Abstract
Klebsiella pneumoniae is a gram negative bacterium of significant clinical importance. This study examines the differential pulmonary host anti-bacterial responses towards two clinical isolates of K. pneumoniae. Intratracheal inoculation with 7×104 CFU of strain 43816 induced 100% mortality in C57BL/6J mice within 5 days post infection, whereas infection with 5×105 CFU of strain IA565 resulted in 100% survival. Infection with strain 43816 resulted in significant pulmonary and peripheral blood bacterial burden and induction of the chemokines MIP-2, KC and MCP-1 by 24 hours post infection. In contrast, IA565-infected mice displayed basal chemokine levels and no detectable bacteria by 24 hours post inoculation were isolated from lungs or peripheral blood. These data indicate an apparent lack of pathogenicity of strain IA565. Since little is known about Klebsiella-specific virulence genes, we have utilized PCR-based genomic DNA and cDNA suppressive subtractive hybridization and identified nine DNA sequences unique to a pathogenic strain of K. pneumoniae 43816. These sequences were highly homologous to enteric bacterial genes regulating iron uptake, fimbrial-mediated adhesion, energy production and conversion, transcriptional regulation, signal transduction, restriction endonuclease activity, and membrane transport.
Keywords: Klebsiella pneumoniae, macrophage phagocytosis, subtractive hybridization, virulence genes
1. Introduction
Klebsiella pneumoniae is a gram-negative opportunistic bacterial pathogen primarily infecting immunocompromised individuals who are hospitalized and suffer from severe underlying diseases[1,2]. K. pneumoniae can cause a range of infections, from mild urinary tract infections to severe septicemia and bacterial pneumonia with mortality rates that may exceed 50% [1,3]. Pulmonary K. pneumoniae infections are often complicated by multilobular involvement, formation of lung abscesses, and dissemination of bacteria from within the pulmonary airspace into the bloodstream [4,5]; all of which are accompanied by the characteristic rapidly progressive clinical course. The recent emergence of antibiotic resistant K. pneumoniae strains [5-7] emphasizes the importance of determining the mechanisms in which K. pneumoniae interacts with the host, bacterial factors that contribute to the disease course, and the role the immune host defense plays in the eventual outcome of the bacterial infection.
Animal models have proven useful in determining host:pathogen interactions during K. pneumoniae infection. Utilizing a murine model of K. pneumoniae pneumonia, IFN-γ, IL-10, IL-12, and TNFα have all been shown to play an important role in mediating lung antibacterial host responses during K. pneumoniae infection [8-12]. The majority of these studies utilized a single clinical isolate of K. pneumoniae, 43816, prompting the question of whether or not measurable immune responses can be observed employing different K. pneumoniae strains. In this study, K. pneumoniae clinical isolates 43816 and IA565 were used in a murine model of bacterial pneumonia. Our results show that strain IA565 is rapidly cleared from the lungs and failed to induce any animal mortality, in sharp contrast to strain 43816.
While significant advances have been made in the understanding of host responses following infection with K. pneumoniae, very little is known about bacterial virulence factors that can contribute to in vivo pathogenicity. However, both 43816 and IA565 strains express the prototypic virulence factors associated with K. pneumoniae pathogenicity; being capsule, lipopolysaccharide, and Type 1 and 3 fimbriae. Of these three, perhaps capsule is the best studied virulence factor. There are over 70 serological types of capsular antigens associated with Klebsiella pneumoniae. Strain 43816 is classified as having a K2 serotype and strain IA565 has a K15 capsular serotype. K. pneumoniae strains displaying in vivo pathogenicity in murine studies most often express capsular serotypes K1 and K2, however this is not an absolute correlation [13]. Capsule switch mutants have been constructed using strains expressing K2 and K21a that have indicated that the genetic background of virulent strains, independent of the capsule serotype, confers significant in vivo murine pathogenicity [14,15]. These studies concluded that pathogenesis of K. pneumoniae was multifactorial and that capsule can only partially account for in vivo murine virulence.
Since our two strains of K. pneumoniae differ significantly in their ability to cause disease using a murine model of pneumonia, this raised the question of whether or not there are any other Klebsiella-specific genes responsible for its virulence uniquely contained within the genome of strain 43816. We utilized PCR-based suppressive subtractive hybridization to identify putative virulence genes in K. pneumoniae and have identified 9 DNA sequences unique to our pathogenic K. pneumoniae strain 43816.
2. Results
2.1 Increased mortality in 43816 infected mice following pulmonary K. pneumoniae infection
To determine the in vivo pathogenicity of strains IA565 and 43816, mice were infected and overall survival was determined over a 7 day course of infection. Strain 43816 has been previously reported to be a virulent strain of K. pneumoniae [16,17]. Intratracheal inoculation of 7 × 104 CFU of strain 43816 into C57BL/6J mice induced mortality within 2-3 days post infection and resulted in 100% mortality by day 5 post infection (Figure 1). Inoculation of animals with this same dose of strain IA565 resulted in 100% survival (data not shown). Interestingly, a higher inoculum dose of 5 × 105 CFU of strain IA565 into mice also resulted in 100% survival (Figure 1, P < 0.01), indicating an apparent lack of pathogenicity of this strain of K. pneumoniae in a murine model of bacterial pneumonia.
Figure 1.
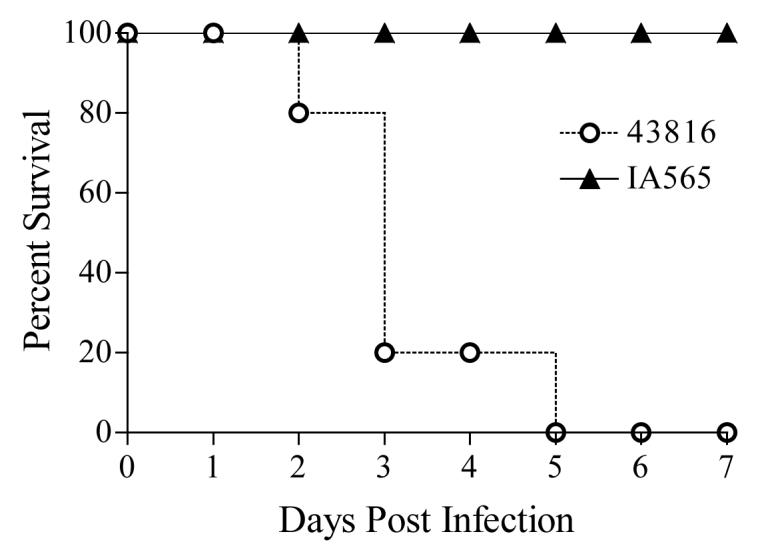
Decreased Mortality in IA565 infected mice following K. pneumoniae infection. Mice were intratracheally inoculated with 7×104 CFU of strain 43816 or 5×105 CFU of strain IA565 and overall survival was determined for 7 days post infection. P < 0.01 for comparison with IA565 infected mice. Survival curves were generated from two independent experiments for strain 43816, with 9 animals total. For strain IA565, survival curves were generated with 5 mice per group.
2.2 Rapid clearance of IA565 from the lung following intratracheal K. pneumoniae infection
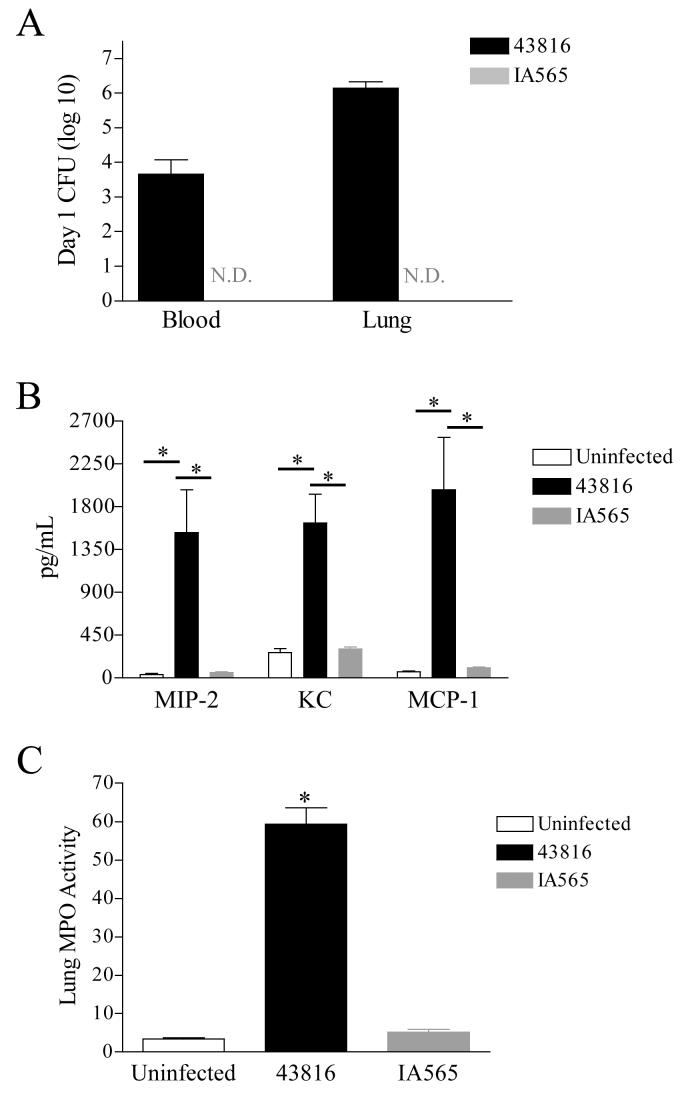
To determine whether the observed lack of mortality correlated with the rapid clearance of strain IA565 bacteria from the lungs, C57BL/6J mice were intratracheally inoculated with 3×105 CFU of IA565 and 43816 and analyzed 24 hours post-infection. Mice infected with strain 43816 displayed high bacterial load in both the lungs and blood (Figure 2A). In contrast, bacterial CFU in the lungs and blood from the IA565 infected mice were below the limit of detection by 24 hours post infection (Figure 2A).
Figure 2.
Bacterial burden, chemokine induction, and MPO activity following infection with strains IA565 or 43816. Mice were infected with 5×105 CFU of strain IA565 or 43816 and analyzed 24 hours post infection. (A) Bacterial burdens from the blood and lung of infected animals. Bacterial numbers for the lung are for the entire tissue, while bacterial numbers for the blood are per milliliter of blood. (B) Chemokine production was assessed from total lung homogenates by ELISA. (C) Lungs were also assessed for MPO activity. Significant activity was observed in the lungs of 43816 infected mice. Data presented as mean ± SEM. Data were generated from two independent experiments with 4-5 animals per group. *, P < 0.001. N.D., none detected.
2.3. Induction of antibacterial host responses in strain 43816 or IA565-infected mice
Both neutrophils and monocyte/macrophages have been shown to be important during pulmonary bacterial infections [18-21]. To determine if the rapid clearance of strain IA565 resulted from enhanced neutrophil and/or macrophage recruitment into the pulmonary airspace, we analyzed lung homogenates for the production of chemokines responsible for recruiting these cell types. Mice were intratracheally infected with 3×105 CFU of each strain and lungs were harvested 24 hours post infection. The neutrophil recruiting chemokines MIP-2 and KC were significantly increased in the lungs of strain 43816 infected mice. MCP-1, capable of recruiting circulating monocytes was also significantly increased in 43816 infected animals (Figure 2B). Correlating with increased MIP-2 and KC, myeloperoxidase (MPO) activity, an indirect measurement of neutrophil numbers, in the lungs of 43816 infected mice was significantly elevated compared to uninfected control animals (Figure 2C, P < 0.001). In contrast, lung homogenates from IA565 infected mice contained baseline levels of MIP-2, KC and MCP-1 and MPO levels were essentially equal to those seen in uninfected controls (Figure 2C).
Since neutrophils have been shown to be important for bacterial pulmonary clearance during an infection, it was surprising that IA565-infected mice showed no induction of neutrophil chemokines MIP-2 and KC or significant levels of MPO activity. This suggests that clearance of strain IA565 from mice is neutrophil independent. To determine the importance of neurophils, mice were injected with the neutrophil depleting anti-Ly-6G monoclonal antibody prior to infection with strain IA565 [22]. Neutrophil depleted mice intratracheally inoculated with 105 or 106 CFU of strain IA565 displayed 100% survival (data not shown), indicating that clearance of strain IA565 is neutrophil independent.
2.4 In vitro and in vivo macrophage phagocytosis of strain 43816 and IA565
Data thus far indicate that strain IA565 is rapidly cleared from the lungs in the absence of recruited circulating neutrophils. To access the ability of strain IA565 and 43816 to be phagocytosed by macrophages, and in vitro phagocytosis assay was performed on freshly isolated, non-elicited peritoneal macrophages (PMϕs) from C57BL/6J mice. In two independent experiments, PMϕs were incubated with opsonized, FITC-labeled IA565 or 43816 bacteria for 30 minutes with a bacterium to macrophage ratio of 100:1. Values were expressed as relative fluorescence units calculated based on fluorescent standards. In one experiment, phagocytosis of strain IA565 and 43816 was essentially equivalent while in the other experiment phagocytosis of strain IA565 was slightly higher than strain 43816 (Figure 3A, P = 0.04). Because of the inconsistencies in these in vitro experiments and the use of peritoneal macrophages, an in vivo pulmonary phagocytosis assay was performed to address the susceptibility or resistance to alveolar macrophage (AMϕ) phagocytosis of both strains. Bronchoalveolar lavages (BALs) were performed two hours post intratracheal inoculation with 1×106 CFU of strain 43816 or IA565 and cytospin slides prepared. Interestingly, the calculated phagocytic index (PI) for strain IA565 was 2.5 fold higher than that for strain 43816 (Figure 3B). Figure 3C and 3D shows a representative image of strain 43816 and IA565, respectively, phagocytosed by AMϕs. These data suggest that resistance to phagocytosis likely contributes to the virulence of strain 43816.
Figure 3.
In vivoand in vitro phagocytosis of strain 43816 and IA565. (A) Non-elicited peritoneal macrophages from C57BL/6J mice were isolated and cultured 100:1 with opsonized FITC-labeled bacteria, * P = 0.04. (B) BAL cells was isolated from mice 2 hours post inoculation with either strain IA565 or 43816. Cytospins of alveolar leukocytes were prepared and 200 cells were counted and any bacteria seen in the cells were recorded. Phagocytic indices are plotted as shown. ± P < 0.001. Representative images of the cytospins are shown for 43816 (C) and IA565 (D) infected mice at 400X. Arrows point to cells containing bacteria.
2.5 Identification of strain 43816 unique DNA sequences absent in strain IA565
Collectively, data thus far indicate that the two strains of K. pneumoniaediffer significantly in their in vivo pathogenicity, even though both strains express prototypical virulence-associated factors. This suggests that strain 43816 contains virulence genes that confer pathogenicity other than the genes responsible for expression of capsular polysaccharide, lipopolysaccharide and fimbriae.
To identify the presence of other putative virulence factors in the pathogenic strain of K. pneumoniae 43816, we employed a PCR-based suppressive subtractive hybridization (SSH) technique to enrich for DNA sequences present in the pathogenic 43816 strain but not the nonpathogenic IA565 strain. This technique is designed to identify genomic differences between two strains of bacteria, most often differences between virulent and non-virulent strains of the same bacteria. Additionally, this approach can also be modified to identify differences in gene expression between two strains by synthesizing cDNA from total RNA [23]. Clontech's PCR-SelectTM Bacterial Genome Subtraction and PCR-SelectTM cDNA subtraction kit protocols were performed on both genomic DNA and total cDNA from strains IA565 and 43816. DNA fragments obtained from both subtractions were then used in a PCR-SelectTM Differential Screening protocol (Clontech, La Jolla, CA) to further eliminate DNA fragments common to both strains. Eighty-three out of 381 sequences from the genomic and cDNA SSH were selected for further analysis because of their homologies to sequences in other enteric bacteria. PCR primers were generated and then K. pneumoniae strain 43816 and IA565 genomic DNA and total cDNA was subjected to PCR to verify if the selected 83 sequences were unique to strain 43816. Of the 83 sequences analyzed, 5 sequences obtained from the cDNA SSH and 4 sequences obtained from the genomic DNA SSH were found to be exclusively present in strain 43816. The 9 sequences were analyzed using the National Center for Biotechnology Information (NCBI) BLAST programs (Table 1). These sequences were highly homologous to enteric bacterial genes regulating iron uptake (iroN), fimbrial-mediated adhesion (sthB, fimD), energy production and conversion (dcoB), transcriptional regulation (Orf350), signal transduction (kvgS), restriction endonuclease activity (Mrr), and membrane transport (ybiT). The role each of these gene sequences play in virulence is currently being investigated.
Table 1.
K. pneumoniae strain 43816 genes identified in this study.
| Accession no. | Homologous gene (accession no.) | % Identity | Predicted Protein |
|---|---|---|---|
| DQ211084a | E. coli iroN (NP_753164) | 89c | siderophore receptor |
| DQ211085b | S. typhimurium iroN (NP_461704) | 78c | siderophore receptor |
| DQ211086b | S. typhimurium sthB (NP_463449) | 59d | putative fimbrial usher protein |
| DQ211087b | E. coli fimD (NP_418737) | 69d | outer membrane protein, export and assembly of type 1 fimbriae |
| DQ211088b | K. pneumoniae KvgS (CAB61240) | 96d | sensor protein KVGS precursor; virulence protein S |
| DQ211089a | E. coli ybiT (NP_752836) | 87c | hypothetical ABC transporter ATP binding protein |
| DQ211090a | S. typhimurium dcoB (NP_459747) | 90c | oxaloacetate decarboxylase beta chain |
| DQ211091a | P. putida Orf350 (NP_863090) | 58d | putative LysR-type transcriptional regulator |
| DQ211092a | S. typhimurium Mrr (NP_463349) | 96a | putative restriction endonuclease |
obtained from cDNA SSH
obtained from genomic DNA SSH
nucleotide sequence homology
protein sequence homology
3. Discussion
The use of K. pneumoniae clinical isolates in various animal models has provided valuable information regarding strain dependence in producing respiratory tract infections [24-26]. In this study, clinical isolates IA565 and 43816 were used to establish an acute pulmonary infection in C57BL/6J mice that closely resembles that of human bacterial pneumonia. K. pneumoniae 43816 induced 100% mortality at doses of 7 × 104 CFU whereas inoculation of 5 × 105 CFU of IA565 failed to induce any mortality (Figure 1), indicating an apparent lack of pathogenicity. This lack of pathogenicity is likely due to the rapid disappearance of bacteria from the lungs of inoculated mice that occurs with minimal induction of innate host responses (Figure 2).
Neutrophils have been shown to be essential for host defense during the clearance of pulmonary infections [10,27,28] including infection with strain 43816. However, the lack of significant MIP-2 and KC production and subsequent MPO activity in the lungs of IA565 infected mice suggests that neutrophil recruitment is not necessary for the clearance of IA565. Indeed, when C57BL/6J mice were depleted of circulating neutrophils, these neutropenic mice displayed 100% survival following inoculation with strain IA565 regardless of inoculation dose (data not shown).
Alveolar macrophages are the primary phagocytes of the innate immune system constituting the first line of defense against pulmonary infectious agents [29]. The observation that the rapid clearance of strain IA565 from the lungs is independent of neutrophil recruitment further supports an important role of resident AMϕs. To determine the relative susceptibility or resistance to phagocytosis, strain IA565 and 43816 were accessed for their ability to be phagocytosed by both PMϕs in vitro and by AMϕs in vivo (Figure 3). The overall conclusion from these studies indicate that strain IA565 is modestly more susceptible to macrophage phagocytosis when compared to strain 43816. We feel, however, that it is unlikely that this modest increase in susceptibility to phagocytosis alone could account for the apparent lack of in vivo pathogenicity of this strain. More importantly, these data also suggest that the pathogenicity of strain 43816 is due to more than the modest resistance to phagocytosis when compared to strain IA565. The enhanced resistance to macrophage phagocytosis seen with strain 43816 is most likely due to the expression of the K2 capsular serotype. It has been well documented that the K2 capsule lacks the mannose –α-2/3-mannose sequence that is recognized by the macrophage mannose receptor. However, when K2 expressing strains have been “switched” to express a non-K2 serotype, these strains retained their in vivo murine pathogenicity while losing their ability to resist macrophage binding [14,15]. These results clearly indicate that the genetic makeup of the parental K2 strains contains genes with the ability to confer in vivo pathogenicity in addition to capsular genes.
Having clearly established that the two strains of K. pneumoniae differ significantly in their in vivo pathogenicity in a mouse model of pneumonia, we initiated a search for putative virulence genes associated with strain 43816 that may contribute to this pathogenicity that are absent in strain IA565. We employed a PCR-based suppressive subtractive hybridization (SSH) technique to look for unique sequences in our pathogenic strain of K. pneumoniae that potentially contribute to virulence in our murine model of bacterial pneumonia. This technique is ideally suited for identifying genomic differences between two strains of bacteria differing in their pathogenicity. Having shown that IA565 is avirulent in our model, we utilized this strain as a source of driver DNA for subtracting out common “housekeeping” DNA sequences shared between the virulent and avirulent strains of K. pneumoniae. While this approach has been successfully used to identify virulence-associated genes in other bacterial species, it should be noted that this technique is not sensitive enough to identify the presence of point mutations in a particular gene in either strain. Using SSH, we have found 9 DNA sequences present in strain 43816 and absent in strain IA565 (Table1). While the role that each of these gene sequences potentially play in K. pneumoniae virulence is currently unknown, one can speculate based on the current understanding of each gene in other bacterial species.
In both the genomic DNA and cDNA SSH technique, sequences DQ211084 and DQ211085 specific for the pathogenic strain of K. pneumoniae 43816 were homologous to iroN from both S. typhimurium and E. coli (Table 1). Iron acquisition via secretion of low molecular weight, high affinity iron chelators termed siderophores has been well documented as a virulence trait in many enteric bacteria [30]. Clinical and environmental isolates of K. pneumoniae have been shown to secrete the siderophore, enterochelin (enterobactin) [31,32]. In E. coli, iroN was recently identified as the siderophore receptor for enterobactin. Additionally, iroN was identified as a virulence factor in a murine model of urinary tract infection [33]. Our findings suggest a putative iron-dependent mechanism for the pathogenicity observed for strain 43816 in our model of bacterial pneumonia.
Sequences DQ211086 and DQ211087 obtained from the genomic DNA SSH technique were found to be homologous to gene products involved in type 1 fimbriae formation in both S. typhimurium and E. coli, sthB and fimD, respectively. Because K. pneumoniae is an extracellular pathogen, a critical step in the infectious process is adherence to host mucosal surfaces. Adherence properties are mediated by fimbrial adhesions. Klebsiella spp. can produce type 1 and/or type 3 fimbrial adhesions [34]. The majority of clinical respiratory isolates of K. pneumoniae reportedly express type 3 fimbriae [35], suggesting the importance of this fimbrial type in virulence. However, a previously constructed mini-Tn5 transposon mutant strain of K. pneumoniae 43816 defective in expression of type 3 fimbriae [36] was equally virulent following intratracheal inoculation as the parental 43816 strain (data not shown). This suggests that in vivo pathogenicity of strain 43816 is not dependent upon type 3 fimbriae expression. However, the role of K. pneumoniae type 1 fimbriae expression in pathogenesis is unknown. Our findings suggest that type 1 fimbriae may play a role in K. pneumoniae pathogenicity in our model of acute bacterial pneumonia.
Sequence DQ211088 had high nucleotide sequence homology to K. pneumoniae gene kvgS which was also previously identified by Lai et. al. as a K. pneumoniae virulent strain specific sequence [37]. The group identified the virulent strain-specific sequence as having high homology to bvgAS, a two-component signal transduction system in Bordetella pertussis previously identified as a virulence factor [38]. They subsequently identified the sequence as gene kvgAS, a two-component system found in K. pneumoniae with homologies to the bvgAS system [39]. The role of the kvgAS system in K. pneumoniae virulence could not be established in their mouse peritonitis model since the pathogenicity of a kvgS deletion mutant was comparable to that of its parental strain. However, the role kvgS plays in our model of bacterial pneumonia is open to further investigation.
Four sequences obtained from the cDNA SSH technique were homologous to genes found in Pseudomonas, Salmonella and E. coli involved in energy production and conversion (DQ211090), transcriptional regulation (DQ211091), restriction endonuclease activity (DQ211092), and membrane transport (DQ211089). The possible involvement of these genes and their putative functions in K. pneumoniae in vivo pathogenicity remains to be established.
The genetic differences in our K. pneumoniae strains, identified in this study, and their role in establishing pneumonia may be extremely useful in determining bacterial factors involved in the disease initiation and progression. To this end, the generation of targeted gene deletion mutants will address the role that these genes play during pathogenesis in our model of murine K. pneumoniae bacterial pneumonia. Ultimately, these data will provide insight into the genetic mechanisms that confer in vivo virulence to K. pneumoniae, thus allowing this organism to be an effective opportunistic pathogen.
4. Materials and Methods
4.1 Animals
C57BL/6J wild-type mice were purchased from The Jackson Laboratory and housed in specific pathogen-free conditions within the animal care facility at the University of Michigan until the day of sacrifice. All experimental animal procedures were approved by the University Committee on Use and Care of Animals at the University of Michigan.
4.2 K. pneumoniae inoculation
K. pneumoniae strain 43816 is a clinical isolate with a K2 serotype [25] (ATCC, Rockville, MD). Strain IA565 is a clinical isolate from the University of Iowa Hospitals and Clinics with a K15 serotype (K-typing performed at the Unit of Gastrointestinal Infections, Statens Serum Institut, Denmark). Both of these strains were grown in tryptic soy broth (Difco, Detroit, MI) overnight at 37°C. Bacterial concentration was determined by measuring the amount of absorbance at 600 nm and compared to a predetermined standard curve. Bacteria were then diluted to the desired concentration for intratracheal inoculation in saline. Mice were anesthetized with ketamine and xylazine. The trachea was exposed, and a 30 μl inoculum was administered via a sterile 26 gauge needle. An aliquot of the inoculated K. pneumoniae suspension was serially diluted onto blood agar plates to determine the actual dose of intratracheally injected bacteria.
Neutrophils were depleted in vivo utilizing the pan-granulocytic antibody RB6-8C5 directed against Ly-6G. Anti-Ly-6G was produced as an ascites in scid mice by TSD BioServices (Germantown, NY). Mice were intraperitoneally injected with 200 μg antibody in a 0.5 ml volume 18 hours prior to infection and again one day post infection.
4.3 Whole lung homogenates for CFU, MPO and Cytokine analysis
At designated time points, the mice were euthanized by inhalation of CO2. Before lung removal, the pulmonary vasculature was perfused with 2 to 3 ml PBS/5 mM EDTA. Excised whole lungs were homogenized using a tissue homogenizer (Biospec Products, Bartlesville, OK) in 1 ml PBS/Complete protease inhibitor cocktail (Boehringer Mannheim Biochemical, Chicago, IL). For lung CFU determination, a small aliquot of lung homogenate was serially diluted and plated on blood agar plates, incubated at 37 °C and colonies counted. The limit of detection of bacterial CFU in the lung was 10 CFU/ml.
Lung myeloperoxidase (MPO) activity, as an indirect measurement of total neutrophil numbers, was quantitated by a method as described previously [40]. Briefly, 100 μL lung homogenate was mixed with 100 μL MPO homogenization buffer (0.5 % hexadecyltrimethylammonium bromide and 5 mM EDTA) and vortexed. The mixture was sonicated and centrifuged at 12,000 g for 15 minutes. The supernatant was then mixed 1:15 with assay buffer and read at 490 nm. MPO units were calculated as the change in absorbance over time.
For total lung cytokine ELISA analyses, lung homogenates were sonicated briefly to ensure complete cellular disruption, then centrifuged at 1,500 g for 10 minutes. The supernatants were collected and assessed for cytokine levels by ELISA. Murine MIP-2, KC, and MCP-1 were quantified using a modification of a sandwich ELISA method [8]. This methodology allows detection of these cytokines at concentrations of 20 pg/ml and higher. Additionally, assays have been shown to be specific for the indicated murine chemokine and show no cross-reactivity with any other murine cytokines tested.
4.4 Peripheral Blood CFU
For determination of peripheral blood bacterial numbers, mice were euthanized and heparinized blood was collected by cardiac puncture at the indicated time points. Serial dilutions were plated onto blood agar plates, incubated at 37 °C and colonies counted. The limit of detection in the blood was 10 CFU/ml.
4.5 In vitro Peritoneal Macrophage Phagocytosis Assay
This protocol, previously described elsewhere [41,42], was used and modified. Non-elicited peritoneal macrophages from C57BL/6J mice was isolated and plated into half-sized 96-well plates. FITC-labeled K. pneumoniae was opsonized by incubation in 5% mouse serum for 15-30 minutes. These bacteria were then added to the macrophage cells in quadruplicate at a bacterium to macrophage ratio of 100:1. After incubation, unbound bacteria were washed off and extracellular fluorescence was quenched with trypan blue. Internalized FITC signal was then analyzed with a fluorometer (Tecan SPECTRAFluorPlus).
4.6 Bronchoalveolar Lavage
Bronchoalveolar lavage (BAL) was performed to obtain BAL cells and fluid. The trachea was exposed and intubated using a 1.27mm OD polyethylene catheter. BAL was performed by instilling 1.6ml PBS/ 5mM EDTA, followed by gentle suction. Approximately 1.5ml of lavage fluid was retrieved per mouse and cytospins were prepared from BAL cells.
4.7 In vivo Alveolar Macrophage Phagocytosis Assay
The protocol for alveolar macrophage in vivo phagocytosis has been previously described [43]. Briefly, mice were injected with approximately 1 × 106 CFU of K. pneumoniae at time zero. At 2 h post infection, BAL was performed to obtain alveolar leukocytes. The lavage fluid was centrifuged at 500 g for 5 min and the pellet resuspended in PBS containing 2% serum. Thirty thousand cells were then spun down onto a glass slide, which was air dried, stained with Diff-Quick, and examined under oil immersion. The initial 200 alveolar macrophages or polymorphonuclear cells were counted to determine the number of whole intracellular bacteria in each cell. The phagocytic index (PI) was calculated as: PI = (percent of macrophages containing at least one bacterium) x (mean number of bacteria per positive cell).
4.8 Statistical Analysis
Statistical significance was determined using the unpaired, two-tailed student t test, ANOVA for multiple group comparisons using the Student-Newman-Keuls post-test, and Fisher's Exact Test. Calculations were performed using InStat 3 (GraphPad Software, San Diego, CA). Statistical analyses of survival curves were performed by the Log Rank Test using the Prism 3 software program (GraphPad Software).
Acknowledgments
This work was supported in part by grants AI49448 (TAM) and AI050011 (SC) from the National Institutes of Health and a Career Investigator Award from the American Lung Association (TAM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bartlett JG, O'Keefe P, Tally FP, Louie TJ, Gorbach SL. Bacteriology of hospital-acquired pneumonia. Arch Intern Med. 1986;146:868–71. [PubMed] [Google Scholar]
- 2.De Champs C, Rich C, Chandezon P, Chanal C, Sirot D, Forestier C. Factors associated with antimicrobial resistance among clinical isolates of Klebsiella pneumoniae: 1-year survey in a French university hospital. Eur J Clin Microbiol Infect Dis. 2004;18:18. doi: 10.1007/s10096-004-1144-2. [DOI] [PubMed] [Google Scholar]
- 3.Hoffman NR, Preston FS., Jr. Friedlander's pneumonia. A report of 11 cases and appraisal of antibiotic therapy. Dis Chest. 1968;53:481–6. doi: 10.1378/chest.53.4.481. [DOI] [PubMed] [Google Scholar]
- 4.Jong GM, Hsiue TR, Chen CR, Chang HY, Chen CW. Rapidly fatal outcome of bacteremic Klebsiella pneumoniae pneumonia in alcoholics. Chest. 1995;107:214–7. doi: 10.1378/chest.107.1.214. [DOI] [PubMed] [Google Scholar]
- 5.Yinnon AM, Butnaru A, Raveh D, Jerassy Z, Rudensky B. Klebsiella bacteraemia: community versus nosocomial infection. Qjm. 1996;89:933–41. doi: 10.1093/qjmed/89.12.933. [DOI] [PubMed] [Google Scholar]
- 6.Davies J. Inactivation of antibiotics and the dissemination of resistance genes. Science. 1994;264:375–82. doi: 10.1126/science.8153624. [DOI] [PubMed] [Google Scholar]
- 7.Meyer KS, Urban C, Eagan JA, Berger BJ, Rahal JJ. Nosocomial outbreak of Klebsiella infection resistant to late-generation cephalosporins. Ann Intern Med. 1993;119:353–8. doi: 10.7326/0003-4819-119-5-199309010-00001. [DOI] [PubMed] [Google Scholar]
- 8.Greenberger MJ, Strieter RM, Kunkel SL, Danforth JM, Goodman RE, Standiford TJ. Neutralization of IL-10 increases survival in a murine model of Klebsiella pneumonia. J Immunol. 1995;155:722–9. [PubMed] [Google Scholar]
- 9.Laichalk LL, Kunkel SL, Strieter RM, Danforth JM, Bailie MB, Standiford TJ. Tumor necrosis factor mediates lung antibacterial host defense in murine Klebsiella pneumonia. Infect Immun. 1996;64:5211–8. doi: 10.1128/iai.64.12.5211-5218.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore TA, Perry ML, Getsoian AG, Newstead MW, Standiford TJ. Divergent role of gamma interferon in a murine model of pulmonary versus systemic Klebsiella pneumoniae infection. Infect Immun. 2002;70:6310–8. doi: 10.1128/IAI.70.11.6310-6318.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Standiford TJ, Strieter RM, Greenberger MJ, Kunkel SL. Expression and regulation of chemokines in acute bacterial pneumonia. Biol Signals. 1996;5:203–8. doi: 10.1159/000109191. [DOI] [PubMed] [Google Scholar]
- 12.Standiford TJ, Huffnagle GB. Cytokines in host defense against pneumonia. J Investig Med. 1997;45:335–45. [PubMed] [Google Scholar]
- 13.Mizuta K, Ohta M, Mori M, Hasegawa T, Nakashima I, Kato N. Virulence for mice of Klebsiella strains belonging to the O1 group: relationship to their capsular (K) types. Infect Immun. 1983;40:56–61. doi: 10.1128/iai.40.1.56-61.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ofek I, Kabha K, Athamna A, Frankel G, Wozniak DJ, Hasty DL, Ohman DE. Genetic exchange of determinants for capsular polysaccharide biosynthesis between Klebsiella pneumoniae strains expressing serotypes K2 and K21a. Infect Immun. 1993;61:4208–16. doi: 10.1128/iai.61.10.4208-4216.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kabha K, Nissimov L, Athamna A, Keisari Y, Parolis H, Parolis LA, Grue RM, Schlepper-Schafer J, Ezekowitz AR, Ohman DE, et al. Relationships among capsular structure, phagocytosis, and mouse virulence in Klebsiella pneumoniae. Infect Immun. 1995;63:847–52. doi: 10.1128/iai.63.3.847-852.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng JC, Moore TA, Newstead MW, Zeng X, Krieg AM, Standiford TJ. CpG oligodeoxynucleotides stimulate protective innate immunity against pulmonary Klebsiella infection. J Immunol. 2004;173:5148–55. doi: 10.4049/jimmunol.173.8.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore TA, Moore BB, Newstead MW, Standiford TJ. Gamma delta-T cells are critical for survival and early proinflammatory cytokine gene expression during murine Klebsiella pneumonia. J Immunol. 2000;165:2643–50. doi: 10.4049/jimmunol.165.5.2643. [DOI] [PubMed] [Google Scholar]
- 18.Fulton SA, Reba SM, Martin TD, Boom WH. Neutrophil-mediated mycobacteriocidal immunity in the lung during Mycobacterium bovis BCG infection in C57BL/6 mice. Infect Immun. 2002;70:5322–7. doi: 10.1128/IAI.70.9.5322-5327.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knapp S, Leemans JC, Florquin S, Branger J, Maris NA, Pater J, van Rooijen N, van der Poll T. Alveolar macrophages have a protective antiinflammatory role during murine pneumococcal pneumonia. Am J Respir Crit Care Med. 2003;167:171–9. doi: 10.1164/rccm.200207-698OC. Epub 2002 Sep 25. [DOI] [PubMed] [Google Scholar]
- 20.Kooguchi K, Hashimoto S, Kobayashi A, Kitamura Y, Kudoh I, WienerKronish J, Sawa T. Role of alveolar macrophages in initiation and regulation of inflammation in Pseudomonas aeruginosa pneumonia. Infect Immun. 1998;66:3164–9. doi: 10.1128/iai.66.7.3164-3169.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tateda K, Moore TA, Deng JC, Newstead MW, Zeng X, Matsukawa A, Swanson MS, Yamaguchi K, Standiford TJ. Early recruitment of neutrophils determines subsequent T1/T2 host responses in a murine model of Legionella pneumophila pneumonia. J Immunol. 2001;166:3355–61. doi: 10.4049/jimmunol.166.5.3355. [DOI] [PubMed] [Google Scholar]
- 22.Moore TA, Newstead MW, Strieter RM, Mehrad B, Beaman BL, Standiford TJ. Bacterial clearance and survival are dependent on CXC chemokine receptor-2 ligands in a murine model of pulmonary Nocardia asteroides infection. J Immunol. 2000;164:908–15. doi: 10.4049/jimmunol.164.2.908. [DOI] [PubMed] [Google Scholar]
- 23.Winstanley C. Spot the difference: applications of subtractive hybridisation to the study of bacterial pathogens. J Med Microbiol. 2002;51:459–67. doi: 10.1099/0022-1317-51-6-459. [DOI] [PubMed] [Google Scholar]
- 24.Domenico P, Johanson WG, Jr., Straus DC. Lobar pneumonia in rats produced by clinical isolates of Klebsiella pneumoniae. Infect Immun. 1982;37:327–35. doi: 10.1128/iai.37.1.327-335.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shankar-Sinha S, Valencia GA, Janes BK, Rosenberg JK, Whitfield C, Bender RA, Standiford TJ, Younger JG. The Klebsiella pneumoniae O Antigen Contributes to Bacteremia and Lethality during Murine Pneumonia. Infect Immun. 2004;72:1423–1430. doi: 10.1128/IAI.72.3.1423-1430.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yadav V, Sharma S, Harjai K, Mohan H, Chhibber S. Induction & resolution of lobar pneumonia following intranasal instillation with Klebsiella pneumoniae in mice. Indian J Med Res. 2003;118:47–52. [PubMed] [Google Scholar]
- 27.Mehrad B, Strieter RM, Moore TA, Tsai WC, Lira SA, Standiford TJ. CXC chemokine receptor-2 ligands are necessary components of neutrophil-mediated host defense in invasive pulmonary aspergillosis. J Immunol. 1999;163:6086–94. [PubMed] [Google Scholar]
- 28.Tsai WC, Strieter RM, Wilkowski JM, Bucknell KA, Burdick MD, Lira SA, Standiford TJ. Lung-specific transgenic expression of KC enhances resistance to Klebsiella pneumoniae in mice. J Immunol. 1998;161:2435–40. [PubMed] [Google Scholar]
- 29.Zhang P, Summer WR, Bagby GJ, Nelson S. Innate immunity and pulmonary host defense. Immunol Rev. 2000;173:39–51. doi: 10.1034/j.1600-065x.2000.917306.x. [DOI] [PubMed] [Google Scholar]
- 30.Payne SM. Iron and virulence in the family Enterobacteriaceae. Critical Reviews in Microbiology. 1988;16:81–111. doi: 10.3109/10408418809104468. [DOI] [PubMed] [Google Scholar]
- 31.Podschun R, Pietsch S, Holler C, Ullmann U. Incidence of Klebsiella species in surface waters and their expression of virulence factors. Appl Environ Microbiol. 2001;67:3325–7. doi: 10.1128/AEM.67.7.3325-3327.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tarkkanen AM, Allen BL, Williams PH, Kauppi M, Haahtela K, Siitonen A, Orskov I, Orskov F, Clegg S, Korhonen TK. Fimbriation, capsulation, and iron-scavenging systems of Klebsiella strains associated with human urinary tract infection. Infect Immun. 1992;60:1187–92. doi: 10.1128/iai.60.3.1187-1192.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russo TA, McFadden CD, Carlino-MacDonald UB, Beanan JM, Barnard TJ, Johnson JR. IroN functions as a siderophore receptor and is a urovirulence factor in an extraintestinal pathogenic isolate of Escherichia coli. Infect Immun. 2002;70:7156–60. doi: 10.1128/IAI.70.12.7156-7160.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11:589–603. doi: 10.1128/cmr.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hornick DB, Allen BL, Horn MA, Clegg S. Fimbrial types among respiratory isolates belonging to the family Enterobacteriaceae. J Clin Microbiol. 1991;29:1795–800. doi: 10.1128/jcm.29.9.1795-1800.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langstraat J, Bohse M, Clegg S. Type 3 fimbrial shaft (MrkA) of Klebsiella pneumoniae, but not the fimbrial adhesin (MrkD), facilitates biofilm formation. Infect Immun. 2001;69:5805–12. doi: 10.1128/IAI.69.9.5805-5812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai YC, Yang SL, Peng HL, Chang HY. Identification of genes present specifically in a virulent strain of Klebsiella pneumoniae. Infect Immun. 2000;68:7149–51. doi: 10.1128/iai.68.12.7149-7151.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boucher PE, Stibitz S. Synergistic binding of RNA polymerase and BvgA phosphate to the pertussis toxin promoter of Bordetella pertussis. J Bacteriol. 1995;177:6486–91. doi: 10.1128/jb.177.22.6486-6491.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lai YC, Lin GT, Yang SL, Chang HY, Peng HL. Identification and characterization of KvgAS, a two-component system in Klebsiella pneumoniae CG43. FEMS Microbiol Lett. 2003;218:121–6. doi: 10.1111/j.1574-6968.2003.tb11507.x. [DOI] [PubMed] [Google Scholar]
- 40.Remick DG, Strieter RM, Eskandari MK, Nguyen DT, Genord MA, Raiford CL, Kunkel SL. Role of tumor necrosis factor-alpha in lipopolysaccharide-induced pathologic alterations. Am J Pathol. 1990;136:49–60. [PMC free article] [PubMed] [Google Scholar]
- 41.Aronoff DM, Canetti C, Peters-Golden M. Prostaglandin E2 inhibits alveolar macrophage phagocytosis through an E-prostanoid 2 receptor-mediated increase in intracellular cyclic AMP. J Immunol. 2004;173:559–65. doi: 10.4049/jimmunol.173.1.559. [DOI] [PubMed] [Google Scholar]
- 42.Wan CP, Park CS, Lau BH. A rapid and simple microfluorometric phagocytosis assay. J Immunol Methods. 1993;162:1–7. doi: 10.1016/0022-1759(93)90400-2. [DOI] [PubMed] [Google Scholar]
- 43.Ojielo CI, Cooke K, Mancuso P, Standiford TJ, Olkiewicz KM, Clouthier S, Corrion L, Ballinger MN, Toews GB, Paine R, 3rd, Moore BB. Defective phagocytosis and clearance of Pseudomonas aeruginosa in the lung following bone marrow transplantation. J Immunol. 2003;171:4416–24. doi: 10.4049/jimmunol.171.8.4416. [DOI] [PubMed] [Google Scholar]