Abstract
The MHC Class II transactivator (CIITA) acts in the cell nucleus as the master regulator of MHC class II (MHC II) gene expression. It is important to study CIITA regulation in multiple myeloma since MHC expression is central to ability of myeloma cells to present antigen and to the ability of the immune system to recognize and destroy this malignancy. Regulation of CIITA by IFN-γ in B lymphocytes occurs through the CIITA type IV promoter (pIV), one of the four potential promoters (pI-pIV) of this gene. To investigate regulation of CIITA by IFN-γ in multiple myeloma cells, first the ability of these cells to respond to IFN-γ was examined. RTPCR analyses show that IFN-γR1, the IFN-γ-binding chain of the IFN-γ receptor, is expressed in myeloma cells and IRF-1 expression increases in response to IFN-γ treatment. Western blotting demonstrates that STAT1 is activated by phosphorylation in response to IFN-γ. RT-PCR and functional promoter analyses show that IFN-γ up regulates the activity of CIITA pIV, as does ectopic expression of IRF-1 or IRF-2. In vivo protein/DNA binding studies demonstrate protein binding at the GAS, E box and IRF-E sites. In vitro studies confirm the binding of IRF-1 and IRF-2 to CIITA pIV. Although multiple myeloma cells express PRDI-BF1/Blimp-1, a factor that represses both the CIITA type III and IV promoters, they retain the capability to up regulate CIITA pIV and MHC II expression in response to IFN-γ treatment. These findings are the first to demonstrate that although PRDI-BF1/Blimp-1 diminishes the constitutive ability of these cells to present antigen by limiting CIITA and MHC II expression, it is possible to enhance this expression through the use of cytokines, like IFN-γ.
Keywords: myeloma; CIITA; IFN-γ, IFN-γR1; STAT1; IRF-1; IRF-2; PRDI-BF1; Blimp-1
1. Introduction
Multiple myeloma, the most common primary malignancy of bone in older adults, is characterized by the clonal expansion of malignant plasma cells. These cells produce monoclonal immunoglobulins that carry unique antigenic determinants and act as tumor-specific antigens for recognition and destruction of these cells by the immune system (Wen et al., 2001; Galea et al., 2002). Immune responses are tightly regulated by CD4+ T helper (TH) lymphocytes (Cresswell, 1994). Activation of TH cells depends on the recognition of antigens presented by MHC class II (MHC II). MHC II-recognizing TH cells secrete cytokines that initiate and sustain CD8+ cytotoxic lymphocyte (CTL) anti-tumor responses. Therefore, MHC II expression is central to the recognition and destruction of tumor cells by CTLs. Since myeloma cells generally express low levels of the MHC Class II transactivator (CIITA), which controls the expression of MHC II, they can escape immune inspection due to their decreased ability to present antigen (Ghosh et al., 2001). Loss of MHC II expression in B cell lymphomas is considered as tumorigenic and an immune escape mechanism (Fuji and Iribe, 1986; Amiot et al., 1998). Consistent with this idea, forced CIITA expression has been shown to restore MHC II expression and decrease the tumorigenicity of some tumor cell types (Shi et al., 2006).
The MHC II transactivator (CIITA) is a non-DNA binding protein factor that is recruited to the enhancer complex of MHC II genes (Harton and Ting, 2000). This factor is the predominant regulator of MHC II expression and CIITA expression acts as a crucial master switch for immune responses. Expression of CIITA is regulated in a complex cell-type specific manner, predominantly at the level of transcription (Ting and Trowsdale, 2002; Leibundgut-Landmann et al., 2004). The human CIITA gene, MHC2TA, has four distinct promoters designated as promoters I-IV and it is controlled by at least three of these (Muhlethaler-Mottet et al., 1997). While the CIITA type I promoter (pI) is mainly expressed in dendritic cells and macrophages, the type III promoter (pIII) is constitutively expressed in B lymphocytes, which constitutively express MHC II (Muhlethaler-Mottet et al., 1997; Landmann et al., 2001; Pai et al., 2002). Recently, we have also described CIITA type IV promoter (pIV) expression that is regulated by IFN-γ in B lymphocytes (Piskurich et al., 2006). Similar to the mechanisms described for other cell types where CIITA pIV expression is IFN-γ-inducible, the mechanism for the regulation of CIITA pIV by IFN-γ in B cells depends on the binding of signal transducer and activator of transcription (STAT)1 to an IFN-γ activation factor DNA-binding sequence (GAS), IFN regulatory factor (IRF)-1 and IRF-2 to an IFN regulatory factor-element (IRF-E), and the ubiquitously expressed factor, USF-1, to an interposed E box site (Muhlethaler-Mottet et al., 1998; Piskurich et al., 1999; Xi et al., 1999; Morris et al., 2002).
Positive regulatory domain I-binding factor 1 (PRDI-BF1), called B lymphocyte-induced maturation protein 1 (Blimp-1) in mice, is a transcriptional repressor that plays a pre-eminent role in the terminal differentiation of B lymphocytes into plasma cells (Turner, Jr. et al., 1994; Lin et al., 1997; Shapiro-Shelef and Calame, 2005). This genetic program of B cell differentiation, which is largely intact in myeloma cells, includes the loss of constitutive MHC II expression via extinction of CIITA pIII activity by PRDI-BF1/Blimp-1 (Piskurich et al., 2000; Ghosh et al., 2001). Since myeloma cells express PRDI-BF1/Blimp-1, they also generally express low constitutive levels of CIITA and MHC II (Nagy et al., 2002). We and others have recently described the IFN-γ-inducible CIITA promoter, pIV, as a new target for repression by PRDIBF1/Blimp-1 (Tooze et al., 2006; Chen et al., 2007). Although they express PRDI-BF1/Blimp-1, increases in MHC II expression in response to IFN-γ have been described both in vivo and in vitro for myeloma cells (Yi et al., 1997; Beatty and Paterson, 2001). While extinction of constitutive MHC II and CIITA pIII expression has been studied in myeloma cells (Ghosh et al., 2001), induction of CIITA by IFN-γ in these cells remains virtually unstudied, despite several reports indicating that multiple myeloma cells can be activated by IFN-γ to increase MHC II expression and act as antigen presenting cells (Yi et al., 1997; Beatty and Paterson, 2001). Here, we demonstrate for the first time that the IFN-γ signaling pathway for gene induction is intact in myeloma cells and that IFN-γ up regulates CIITA type IV promoter activity. This is important because it describes a mechanism by which cytokines, like IFN-γ, are potentially useful in therapies aimed at enhancing the ability of the immune system to recognize myeloma cells. Knowledge of this response is important to the discovery of ways to increase the immunogenicity and decrease the tumorigenicity of these cells by increasing their expression of CIITA and MHC II.
2. Materials and methods
2.1. Culture and stimulation of myeloma and B cell lines
The human myeloma cells lines, U266 and RPMI 8266, and Raji B lymphocytes were grown according to American Type Culture Collection specifications in RPMI 1640 with 10% fetal calf serum, 2mM L-glutamine, and 100 U/ml penicillin and streptomycin. NCI-H929 human myeloma cells were grown similarly, except 2-mercaptoethanol (50 μm) was added to the culture medium. Cells were treated with recombinant human IFN-γ (500 U/ml, R&D Systems) as indicated below.
2.3. Constructs
The pIV, pmIV(GAS) and pmIV(IRF-E) MHC2TA promoter reporter constructs have been described previously (Piskurich et al., 1998; Piskurich et al., 1999). These constructs were formerly called pIVCIITA.Luc, pmGAS.IVCIITA.Luc (contains the GAS site mutations, ggagtcTAAA, with mutations designated by lowercase type), and pmIRF.IVCIITA.Luc (contains the IRF-E site mutations, GAActTagAAGG), respectively. The human IRF-1 and IRF-2 expression plasmids were a kind gift of Ying Cha Henderson (Cha and Deisseroth, 2004).
2.4. RT-PCR
Total cellular RNA was isolated using TRIzol (Invitrogen) according to the manufacturer's instructions from 5 × 106 cells, and then digested with RQ1 RNase-free DNase (Promega) according to manufacturer's directions. To prepare the first-strand cDNA, RNA (1 μg) was reverse-transcribed in a reaction volume of 20 μl using Superscript II Reverse Transcriptase and random primers (Invitrogen). Of the cDNA, 1 μl was amplified with AmpliTaq Gold DNA polymerase (Applied Biosystems). Conditions for the PCR and sizes of the amplified products were as follows: IFN-γR1, 5'-GTTAAAGCCAGGGTTGGACA-3' and 5'-ATCGACTTCCTGCTCGTCTC-3', 60°C, 27 cycles, 177 bp; IRF-1, 5'-GAAGTCCAGCCGAGATGC-3' and 5'-CGGCACAACTTCCACTG-3', 60°C, 24 cycles, 235 bp; IRF-2, 5'-CCACTGAGAGCGACGAGC-3' and 5'-GTTGGAAGTGACGAAGGACG-3', 60°C, 25 cycles, 236 bp; CIITA pIV, 5'-AGGGAGAGGCCACCAGCAG-3' and 5'-GAACTGGTCGCAGTTGATG-3', 60°C, 37 cycles, 227 bp; MHC II (HLADRA), 5'-TGTTTGACTTTGATGGTGATGAG-3' and 5'-AATAATGATGCCCA CCAGACC-3', 55°C, 28 cycles, 558 bp; PRDI-BF1, 5'-TTCAAGTATGCCACCAACAG-3' and 5'-AATGTTAGAACGGTAGAGGTC-3', 55°C, 25 cycles, 549 bp; PRDI-BF1β, 5'-GTGGTGGGTTAATCGGTTTG-3' and 5'-ATAGCGCATCCAGTTGCTTT-3', 55°C, 30 cycles, 172 bp; GAPDH, 5'-ACCACAGTCCATGCCATCAC-3' and 5'-TCCACCACCCTGTTGCTGTA-3', 60°C, 25 cycles, 452 bp. In all cases, PCR was performed as demonstrated in Figure 1B using serial dilutions of the template and a range of cycle numbers were tested to verify that amplifications were in the linear range for the assay. Mock preparations of cDNA, where reverse transcriptase was omitted, were tested by PCR and did not give specific bands. PCR products (50% of the total reaction volume) were resolved on 1.5% agarose gels, stained with ethidium bromide and scanned digitally using an electrophoresis documentation and analysis system (EDAS 290, Kodak).
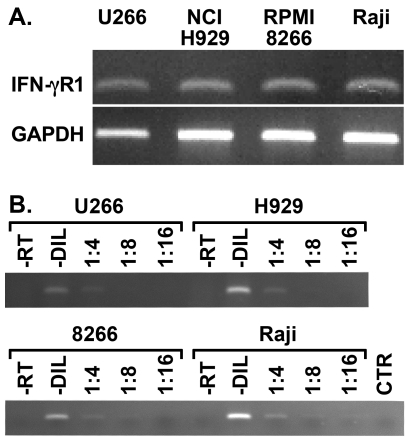
Fig. 1.
Myeloma cells express IFN-γR1. (A) Equal amounts of RNA from U266, NCI-H929, and RPMI 8266 human myeloma cells and Raji B cells were subjected to RT-PCR using primers specific for human IFN-γR1. PCR using primers for GAPDH was performed as an internal standard for cDNA loading. This experiment has been repeated twice with similar results. (B) PCR for IFN-γR1 was performed using serial dilutions of the cDNA template to verify that amplifications were in the linear range for the assay (-DIL, undiluted). Mock preparations of cDNA where reverse transcriptase was omitted (-RT), and reaction mixtures that did not contain cDNA (CTR) were also tested by PCR and did not give specific bands.
2.5 Western blot analysis
U266, RPMI 8266, and NCI-H929 myeloma cells were treated with IFN-γ for 0, or 24 h before harvesting. Cells were lysed in Mammalian Protein Extraction Reagent (M-PER®) and the protein concentration of the extracts was determined using the BCA™ Protein Assay Kit, according to the manufacturer's directions (Pierce). For the analysis, 0.3 μg/lane of whole cell extracts were resolved on 4-20% SDS-polyacrylamide gels and transferred to polyvinylidene difluoride membranes (Bio-Rad Laboratories). Immunostaining was performed for 2 h at room temperature. Proteins were detected using rabbit anti-phospho-STAT1 (Tyr701) IgG (Upstate Biotechnologies). Bound antibody was visualized using anti-rabbit IgG horseradish peroxidase-conjugated secondary antibody (Promega), followed by detection using SuperSignal® West Pico chemiluminescence substrate (Pierce). Membranes were stripped and reprobed with anti-STAT1 (Upstate Biotechnologies). This was repeated with anti-β-actin (Abcam) as a loading control.
2.6 Transfections and luciferase assays
U266 and RPMI 8266 myeloma cells were transiently transfected by electroporation and luciferase assays were performed as previously described (Piskurich et al., 1998) with the following modifications: 6 h after electroporation, cells were treated with human recombinant IFN-γ for 24 h and then harvested; luciferase activity was measured using a Tropix TR717 luminometer (PE Applied Biosystems); as an internal control, each transfection included 300 ng of the pRL-TK Vector (Promega) which directs the expression of Renilla luciferase. Renilla luciferase activity was measured using the Renilla Luciferase Assay System (Promega) according to the manufacturer and was used to normalize the luciferase activity for transfection efficiency.
2.7. Electrophoretic mobility shift assays (EMSAs)
Nuclear extracts were prepared from 2 × 106 U266 myeloma cells, treated with IFN-γ for 0 or 24 h, according to the manufacturer's directions using NE-PER® Nuclear and Cytoplasmic Extraction Reagents (Pierce). The CIITA pIV oligonucleotide probe was as follows: 5'-CTGCAGAAAGAAAGTGAAAGGGAAAAAGAACT-3'. EMSAs were performed as previously described (Piskurich et al., 2006). Antibodies were purchased from Santa Cruz Biotechnology.
2.8. In vivo genomic footprinting
The dimethyl sulfate (DMS) treatment, genomic DNA preparation, amplification of human CIITA (MHC2TA) pIV using ligation-mediated PCR, electrophoresis and visualization were all performed as previously described (Piskurich et al., 1999). Briefly, 1 × 107 U266 or NCI-H929 cells were treated with IFN-γ for 0 or 24 h before DMS treatment and genomic DNA isolation. Three MHC2TA locus-specific primers were used to amplify cleaved fragments from the upper strand of pIV. Ligation-mediated PCR was also performed on the lower strand, but no significant protections or enhancements were observed.
3. Results
3.1. Myeloma cells express IFN-γR1, and STAT1 is phosphorylated in response to IFN-γ
Evidence that myeloma cells are capable of responding to IFN-γ is conflicting. Previous reports demonstrate that MHC II expression is increased in IFN-γ-stimulated myeloma cells and that these cells present antigen after treatment with this cytokine (Yi et al., 1997; Beatty and Paterson, 2001). In contrast, a recent report shows microarray data which indicate that expression of RNA for a portion of the IFN-γ receptor, known as IFN-γR1 or the IFN-γR α chain, is decreased in PRDI-BF1/Blimp-1-expressing cells including myeloma cells (Shaffer et al., 2002). To examine the expression of IFN-γR1 transcripts, primers were designed and RT-PCR was performed on three human myeloma cell lines and Raji human B lymphocytes as a positive control (Fig. 1). To control for cDNA loading, RT-PCR was also performed using primers for GAPDH. This RT-PCR analysis demonstrates that IFN-γR1 RNA is present in these myeloma cells. In addition, PCR using serial dilutions of template did not show decreased IFN-γR1 RNA for the myeloma cells compared to Raji B cells, which serve as a positive control since they express IFN-γR1 but do not express PRDI-BF1/Blimp-1.
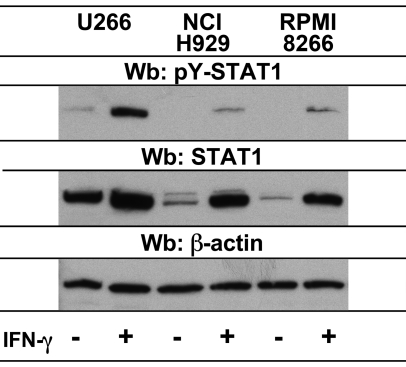
The model for cell signaling by IFN-γ includes the binding of IFN-γ to IFN-γR1, which prompts the association of the IFN-γR1 and IRN-γR2 subunits and activation of the receptor-associated Janus family of protein tyrosine kinases, (Jak)1 and Jak2 (Schroder et al., 2004). Jak kinases then phosphorylate IFN-γR1 generating a docking site for STAT1, which is then activated by tyrosine phosphorylation (Kotenko and Pestka, 2000). As an indicator of the activity of the IFN-γ signaling pathway in myeloma cells, Western blots were performed to detect tyrosine phosphorylation of STAT1 in response to IFN-γ (Fig. 2). In all cell lines tested, STAT1 was phosphorylated in response to IFN-γ treatment both at 24 hours and at earlier timepoints (data not shown). U266 cells expressed more STAT1 protein than the other cells, consistent with the higher amount of phosphorylated STAT1 observed for these cells. STAT1 protein levels were increased for all cells at the 24 hour timepoint, which is consistent with the ability of IFN-γ to regulate the human STAT1 gene (Wong et al., 2002). Taken together, these experiments demonstrate that myeloma cells are capable of responding to IFN-γ, since they express IFN-γR1 and the pathway leading to STAT1 activation is intact in these cells.
Fig. 2.
STAT1 is phosphorylated in myeloma cells in response to IFN-γ treatment. U266, NCI-H929, and RPMI 8266 human myeloma cells were either untreated or treated for 24 h with recombinant human IFN-γ. Whole cell extracts were analyzed by Western blot. Membranes were probed with an antibody to phospho-STAT1 (Tyr701) to detect the activation of STAT1 by phosphorylation in response to treatment. Blots were stripped and reprobed with anti-STAT1. This was repeated with anti-β-actin to analyze protein loading.
3.2. MHC II and CIITA type IV promoter-specific RNAs increase in myeloma cells in response to IFN-γtreatment
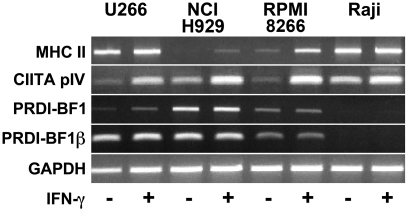
Although there have been several reports of increased MHC II expression in myeloma cells in response to IFN-γ, regulation of CIITA by IFN-γ in these cells was not previously studied. We and others have recently reported repression of CIITA pIV activity by PRDI-BF1/Blimp-1 and also by PRDI-BF1β, a truncated isoform of PRDI-BF1 that is co-expressed with the full-length form in myeloma cells (Gyory et al., 2003; Chen et al., 2007). Importantly, an increase in CIITA pIV activity in response to IFN-γ was still detected in Raji B cells when they were subjected to ectopic expression of PRDI-BF1/Blimp-1. This data suggested that IFN-γ might retain the ability to up regulate CIITA pIV expression in myeloma cells, which express PRDI-BF1/Blimp-1. Since each CIITA promoter controls the transcription of a unique first exon, transcripts representing the activity CIITA pIV have a unique sequence at their 5' termini and can be selectively amplified by RT-PCR. To detect increases in response to IFN-γ in myeloma cells, RT-PCR analyses using primers specific for MHC II and CIITA pIV were performed. PCR using either primers specific for PDRI-BF1 or PRDI-BF1β were included to verify that the myeloma cells in this study express PRDI-BF1/Blimp-1. The sizes for the PCR products are listed above. Increases in both MHC II and CIITA pIV-specific RNAs were detected in all of the cell lines examined (Fig. 3). Basal CIITA pIV expression was slightly lower for the myeloma cells compared to Raji B cells. We have previously demonstrated low constitutive levels of CIITA pIV expression in both Raji and primary human B cells (Piskurich et al., 2006). These results demonstrate for the first time that CIITA pIV is active in myeloma cells and that its activity can be increased by IFN-γ.
Fig. 3.
Increased levels of MHC II and CIITA pIV-specific RNAs are observed in myeloma cells in response to IFN-γ. Equal amounts of RNA from U266, NCI-H929, and RPMI 8266 human myeloma cells and Raji B cells, treated for 0 or 24 h with recombinant human IFN-γ, were subjected to RT-PCR using primers specific for HLADRα (MHC II), CIITA pIV, PRDI-BF1 or PRDI-BF1β, a truncated form of PRDI-BF1 co-expressed with full-length PRDI-BF1 in myeloma cells. PCR using primers for GAPDH was performed as an internal standard for cDNA loading. Mock preparations of cDNA where reverse transcriptase was omitted, and reaction mixtures that did not contain cDNA were also tested by PCR and did not give specific bands. This experiment has been repeated twice with similar results.
3.3. IFN-γincreases CIITA type IV promoter activity in myeloma cells
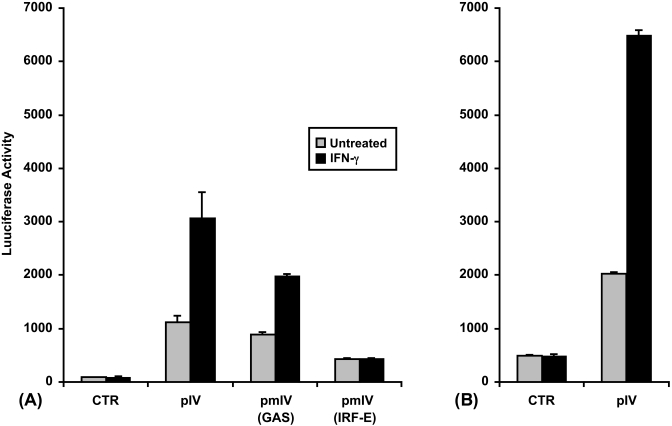
To functionally test this response, U266 and also RPMI 8266 myeloma cells were transfected by electroporation with reporter plasmids where the luciferase gene is under the control of CIITA pIV. The base vector for the CIITA promoter constructs, pGL2-Basic plasmid (Promega), was used as a negative control. A 2 to 3-fold increase in the activity of CIITA pIV was seen in these cells in response to treatment with IFN-γ (Figs. 4A and B). The mechanism described for CIITA pIV regulation by IFN-γ in other cell types includes the binding of STAT1 to a GAS sequence element, in conjunction with the binding of IRF-1 and IRF-2 to an IRF-E sequence motif (Muhlethaler-Mottet et al., 1998; Piskurich et al., 1999; Xi et al., 1999; Morris et al., 2002; Piskurich et al., 2006). Therefore, the activity of the unmutated CIITA pIV luciferase reporter construct was compared to the activity of CIITA pIV constructs with mutated GAS or IRF-E sites. The IRF-E mutation completely abolishes the induction by IFN-γ, while the GAS mutation only partially effects this activation (Fig. 4A). It is likely that both sites are required for full induction, a finding that is consistent with previous studies in other cell types by ourselves and others (Dong et al., 1999; Piskurich et al., 2006). The low basal activity for the IRF-E mutation was observed previously in B cells and fibroblasts, and therefore is not specific for myeloma cells (Piskurich et al., 2006; Chen et al., 2007). These data are consistent with the RT-PCR analysis which demonstrates that CIITA pIV is up regulated by IFN-γ in myeloma cells.
Fig. 4.
An increase in CIITA type IV promoter activity is observed in myeloma cells in response to IFN-γ. (A) Transient transfections of U266 human myeloma cells were performed by electroporation using 10 μg of a luciferase reporter construct containing unmutated CIITA pIV (pIV), pIV with a mutated STAT1-binding site (GAS) [pmIV(GAS)], pIV with a mutated IRF-1-binding site (IRF-E) [pmIV(IRF-E)] or 10 μg of the negative control, pGL2-Basic (CTR). (B) Transient transfections of RPMI 8266 human myeloma cells were performed by electroporation using 10 μg of a luciferase reporter construct containing unmutated CIITA pIV (pIV) or 10 μg of the negative control, pGL2-Basic (CTR). After a 6 h recovery period, half of the cultures were treated with IFN-γ for 24 h before harvesting and measurement of luciferase activity. Bars show the s.e.m., n = 3. At least three independent experiments have been performed with similar results.
3.4. IRF-1 and IRF-2 activate CIITA type IV promoter activity in myeloma cells
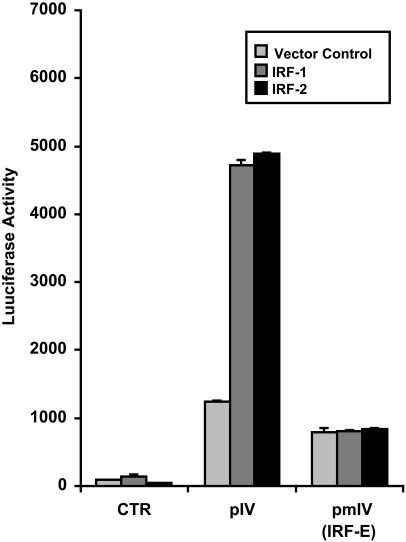
Previous reports showed that the CIITA pIV IRF-E site is bound and activated by IRF-1 and also IRF-2 (Muhlethaler-Mottet et al., 1998; Piskurich et al., 1999; Xi et al., 1999; Morris et al., 2002; Piskurich et al., 2006). To investigate the ability of IRF-1 and IRF-2 to activate CIITA pIV in myeloma cells, the effects of ectopic over expression of IRF-1 or IRF-2 on CIITA pIV activity were examined. U266 myeloma cells were co-transfected with either the unmutated CIITA pIV luciferase reporter construct or the construct with a mutated IRF-E site, and with a plasmid that constitutively expresses IRF-1. Ectopic expression of IRF-1 alone without IFN-γ treatment was sufficient to activate CIITA pIV activity, a finding similar to previous experiments performed in P19 carcinoma cells, embryonic fibroblasts and B cells (Piskurich et al., 1999; Xi et al., 1999; Xi and Blanck, 2003; Piskurich et al., 2006). IRF-1 over expression resulted in a 3 to 4-fold induction of activity for the unmutated CIITA pIV (Fig. 5). In contrast, the CIITA pIV construct with the mutated IRF-E site and the negative control plasmid were not induced. Ectopic expression of IRF-2 resulted in a similar activation of CIITA pIV activity for the unmutated CIITA pIV construct. While IRF-1 and IRF-2 can cooperatively activate CIITA pIV by co-occupying the IRF-E site, activation of this promoter by IRF-1 or IRF-2 alone has also been previously reported (Xi et al., 1999; Piskurich et al., 2006). These data support the finding that the IRF-E site is necessary for the induction of CIITA pIV in myeloma cells. Additionally, they demonstrate for the first time that IRF-1 and IRF-2 activate this promoter in myeloma cells.
Fig. 5.
IRF-1 and IRF-2 activate CIITA pIV activity in myeloma cells. Transient co-transfections of U266 human myeloma cells were performed by electroporation using 10 μg of IRF-1, IRF-2 or the empty expression plasmid vector, and 10 μg of a luciferase reporter construct containing either unmutated CIITA pIV (pIV) or CIITA pIV with a mutated IRF-1-binding site (IRF-E) [pmIV(IRF-E)] or 10 μg of the negative control, pGL2-Basic (CTR). After 24 h, cells were harvested and luciferase activity was measured. Bars show the s.e.m., n = 3. At least three independent experiments have been performed with similar results.
3.5. Expression of IRF-1 and IRF-2 in myeloma cells
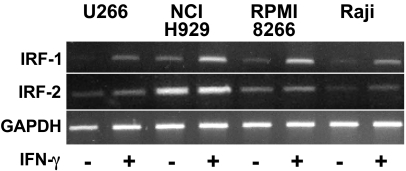
Expression of IRF-1 is highly regulated at the level of transcription by IFN-γ (Pine et al., 1990). While IRF-2 is constitutively expressed in many cell types, its expression can also be up regulated by IFN-γ through IRF-1, which activates its transcription (Cha and Deisseroth, 2004; Harada et al., 1994). To further examine the down stream activity of the IFN-γ signaling pathway and to verify the expression of IRF-1 and IRF-2 in myeloma cells, RT-PCR analyses to detect IRF-1 and IRF-2 transcripts were performed. The sizes for the PCR products are listed above. Increases in IRF-1 expression in response to IFN-γ were see in all cell lines examined (Fig. 6). In contrast, IRF-2 was expressed at similar levels both before and after treatment, although a small increase was detected in response to treatment. These results demonstrate that IFN-γ-stimulated myeloma cells express both IRF-1 and IRF-2, which is consistent with the ability of CIITA pIV activity to be up regulated by IFN-γ in these cells.
Fig. 6.
IRF-1 and IRF-2 expression in myeloma cells. Equal amounts of RNA from U266, NCI-H929, and RPMI 8266 human myeloma cells and Raji B cells, treated for 0 or 24 h with IFN-γ, was subjected to RT-PCR using primers specific for human IRF-1 or IRF-2. PCR using primers for GAPDH was performed as an internal standard for cDNA loading. Mock preparations of cDNA, where reverse transcriptase was omitted, and reaction mixtures that did not contain cDNA were also tested by PCR and did not give specific bands. This experiment has been repeated twice with similar results.
3.6. IRF-1 and IRF-2 bind to the CIITA type IV promoter in myeloma cells
To determine whether IRF-1 and/or IRF-2 bind to CIITA pIV in response to IFN-γ, electrophoretic mobility shift assays (EMSAs) using U266 myeloma cells were performed. One specific complex formed on the CIITA pIV site in response to IFN-γ treatment (Fig. 7, compare lanes 1 and 2). Preincubation of nuclear extracts from IFN-γ-treated cells with anti-IRF-1 antibody resulted in a supershifted band (lane 4), while preincubation with normal serum did not (lane 3). Preincubation of extracts from treated cells with an antibody to IRF-2 resulted in a partially supershifted band (lane 5). The EMSA banding pattern with anti-IRF-2 and the CIITA pIV probe was observed previously for other cell types and is not specific for myeloma cells (Xi et al., 1999). The upper bands in this analysis are non-specific, based on oligonucleotide competition assays (data not shown). We have previously demonstrated in EMSAs using extracts from untreated U266 cells that pretreatment with anti-IRF-1 or anti-IRF-2 does not result in supershifted bands (Chen et al., 2007). This analysis demonstrates that IRF-1 and IRF-2 in extracts from IFN-γ-treated myeloma cells bind CIITA pIV, and these findings are consistent with IRF-1 and IRF-2 binding and activating CIITA pIV in myeloma cells. This result lends further support to the functional promoter analyses, which demonstrated that the IRF-E site is necessary for up regulation of CIITA pIV activity by IFN-γ.
Fig. 7.
IRF-1 and IRF-2 bind to CIITA pIV in myeloma cells. EMSA analysis demonstrates that a specific protein complex is induced by IFN-γ treatment (lane 1 versus lane 2) (arrow). Nuclear extracts were from U266 human myeloma cells treated with IFN-γ for 0 (UNT) or 24 h (IFN-γ). The probe spans the CIITA pIV IRF-E. Incubation with anti-IRF-1 or anti-IRF-2 induces a supershifted or partially supershifted complex, respectively (see asterisks). This experiment has been repeated with similar results. Antibodies are indicated at the top. Abbreviations: Ab., antibody; CTR, normal control serum; N.E., nuclear extract.
3.7. Genomic footprint analysis detects protein-DNA interactions at the GAS, E box and IRF-E sites of the CIITA type IV promoter in myeloma cells
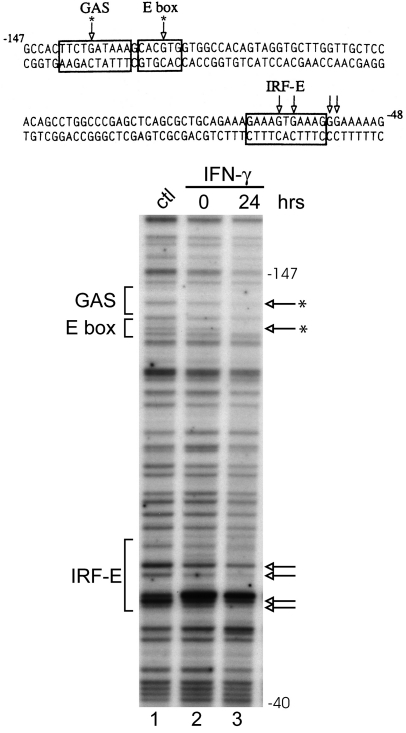
Since the IFN-γ-responsive region of CIITA pIV consists of a relatively small (approximately 100 bp) region of DNA located upstream of the pIV transcriptional start site (Muhlethaler-Mottet et al., 1997), genomic footprinting is the method of choice to identify protein-DNA interactions in vivo at physiologically relevant sequences. DMS-treated genomic DNA was obtained from U266 and NCI-H929 cells that were either uninduced or induced with IFN-γ for 24 h. Significant protections indicating protein-DNA contacts were observed in myeloma cells at the GAS, E box and IRF-E sites (Fig. 8). Similar results were obtained for both cell lines, and the results for NCIH929 cells are shown. Treatment with IFN-γ produced significant changes in the protein-DNA contacts at the GAS and E box sites, with the central guanine residue of each site becoming protected after treatment (see asterisks). In contrast, similar protein-DNA contacts were observed at the IRF-E site both before and after induction. These results are consistent with a change in protein binding in response to IFN-γ at CIITA pIV in myeloma cells and lend additional support to the finding that IFN-γ modulates CIITA pIV activity in these cells.
Fig. 8.
In vivo footprint of CIITA pIV in myeloma cells reveals protein-DNA contacts within the GAS, IRF-E and E box sites. The sequence of the promoter region is shown with boxes indicating the relevant cis-elements. Genomic footprints of the upper strand are shown in the lower panel. Lane 1 shows genomic DNA that was methylated in vitro to reveal the complete guanine ladder. Lanes 2 and 3 show the results of IFN-γ treatment using cells treated with DMS in culture. Open arrows indicate bases protected from modification. While a number of bases in the IRF-E site are protected both before and after treatment (indicated by arrows), central bases in the GAS and E box sites become protected after IFN-γ treatment (see asterisks). Similar results were seen for U266 cells (data not shown).
4. Discussion
Multiple myeloma is one of the most common hematological malignancies. Despite a variety of therapeutic approaches, less than 20% of multiple myeloma patients achieve long-term survival, relapses are common and cures are rare. IFN-α, growth inhibitory for many human multiple myeloma cell lines, has been used as a maintenance therapy although it is not effective in all cases (Schaar et al., 2005). In addition to IFN-α, IFN-γ also inhibits myeloma cell growth and could be useful as a treatment (Portier et al., 1993; Yi et al., 1997). IFN-γ inhibits osteoclasts and therefore could potentially slow the bone resorption that is seen in multiple myeloma patients (Takahashi et al., 1986). This cytokine also induces CD20 expression in myeloma cells, which could facilitate rituximab therapy (Treon et al., 2002). In addition to these effects, recent studies show that IFN-γ enhances MHC II and immunoproteosome expression in multiple myeloma cells, therefore it could also enhance the ability of the immune system to recognize this malignancy (Yi et al., 1997; Beatty and Paterson, 2001; Altun et al., 2005). While the potential of IFN-γ as a therapy for multiple myeloma is promising, the ability of the signaling pathway for IFN-γ to function in these cells remained largely unexplored. Recent data from microarrays indicated that even the expression of the IFN-γ-binding chain of the IFN-γ receptor called IFN-γR1, or alternatively the IFN-γR α chain or CD119, itself might be decreased at the level of transcription in these cells (Shaffer et al., 2002). Importantly, in our study no decrease in the expression of this receptor was observed relative to the expression in Raji B lymphocytes.
The binding of IFN-γ to its receptor results in the activation of receptor-associated Jaks and the phosphorylation of STAT1. Phosphorylated STAT1 forms homodimers, which migrate to the cell nucleus and activate transcription. Several inhibitors of IFN-γ signaling work downstream of cytokine binding. One of these inhibitors, SHP1, an SH2 domain-containing protein tyrosine phosphatase, down regulates IFN-γ signaling by counteracting Jak activity (Wu et al., 2003). It is noteworthy that for all of the myeloma cells lines in this study, STAT1 was phosphorylated in response to IFN-γ even after 24 h of cytokine treatment. This finding is consistent with a previous finding that the gene for SHP1 is often hypermethylated and inactive in multiple myeloma cells (Chim et al., 2004).
Because of its high constitutive activity in B lymphocytes, the principal target for studies of CIITA expression in this cell type has been the CIITA type III promoter. However, PRDI-BF1/Blimp-1 strongly suppresses CIITA pIII in both normal plasma cells and multiple myeloma cells (Piskurich et al., 2000; Ghosh et al., 2001). We have recently demonstrated CIITA type IV promoter activity that is up regulated by IFN-γ in B lymphocytes (Piskurich et al., 2006). We and others have also recently shown that this promoter, like pIII, is a candidate for repression by PRDI-BF1/Blimp-1 (Tooze et al., 2006; Chen et al., 2007). Since CIITA regulation is complex and cell-type specific, understanding its regulation in multiple myeloma requires the study of mechanisms of CIITA transcription as they relate specifically to these cells. While DNA methylation of both CIITA pIII and pIV limits CIITA expression in many human developmental and hematopoietic tumors, including myeloid and T cell malignancies (van den Elsen et al., 2003; Holling et al., 2004; Morimoto et al., 2004), myeloma cells are an exception in that the CIITA pIV is unmethylated in most cell lines that have been examined (Morimoto et al., 2004). In this report, we demonstrate for the first time that CIITA pIV is active in myeloma cells and that its activity is up regulated by IFN-γ even though these cells express PRDI-BF1/Blimp-1. The activity for CIITA pIV is approximately 2-fold lower than we have observed previously for B lymphocytes (Piskurich et al., 2006), which might be due to the expression of PRDI-BF1/Blimp-1 by these cells. IL-6 plays a role in the progression of multiple myeloma by regulating the proliferation and survival of multiple myeloma cells (Klein et al., 1995). In contrast to a study in which macrophages secreting IL-6 showed decreased CIITA expression (Nagabhushanam et al., 2003), no decrease in CIITA expression was seen for U266 cells which secrete IL-6 compared to RPMI 8266 and NCI-H929 cells which do not.
Consistent with the ability of IFN-γ to increase the activity of CIITA pIV in myeloma cells, in vivo footprinting analysis revealed a protein-DNA contact at the STAT1-binding GAS site and the enhancement of a protein-DNA contact at the E box site in response to treatment with this cytokine. These contacts closely resemble those we have previously observed in response to IFN-γ treatment in a similar study of this region performed in Raji B lymphocytes (Piskurich et al., 2006). This footprinting assay also detected protein-DNA interactions in myeloma cells at the CIITA pIV IRF-E site that are similar before and after treatment with IFN-γ. We and others have recently demonstrated that the CIITA pIV IRF-E site is a binding site for the PRD-IBF1/Blimp-1 repressor (Tooze et al., 2006; Chen et al., 2007). It is possible that PRD-IBF1/Blimp-1 binds the IRF-E site in untreated myeloma cells and that this binding is replaced after treatment with IFN-γ by IRF-1 and IRF-2, which make similar contacts at this site and activate CIITA pIV expression. The sequence of the CIITA pIV IRF-E site is a good consensus site for either PRDI-BF1/Blimp-1 or IRF-1 and IRF-2 binding (Fig. 8). It has been suggested that PRDI-BF1/Blimp-1 could compete in vivo with IRF proteins for binding (Kuo and Calame, 2004). Such a competition could be reflected by the lower fold activation that we typically observe in CIITA pIV promoter analyses performed in myeloma cells versus Raji B cells. However, our experiments indicate that competition for this site by PRDI-BF1/Blimp-1 does not completely block activation of this promoter by IFN-γ in myeloma cells.
In summary, we have investigated the expression of the CIITA type IV promoter in human myeloma cells and its regulation by IFN-γ. Although our previous findings show that this promoter is a candidate for repression by PRDI-BF1/Blimp-1, we demonstrate here for the first time that CIITA pIV is active and exhibits increased activity in response to IFN-γ in multiple myeloma cells. It is therefore possible that cytokines, like IFN-γ, might be useful therapeutically to increase the expression of CIITA in some B cell malignancies, like multiple myeloma, to activate the ability of these tumor cells to function as antigen presenting cells and to be recognized and destroyed by the immune system.
Acknowledgements
This investigation was supported by research grants from the National Cancer Institute (RO1CA102203 and RO1CA114504) and the MEDCEN Community Health Foundation of Central Georgia.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altun M, Galardy PJ, Shringarpure R, Hideshima T, LeBlanc R, Anderson KC, Ploegh HL, Kessler BM. Effects of PS-341 on the activity and composition of proteasomes in multiple myeloma cells. Cancer Res. 2005;65:7896–7901. doi: 10.1158/0008-5472.CAN-05-0506. [DOI] [PubMed] [Google Scholar]
- Amiot L, Onno M, Lamy T, Dauriac C, Le Prise PY, Fauchet R, Drenou B. Loss of HLA molecules in B lymphomas is associated with an aggressive clinical course. Br. J. Haematol. 1998;100:655–663. doi: 10.1046/j.1365-2141.1998.00631.x. [DOI] [PubMed] [Google Scholar]
- Beatty GL, Paterson Y. Regulation of tumor growth by IFN-gamma in cancer immunotherapy. Immunol. Res. 2001;24:201–210. doi: 10.1385/IR:24:2:201. [DOI] [PubMed] [Google Scholar]
- Cha Y, Deisseroth AB. Human interferon regulatory factor 2 gene: intron-exon organization and functional analysis of 5′-flanking region. J. Biol. Chem. 2004;269:5279–5287. [PubMed] [Google Scholar]
- Chen H, Gilbert CA, Hudson JA, Bolick SC, Wright KL, Piskurich JF. Positive regulatory domain I-binding factor 1 mediates repression of the MHC class II transactivator (CIITA) type IV promoter. Mol. Immunol. 2007;44:1461–1470. doi: 10.1016/j.molimm.2006.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chim CS, Fung TK, Cheung WC, Liang R, Kwong YL. SOCS1 and SHP1 hypermethylation in multiple myeloma: implications for epigenetic activation of the Jak/STAT pathway. Blood. 2004;103:4630–4635. doi: 10.1182/blood-2003-06-2007. [DOI] [PubMed] [Google Scholar]
- Cresswell P. Assembly, transport, and function of MHC class II molecules. Annu. Rev. Immunol. 1994;12:259–293. doi: 10.1146/annurev.iy.12.040194.001355. [DOI] [PubMed] [Google Scholar]
- Dong Y, Rohn WM, Benveniste EN. IFN-gamma regulation of the type IV class II transactivator promoter in astrocytes. J. Immunol. 1999;162:4731–4739. [PubMed] [Google Scholar]
- Fuji H, Iribe H. Clonal variation in tumorigenicity of L1210 lymphoma cells: nontumorigenic variants with an enhanced expression of tumor-associated antigen and Ia antigens. Cancer Res. 1986;46:5541–5547. [PubMed] [Google Scholar]
- Galea HR, Denizot Y, Cogne M. Light chain myeloma plasma cells induce a strong cell-mediated immune response mainly directed against the monoclonal light chain determinants in a murine experimental model. Cancer Immunol. Immunother. 2002;51:229–234. doi: 10.1007/s00262-002-0274-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh N, Gyory I, Wright G, Wood J, Wright KL. Positive regulatory domain I binding factor 1 silences class II transactivator expression in multiple myeloma cells. J. Biol. Chem. 2001;276:15264–15268. doi: 10.1074/jbc.M100862200. [DOI] [PubMed] [Google Scholar]
- Gyory I, Fejer G, Ghosh N, Seto E, Wright KL. Identification of a functionally impaired positive regulatory domain I binding factor 1 transcription repressor in myeloma cell lines. J. Immunol. 2003;170:3125–3133. doi: 10.4049/jimmunol.170.6.3125. [DOI] [PubMed] [Google Scholar]
- Harada H, Takahashi E, Itoh S, Harada K, Hori TA, Taniguchi T. Structure and regulation of the human interferon regulatory factor 1 (IRF-1) and IRF-2 genes: implications for a gene network in the interferon system. Mol. Cell Biol. 1994;14:1500–1509. doi: 10.1128/mcb.14.2.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harton JA, Ting JP. Class II transactivator: mastering the art of major histocompatibility complex expression. Mol. Cell Biol. 2000;20:6185–6194. doi: 10.1128/mcb.20.17.6185-6194.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holling TM, Schooten E, Langerak AW, van den Elsen PJ. Regulation of MHC class II expression in human T-cell malignancies. Blood. 2004;103:1438–1444. doi: 10.1182/blood-2003-05-1491. [DOI] [PubMed] [Google Scholar]
- Klein B, Zhang XG, Lu ZY, Bataille R. Interleukin-6 in human multiple myeloma. Blood. 1995;85:863–872. [PubMed] [Google Scholar]
- Kotenko SV, Pestka S. Jak-Stat signal transduction pathway through the eyes of cytokine class II receptor complexes. Oncogene. 2000;19:2557–2565. doi: 10.1038/sj.onc.1203524. [DOI] [PubMed] [Google Scholar]
- Kuo TC, Calame KL. B lymphocyte-induced maturation protein (Blimp)-1, IFN regulatory factor (IRF)-1, and IRF-2 can bind to the same regulatory sites. J. Immunol. 2004;173:5556–5563. doi: 10.4049/jimmunol.173.9.5556. [DOI] [PubMed] [Google Scholar]
- Landmann S, Muhlethaler-Mottet A, Bernasconi L, Suter T, Waldburger JM, Masternak K, Arrighi JF, Hauser C, Fontana A, Reith W. Maturation of dendritic cells is accompanied by rapid transcriptional silencing of class II transactivator (CIITA) expression. J. Exp. Med. 2001;194:379–391. doi: 10.1084/jem.194.4.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibundgut-Landmann S, Waldburger JM, Krawczyk M, Otten LA, Suter T, Fontana A, Acha-Orbea H, Reith W. Mini-review: Specificity and expression of CIITA, the master regulator of MHC class II genes. Eur. J. Immunol. 2004;34:1513–1525. doi: 10.1002/eji.200424964. [DOI] [PubMed] [Google Scholar]
- Lin Y, Wong K, Calame K. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science. 1997;276:596–599. doi: 10.1126/science.276.5312.596. [DOI] [PubMed] [Google Scholar]
- Morimoto Y, Toyota M, Satoh A, Murai M, Mita H, Suzuki H, Takamura Y, Ikeda H, Ishida T, Sato N, Tokino T, Imai K. Inactivation of class II transactivator by DNA methylation and histone deacetylation associated with absence of HLA-DR induction by interferon-gamma in haematopoietic tumour cells. Br. J. Cancer. 2004;90:844–852. doi: 10.1038/sj.bjc.6601602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AC, Beresford GW, Mooney MR, Boss JM. Kinetics of a Gamma Interferon Response: Expression and Assembly of CIITA Promoter IV and Inhibition by Methylation. Mol. Cell Biol. 2002;22:4781–4791. doi: 10.1128/MCB.22.13.4781-4791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlethaler-Mottet A, Di BW, Otten LA, Mach B. Activation of the MHC class II transactivator CIITA by interferon-gamma requires cooperative interaction between Stat1 and USF-1. Immunity. 1998;8:157–166. doi: 10.1016/s1074-7613(00)80468-9. [DOI] [PubMed] [Google Scholar]
- Muhlethaler-Mottet A, Otten LA, Steimle V, Mach B. Expression of MHC class II molecules in different cellular and functional compartments is controlled by differential usage of multiple promoters of the transactivator CIITA. EMBO J. 1997;16:2851–2860. doi: 10.1093/emboj/16.10.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagabhushanam V, Solache A, Ting LM, Escaron CJ, Zhang JY, Ernst JD. Innate inhibition of adaptive immunity: Mycobacterium tuberculosis-induced IL-6 inhibits macrophage responses to IFN-gamma. J. Immunol. 2003;171:4750–4757. doi: 10.4049/jimmunol.171.9.4750. [DOI] [PubMed] [Google Scholar]
- Nagy M, Chapuis B, Matthes T. Expression of transcription factors Pu.1, Spi-B, Blimp-1, BSAP and oct-2 in normal human plasma cells and in multiple myeloma cells. Br. J. Haematol. 2002;116:429–435. doi: 10.1046/j.1365-2141.2002.03271.x. [DOI] [PubMed] [Google Scholar]
- Pai RK, Askew D, Boom WH, Harding CV. Regulation of class II MHC expression in APCs: roles of types I, III, and IV class II transactivator. J. Immunol. 2002;169:1326–1333. doi: 10.4049/jimmunol.169.3.1326. [DOI] [PubMed] [Google Scholar]
- Pine R, Decker T, Kessler DS, Levy DE, Darnell JE., Jr. Purification and cloning of interferon-stimulated gene factor 2 (ISGF2): ISGF2 (IRF-1) can bind to the promoters of both beta interferon- and interferon-stimulated genes but is not a primary transcriptional activator of either. Mol. Cell Biol. 1990;10:2448–2457. doi: 10.1128/mcb.10.6.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurich JF, Gilbert CA, Ashley BD, Zhao M, Chen H, Wu J, Bolick SC, Wright KL. Expression of the MHC class II transactivator (CIITA) type IV promoter in B lymphocytes and regulation by IFN-gamma. Mol. Immunol. 2006;43:519–528. doi: 10.1016/j.molimm.2005.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurich JF, Lin KI, Lin Y, Wang Y, Ting JP, Calame K. BLIMP-I mediates extinction of major histocompatibility class II transactivator expression in plasma cells. Nat. Immunol. 2000;1:526–532. doi: 10.1038/82788. [DOI] [PubMed] [Google Scholar]
- Piskurich JF, Linhoff MW, Wang Y, Ting JP. Two distinct gamma interferon-inducible promoters of the major histocompatibility complex class II transactivator gene are differentially regulated by STAT1, interferon regulatory factor 1, and transforming growth factor beta. Mol. Cell Biol. 1999;19:431–440. doi: 10.1128/mcb.19.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurich JF, Wang Y, Linhoff MW, White LC, Ting JP. Identification of distinct regions of 5′ flanking DNA that mediate constitutive, IFN-gamma, STAT1, and TGF-beta-regulated expression of the class II transactivator gene. J. Immunol. 1998;160:233–240. [PubMed] [Google Scholar]
- Portier M, Zhang XG, Caron E, Lu ZY, Bataille R, Klein B. gamma-Interferon in multiple myeloma: inhibition of interleukin-6 (IL-6)-dependent myeloma cell growth and downregulation of IL-6-receptor expression in vitro. Blood. 1993;81:3076–3082. [PubMed] [Google Scholar]
- Schaar CG, Kluin-Nelemans HC, Te Marvelde C, le Cessie S, Breed WP, Fibbe WE, van Deijk WA, Fickers MM, Roozendaal KJ, Wijermans PW. Interferon-alpha as maintenance therapy in patients with multiple myeloma. Ann. Oncol. 2005;16:634–639. doi: 10.1093/annonc/mdi125. [DOI] [PubMed] [Google Scholar]
- Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J. Leukoc. Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, Giltnane JM, Yang L, Zhao H, Calame K, Staudt LM. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- Shapiro-Shelef M, Calame K. Regulation of plasma-cell development. Nat. Rev. Immunol. 2005;5:230–242. doi: 10.1038/nri1572. [DOI] [PubMed] [Google Scholar]
- Shi B, Vinyals A, Alia P, Broceno C, Chen F, Adrover M, Gelpi C, Price JE, Fabra A. Differential expression of MHC class II molecules in highly metastatic breast cancer cells is mediated by the regulation of the CIITA transcription. Implication of CIITA in tumor and metastasis development. Int. J. Biochem. Cell Biol. 2006;38:544–562. doi: 10.1016/j.biocel.2005.07.012. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Mundy GR, Roodman GD. Recombinant human interferon-gamma inhibits formation of human osteoclast-like cells. J. Immunol. 1986;137:3544–3549. [PubMed] [Google Scholar]
- Ting JP, Trowsdale J. Genetic control of MHC class II expression. Cell. 2002;109:S21–S33. doi: 10.1016/s0092-8674(02)00696-7. [DOI] [PubMed] [Google Scholar]
- Tooze RM, Stephenson S, Doody GM. Repression of IFN-gamma induction of class II transactivator: a role for PRDM1/Blimp-1 in regulation of cytokine signaling. J. Immunol. 2006;177:4584–4593. doi: 10.4049/jimmunol.177.7.4584. [DOI] [PubMed] [Google Scholar]
- Treon SP, Pilarski LM, Belch AR, Kelliher A, Preffer FI, Shima Y, Mitsiades CS, Mitsiades NS, Szczepek AJ, Ellman L, Harmon D, Grossbard ML, Anderson KC. CD20-directed serotherapy in patients with multiple myeloma: biologic considerations and therapeutic applications. J. Immunother. 2002;25:72–81. doi: 10.1097/00002371-200201000-00008. [DOI] [PubMed] [Google Scholar]
- Turner CA, Jr., Mack DH, Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77:297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- van den Elsen PJ, Holling TM, van der Stoep N, Boss JM. DNA methylation and expression of major histocompatibility complex class I and class II transactivator genes in human developmental tumor cells and in T cell malignancies. Clin. Immunol. 2003;109:46–52. doi: 10.1016/s1521-6616(03)00200-6. [DOI] [PubMed] [Google Scholar]
- Wen YJ, Barlogie B, Yi Q. Idiotype-specific cytotoxic T lymphocytes in multiple myeloma: evidence for their capacity to lyse autologous primary tumor cells. Blood. 2001;97:1750–1755. doi: 10.1182/blood.v97.6.1750. [DOI] [PubMed] [Google Scholar]
- Wong LH, Sim H, Chatterjee-Kishore M, Hatzinisiriou I, Devenish RJ, Stark G, Ralph SJ. Isolation and characterization of a human STAT1 gene regulatory element. Inducibility by interferon (IFN) types I and II and role of IFN regulatory factor-1. J. Biol. Chem. 2002;277:19408–19417. doi: 10.1074/jbc.M111302200. [DOI] [PubMed] [Google Scholar]
- Wu C, Sun M, Liu L, Zhou GW. The function of the protein tyrosine phosphatase SHP-1 in cancer. Gene. 2003;306:1–12. doi: 10.1016/s0378-1119(03)00400-1. [DOI] [PubMed] [Google Scholar]
- Xi H, Blanck G. The IRF-2 DNA binding domain facilitates the activation of the class II transactivator (CIITA) type IV promoter by IRF-1. Mol. Immunol. 2003;39:677–684. doi: 10.1016/s0161-5890(02)00214-6. [DOI] [PubMed] [Google Scholar]
- Xi H, Eason DD, Ghosh D, Dovhey S, Wright KL, Blanck G. Co-occupancy of the interferon regulatory element of the class II transactivator (CIITA) type IV promoter by interferon regulatory factors 1 and 2. Oncogene. 1999;18:5889–5903. doi: 10.1038/sj.onc.1202969. [DOI] [PubMed] [Google Scholar]
- Yi Q, Dabadghao S, Osterborg A, Bergenbrant S, Holm G. Myeloma bone marrow plasma cells: evidence for their capacity as antigen-presenting cells. Blood. 1997;90:1960–1967. [PubMed] [Google Scholar]