1202–1208. doi: 10.1046/j.1460-9568.1998.00163.x. [DOI] [PubMed] [Google Scholar]
- CONDORELLI DF, PARENTI R, SPINELLA F, TROVATO SALINARO A, MUDO G. Expression of Cx36 in mammalian brain neurons. Eur J Neurosci. 2003;10:1202–1208. doi: 10.1046/j.1460-9568.1998.00163.x. [DOI] [PubMed] [Google Scholar]
- DEANS MR, GIBSON JR, SELLITTO C, CONNORS BW, PAUL D. Synchronous activity of inhibitory networks in neocortex requires electrical synapses containing connexin36. Neuron. 2001;31:477–485. doi: 10.1016/s0896-6273(01)00373-7. [DOI] [PubMed] [Google Scholar]
- DINCHUK JE, JOHNSON TJA, RASH JE. Postreplication labeling of E-leaflet molecules: Membrane immunoglobulins localized in sectioned labeled replicas examined by TEM and HVEM. J Electron Microsc Tech. 1987;7:1–16. doi: 10.1002/jemt.1060070102. [DOI] [PubMed] [Google Scholar]
- DUFFY HS, DELMAR M, SPRAY DC. Formation of the gap junction nexus: Binding partners for connexins. J Physiol Paris. 2002;96:243–249. doi: 10.1016/s0928-4257(02)00012-8. [DOI] [PubMed] [Google Scholar]
- FUJIMOTO K. Freeze-fracture replica electron microscopy combined with SDS digestion for cytochemical labeling of integral membrane proteins. Application to the immunogold labeling of intercellular junctional complexes. J Cell Science. 1995;108:3443–3449. doi: 10.1242/jcs.108.11.3443. [DOI] [PubMed] [Google Scholar]
- FUJIMOTO K. SDS-digested freeze-fracture replica labeling electron microscopy to study the two-dimensional distribution of integral membrane proteins and phospholipids in biomembranes: Practical procedure, interpretation and application. Histochem Cell Biol. 1997;107:87–96. doi: 10.1007/s004180050092. [DOI] [PubMed] [Google Scholar]
- FURSHPAN EJ. Electrical transmission at an excitatory synapse in a vertebrate brain. Science. 1964;144:878–880. doi: 10.1126/science.144.3620.878. [DOI] [PubMed] [Google Scholar]
- GIEPMANS BN, MOOLENAAR WH. The gap junction protein connexin43 interacts with the second PDZ domain of the zonula occludens-1 protein. Curr Biol. 1998;8:931–934. doi: 10.1016/s0960-9822(07)00375-2. [DOI] [PubMed] [Google Scholar]
- GIOT L, BADER JS, BROUWER C, CHAUDHURI A, KUANG B, LI Y, HAO YL, OOI CE, GODWIN B, VITOLS E, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- GOLD GH, DOWLING JE. Photoreceptor coupling in retina of the toad, Bufo Marinus. I Anatomy. J Neurophysiol. 1979;42:292–310. doi: 10.1152/jn.1979.42.1.292. [DOI] [PubMed] [Google Scholar]
- GONZALEZ-MARISCAL L, BETANZOS A, AVILA-FLORES A. MAGUK proteins: Structure and role in the tight junction. Cell Dev Biol. 2000;11:315–324. doi: 10.1006/scdb.2000.0178. [DOI] [PubMed] [Google Scholar]
- GONZALES-MARISCAL L, BETANZOS A, NAVA P, JARAMILLO BE. Tight junction proteins. Prog Biophys Mol Biol. 2003;81:1–44. doi: 10.1016/s0079-6107(02)00037-8. [DOI] [PubMed] [Google Scholar]
- HARRIS KM, LANDIS DMD. Membrane structure at synaptic junctions in area CA1 of the rat hippocampus. Neuroscience. 1986;19:857–872. doi: 10.1016/0306-4522(86)90304-0. [DOI] [PubMed] [Google Scholar]
- HOF PR, YOUNG WG, BLOOM FE, BELICHENKO PV, CELIO MR. Comparative cytoarchitectonic atlas of the C57BL/6 and 129/Sv mouse brains. NY: Elsevier; 2000. [Google Scholar]
- HORMUZDI SJ, PAIS I, LEBEAU FEN, TOWERS SK, ROZOV A, BUHL EH, WHITTINGTON MA, MONYER H. Impaired electrical signaling disrupts gamma frequency oscillations in connexin 36-deficient mice. Neuron. 2001;31:487–495. doi: 10.1016/s0896-6273(01)00387-7. [DOI] [PubMed] [Google Scholar]
- INAGAKI M, IRIE K, DEGUCHI-TAWARADA M, IKEDA W, OHTSUKA T, TAKEUCHI M, TAKAI Y. Nectin-dependent localization of ZO-1 at puncta adhaerentia junctions between the mossy fiber terminals and the dendrites of the pyramidal cells in the CA3 area of adult mouse hippocampus. J Comp Neurol. 2003;460:514–524. doi: 10.1002/cne.10653. [DOI] [PubMed] [Google Scholar]
- INTERNAT. HUMAN GENOME SEQ. CONS. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- KAUSALYA PJ, REICHERT M, HUNZIKER W. Connexin45 directly binds to ZO-1 and localizes to the tight junction region in epithelial MDCK cells. FEBS Letters. 2001;505:92–96. doi: 10.1016/s0014-5793(01)02786-7. [DOI] [PubMed] [Google Scholar]
- LAING JG, MANLEY-MARKOWSKI RN, KOVAL M, CIVITELLI R, STEINBERG TH. Connexin45 interacts with zonula occludens-1 and connexin43 in osteoblastic cells. J Biol Chem. 2001;276:23051–23055. doi: 10.1074/jbc.M100303200. [DOI] [PubMed] [Google Scholar]
- LANDISMAN CE, LONG MA, BEIERLEIN M, DEANS MR, PAUL D, CONNORS BW. Electrical synapses in the thalamic reticular nucleus. J Neurosci. 2002;22:1002–1009. doi: 10.1523/JNEUROSCI.22-03-01002.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI X, OLSON C, LU S, KAMASAWA N, YASUMURA T, RASH JE, NAGY JI. Neuronal connexin36 association with zonula occludens-1 protein (ZO-1) in mouse brain and interaction with the first PDZ domain of ZO-1. Eur J Neurosci. 2004a;19:2132–2146. doi: 10.1111/j.l460-9568.2004.03283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI X, OLSON C, LU S, NAGY JI. Connexin36 association with zonula occludens-1 (ZO-1) in HeLa cells, β -TC-3 cells, pancreas and adrenal gland. 2004b. Submitted. [DOI] [PubMed] [Google Scholar]
- MIRAGALL F, KRAUSE D, DE VRIES U, DERMIETZEL R. Expression of the tight junction protein ZO-1 in the olfactory system: Presence of ZO-1 on olfactory sensory neurons and glial cells. J Comp Neurol. 1994;341:433–448. doi: 10.1002/cne.903410402. [DOI] [PubMed] [Google Scholar]
- MULLER JF, MASCAGNI F, BETETTE RLN, MCDONALD AJ. Synaptic connectivity of parvalbumin—immunoreactive neurons in the basolateral amygdala. Soc Neurosci Abs. 2002;28:430–7. [Google Scholar]
- NAGY JI, DERMIETZEL R. Gap junctions and connexins in the mammalian central nervous system. In: HERTZBERG EL, editor. Advances in Molecular and Cell Biology. Vol. 30 Greenwich: JAI Press ; 2000. pp. 323–396. [Google Scholar]
- NIELSEN PA, BARUCH A, GIEPMANS BN, KUMAR NM. Characterization of the association of connexins and ZO-1 in the lens. Cell Commun Adhes. 2001;8:213–217. doi: 10.3109/15419060109080726. [DOI] [PubMed] [Google Scholar]
- NIELSEN PA, BARUCH A, SHESTAPOLOV VI, GIEPMANS BN, BENEDETTI EL, KUMAR NM. Lens connexins Cx46 and Cx50 interact with Zonula Occludens Protein-1 (ZO-1) Mol Biol Cell. 2003;14 :2470–2481. doi: 10.1091/mbc.E02-10-0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIELSEN PA, BEAHM DL, GIEPMANS BN, BARUCH A, HALL JE, KUMAR NM. Molecular cloning, functional expression, and tissue distribution of a novel human gap junction-forming protein, connexin-31.9. Interaction with zona occludens protein-1. J Biol Chem. 2002;277:38272–38283. doi: 10.1074/jbc.M205348200. [DOI] [PubMed] [Google Scholar]
- O’BRIEN J, BRUZZONE R, WHITE TW, AL-UBAIDI MR, RIPPS H. Cloning and expression of two related connexins from the perch retina define distinct subgroups of the connexin family. J Neurosci. 1998;18:7625–7637. doi: 10.1523/JNEUROSCI.18-19-07625.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OTTERSEN OP, LANDSEND AS. Organization of glutamate receptors at the synapse. Eur J Neurosci. 2003;9:2219–2224. doi: 10.1111/j.1460-9568.1997.tb01640.x. [DOI] [PubMed] [Google Scholar]
- PEREDA A, FABER DS. Activity dependent short-term plasticity of intercellular coupling. J Neurosci. 1996;16 :983–992. doi: 10.1523/JNEUROSCI.16-03-00983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEREDA A, O’BRIEN J, NAGY JI, BUKAUSKAS F, DAVIDSON KGV, KAMASAWA N, YASUMURA T, RASH JE. Connexin35 mediates electrical transmission at mixed synapses on Mauthner cells. J Neurosci. 2003;23:7489–7503. doi: 10.1523/JNEUROSCI.23-20-07489.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEREDA AE, BELL T, CHANG B, CZERNIC A, NAIRN A, SODERLING T, FABER DS. Ca2+/calmodulin-dependent kinase II mediates simultaneous enhancement of gap junctional conductance and glutamatergic transmission. Pro Natl Acad Sci. 1998;95:13272–13277. doi: 10.1073/pnas.95.22.13272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PHILLIPS TE, BOYNE AF. Liquid nitrogen-based quick freezing: Experiments with bounce-free delivery of cholinergic nerve terminals to a metal surface. J Electron Microsc Tech. 1984;1:9–29. [Google Scholar]
- RASH JE, DILLMAN R, MORITA M, WHALEN LR, GUTHRIE PB, FAY-GUTHRIE D, WHEELER DW. Grid-mapped freeze fracture: Correlative confocal laser scanning microscopy and freeze-fracture electron microscopy of preselected neurons in spinal cord slices. In: SHOTTON DM, SEVERS NJ, editors. Techniques in Modern Biomedical Microscopy. 2 Rapid Freezing, Freeze-Fracture and Deep Etching. Wiley-Liss; 1995. pp. 127–150. [Google Scholar]
- RASH JE, DILLMAN RK, BILHARTZ BL, DUFFY HS, WHALEN LR, YASUMURA T. Mixed synapses discovered and mapped throughout mammalian spinal cord. Proc Natl Acad Sci USA. 1996;93:4235–4239. doi: 10.1073/pnas.93.9.4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RASH JE, DUFFY HS, DUDEK FE, BILHARTZ BL, WHALEN LR, YASUMURA T. Grid-mapped freeze-fracture analysis of gap junctions in gray and white matter of adult rat central nervous system, with evidence for a “panglial syncytium” that is not coupled to neurons. J Comp Neurol. 1997;388:265–292. doi: 10.1002/(sici)1096-9861(19971117)388:2<265::aid-cne6>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- RASH JE, HUDSON CS, ELLISMAN MH. Ultrastructure of acetylcholine receptors at the mammalian neuromuscular junction. In: STRAUB RW, BOLIS L, editors. Cell Membrane Receptors for Drugs and Hormones: A Multidisciplinary Approach. New York: Raven Press; 1978. pp. 47–68. [Google Scholar]
- RASH JE, STAINES WA, YASUMURA T, PATEL D, HUDSON CS, STELMACK GL, NAGY JI. Immunogold evidence that neuronal gap junctions in adult rat brain and spinal cord contain connexin36 (Cx36) but not Cx32 or Cx43. Proc Natl Acad Sci (USA) 2000;97:7573–7578. doi: 10.1073/pnas.97.13.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RASH JE, YASUMURA T. Direct immunogold labeling of connexins and aquaporin4 in freeze-fracture replicas of liver, brain and spinal cord: Factors limiting quantitative analysis. Cell Tissue Res. 1999;296:307–321. doi: 10.1007/s004410051291. [DOI] [PubMed] [Google Scholar]
- RASH JE, YASUMURA T, DAVIDSON K, FURMAN CS, DUDEK FE, NAGY JI. Identification of cells expressing Cx43, Cx30, Cx26, Cx32 and Cx36 in gap junctions of rat brain and spinal cord. Cell Commun Adhes. 2001a;8:315–320. doi: 10.3109/15419060109080745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RASH JE, YASUMURA T, DUDEK FE. Ultrastructure, histological distribution, and freeze-fracture immunocytochemistry of gap junctions in rat brain and spinal cord. Cell Biol Internat. 1998a;22:731–749. doi: 10.1006/cbir.1998.0392. [DOI] [PubMed] [Google Scholar]
- RASH JE, YASUMURA T, DUDEK FE, NAGY JI. Cell-specific expression of connexins, and evidence for restricted gap junctional coupling between glial cells and between neurons. J Neurosci. 2001b;21:1983–2001. doi: 10.1523/JNEUROSCI.21-06-01983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RASH JE, YASUMURA T, HUDSON CS, AGRE P, NIELSEN S. Direct immunogold labeling of Aquaporin-4 in “square arrays” of astrocyte and ependymocyte plasma membranes in rat brain and spinal cord. Pro Natl Acad Sci. 1998b;95:11981–11986. doi: 10.1073/pnas.95.20.11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAVIOLA E, GILULA NB. Intramembrane organization of specialized contacts in the outer plexiform layer of the retina. J Cell Biol. 1975;65:192–222. doi: 10.1083/jcb.65.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROBERTSON JD. The occurrence of a subunit pattern in the unit membranes of club endings in Mauthner cell synapses in goldfish brains. J Cell Biol. 1963;19:201–221. doi: 10.1083/jcb.19.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SMITH M, PEREDA A. Chemical synaptic activity modulates nearby electrical synapses. Pro Natl Acad Sci. 2003;100:4849–4854. doi: 10.1073/pnas.0734299100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TUTTLE R, MASUKO S, NAKAJIMA Y. Freeze-fracture study of the large myelinated club ending synapse on the goldfish Mauthner cell: Specialized reference to the quantitative analysis of gap junctions. J Comp Neurol. 1986;246:202–211. doi: 10.1002/cne.902460206. [DOI] [PubMed] [Google Scholar]
- VENTER JC, ADAMS MD, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- VERVEER PJ, HARPUR AG, BASTIAENS PIH. Imaging protein interactions by FRET microscopy. In: GOLEMIS E, editor. Protein-Protein Interactions: A Molecular Cloning Manual. New York: Cold Spring Harbor Laboratory Press; 2003. pp. 181–213. [Google Scholar]
- VORBRODT A, DOBROGOWSKI DH. Molecular anatomy of intercellular junctions in brain endothelial and epithelial barriers: Electron microscopist’s view. Brain Res Rev. 2003;42:221–242. doi: 10.1016/s0165-0173(03)00177-2. [DOI] [PubMed] [Google Scholar]
- WILLECKE K, EIBERGER J, DEGEN J, ECKARDT D, ROMUALDI A, GUELDENAGEL M, DEUTSCH U, SOEHL G. Structural and functional diversity of connexin genes in the mouse and human genome. Biol Chem. 2002;383:725–737. doi: 10.1515/BC.2002.076. [DOI] [PubMed] [Google Scholar]
- WINKLER H, WILDHABER H, GROSS H. Decoration on the surface of a regular protein layer. Ultramicroscopy. 2002;16:331–339. [Google Scholar]
- YANG XD, KORN H, FABER DS. Long-term potentiation of electrotonic coupling at mixed synapses. Nature. 1990;348:542–545. doi: 10.1038/348542a0. [DOI] [PubMed] [Google Scholar]
- ZINGSHEIM HP, ABERMAN R, BACHMANN L. Shadow casting and heat damage. Proc 7th Int Cong Elec Microsc (Grenoble) 1970;1:411–412. (Abstract) [Google Scholar]