Abstract
Recurrent chromosomal translocations in neoplasms often generate hybrid genes that play critical roles in tumorigenesis. Desmoplastic small round-cell tumor (DSRCT) is an aggressive malignancy associated with the chromosomal translocation t(11;22)(p13;q12). This translocation generates a chimeric transcription factor, EWS-WT1, which consists of the transcriptional activation domain of the Ewing’s sarcoma (EWS) protein and the DNA binding domain of the Wilms’ tumor 1 (WT1) protein. One of the splice variants, EWS-WT1(-KTS) lacks three amino acid residues (Lys-Thr-Ser) in the DNA binding domain and transforms NIH3T3 cells. Therefore, it is likely that aberrant gene expression caused by EWS-WT1(-KTS) is involved in the malignant phenotype of DSRCT. Microarray analysis of 9600 human genes revealed that a gene encoding a tetraspanin-family protein, T-cell acute lymphoblastic leukemia-associated antigen 1 (TALLA-1), was induced in EWS-WT1(-KTS)-expressing cell clones. This induction was EWS-WT1(-KTS)-specific, and more importantly, TALLA-1 protein was expressed in the three independent cases of DSRCT. Tetraspanin-family genes encode transmembrane proteins that regulate various cell processes such as cell adhesion, migration and metastasis. Our findings provide a novel insight into the malignant phenotype of DSRCT, suggesting that TALLA-1 is a useful marker for diagnosis and a potential target for the therapy of DSRCT.
Specific recurrent chromosomal translocation in malignant tumors is thought to play a causative role in tumorigenesis. Translocation frequently generates a chimeric gene, and the functions of such chimeric genes have been studied extensively to clarify the molecular mechanisms underlying tumorigenesis. Desmoplastic small round-cell tumor (DSRCT) is an aggressive neoplasm with distinctive histological and immunophenotypic features that suggest a multilineage origin. 1-3 The tumor often develops in the abdominal and pelvic peritoneum of adolescent males. Recent integrated strategies including surgery, multiagent chemotherapy, and radiation therapy have improved the prognosis considerably, although progression-free survival remains poor. 3 Thus, identification of tumor-associated antigens for DSRCT would be useful for the development of anti-tumor drugs. DSRCT has a unique chromosomal translocation t(11;22)(p13;q12) that generates a chimeric gene comprising part of the Ewing’s sarcoma (EWS) gene on chromosome 22 and the Wilms’ tumor 1 (WT1) gene on chromosome 11. 4,5 The EWS-WT1 chimeric protein consists of the N-terminal transcriptional regulatory domain of EWS and the C-terminal three zinc-finger motifs of WT1 as the DNA binding domain. Therefore, the fusion protein acts as a transcriptional activator. Alternative splicing in the WT1 region generates two types of EWS-WT1 protein. One isoform, EWS-WT1(-KTS), which lacks three amino acid residues (Lys-Thr-Ser) between the third and the fourth zinc fingers of WT1, confers NIH3T3 cells with anchorage-independent growth and tumorigenicity in nude mice. 6 In contrast, EWS-WT1(+KTS), which contains the three amino acid residues, does not have such transforming activity. Thus, the EWS-WT1 protein is a molecular marker for DSRCT and is believed to be involved in tumorigenesis.
Recent reports have shown that EWS-WT1(-KTS) activates a number of genes, including those encoding platelet-derived growth factor-A, 7 insulin-like growth factor-1 receptor, 8,9 interleukin-2/15 receptor β-chain (IL2/15Rβ), 10 and brain-specific angiogenesis inhibitor 1-associated protein 3 (BAIAP3). 11 In the present study, we used DNA microarrays to carry out a comprehensive analysis of the downstream genes that are up-regulated by EWS-WT1(-KTS). We found that a tetraspanin-family protein, T-cell acute lymphoblastic leukemia-associated antigen 1 (TALLA-1, also referred to as A15, 12 CCG-B7, 13 and TM4SF2), was induced specifically by EWS-WT1(-KTS) and TALLA1 protein was expressed in the three independent cases of DSRCT. Tetraspanin-family proteins contain four transmembrane domains and two extracellular loops. This evolutionally conserved gene family contains more than 30 known members in mammals, 37 in Drosophila and 20 in Caenorhabditis elegans. 14 They regulate a variety of normal and pathological processes such as cell adhesion, motility, egg-sperm fusion, virus-induced syncytium formation, and cancer metastasis through formation of a network of multimolecular complexes via tetraspanin-tetraspanin and tetraspanin-protein interactions. 14,15 We discuss the significance of TALLA-1 in the malignant phenotype of tumors.
Materials and Methods
Microarray Analysis
Synthetic polynucleotides (80-mers) representing 9600 human genes (MicroDiagnostic, Tokyo, Japan) were arrayed with a custom-made arrayer. Poly(A)+ RNA was prepared from cells with TRIzol reagent (Invitrogen, Carlsbad, CA) and Poly(A)Purist Kit (Ambion, Austin, TX). Two micrograms of poly(A)+ RNA were labeled with Cyanine 5-dUTP or Cyanine 3-dUTP. Hybridization and subsequent washes of arrays were performed with a Labeling & Hybridization Kit (MicroDiagnostic). Hybridization signals were measured with a GenePix 400A scanner (Axon Instruments, Union City, CA) and then processed into primary expression ratios (ratios of Cyanine 5-labeled to Cyanine 3-labeled samples) by the GenePix Pro software (Axon Instruments). A secondary ratio of expression of each gene was calculated by averaging the primary expression ratio obtained from an experiment with Cyanine 5-labeled target and Cyanine 3-labeled control sample and the reciprocal of the primary expression ratio obtained from an experiment with Cyanine 5-labeled control and Cyanine 3-labeled target. The secondary expression ratios calculated from the pair of experiments were converted into log2 values as the final expression ratios.
Cells
A human osteosarcoma cell line, U2OS, was maintained in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS). A human DSRCT cell line, JN-DSRCT-1, 16 was maintained in DMEM:Nutrient Mixture F12 (1:1) mixture with 10% FBS. To establish clones with stable expression of EWS-WT1(-KTS), U2OS cells (1 × 105) were transfected with 2 μg of an EWS-WT1(-KTS) expression vector, pcDNA-EWS-WT1(-KTS), and 6 μl of Lipofectamine (Invitrogen). Clones were isolated in the presence of 800 μg/ml of G418 and used for subsequent microarray analysis. A mixture of mock-transfected cells was maintained in the presence of G418 and used as control.
Transfection
A nucleofector device (Amaxa Biosystems, Cologne, Germany) was used for highly efficient gene transfer. This device delivers DNA directly into nuclei, allowing exogenous gene expression after a short incubation period of only 2 to 8 hours. U2OS cells (2 × 106) were transfected with 2 μg of an expression vector with R solution and the pre-set U-16 program. The transfection efficiency was monitored by flow cytometric analysis of pCMV-GFP transfected cells.
Western Blotting
Proteins solubilized in Laemmli sample buffer were resolved on 10% sodium dodecyl sulfate (SDS)-polyacrylamide gels and blotted onto Immobilon membranes (Millipore Co., Bedford, MA). Filters were probed for WT1 and EWS-WT1 with anti-WT1 antibody (C19; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and for EWS-FLI1 with anti-FLI1 antibody (C19; Santa Cruz Biotechnology). Bands were visualized with Renaissance Chemiluminescence Reagent Plus (NEN Life Sci Products, Boston, MA).
Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
RNA was prepared with TRIzol reagent from cultured cells or tumor tissues. First-strand cDNA was synthesized using SuperScript first-strand synthesis system for RT-PCR (Invitrogen). Taq polymerase was purchased from Sigma (St. Louis, MO). Primers are 5′-TACACGGACGCTATGCAGAC-3′ and 5′-GATTCCAAACGCCACTCCAG-3′ for Talla-1, 5′-TACCCCATGCAGCCAGTCAC-3′ and 5′-TTTGAGCTGGTCTGAACGAG-3′ for EWS-WT1, and 5′-CCAGCCGAGCCACATCGCTC-3′ and 5′-ATGAGCCCCAGCCTTCTCCAT-3′ for GAPDH.
Northern Blotting
Northern blotting was done with the NorthernMax Kit (Ambion). 32P-labeled probe was prepared with a Multiprime DNA Labeling System (Amersham Biosciences, Arlington Heights, IL).
Flow Cytometry
Cells were incubated with ascites diluted 100-fold containing monoclonal anti-TALLA-1 antibody (B2D) and fluorescent anti-mouse IgG (Alexa Fluor 488; Molecular Probes, Eugene, OR), and then subjected to flow cytometric analysis (Epics XL; Beckman Coulter, Fullerton, CA).
Immunofluorescence
Cells seeded on cover glasses were fixed with 4% paraformaldehyde for 15 minutes at 37°C, and permeabilized with 0.2% Triton X-100/PBS for 5 minutes at room temperature. EWS-WT1 and TALLA-1 were detected with the same antibodies diluted 100-fold as used for Western blotting and flow cytometry. Proteins were visualized with fluorescent secondary antibody (Molecular Probes).
Immunohistochemistry
Frozen blocks and sections of DSRCT were kindly provided by the Pediatric Division of Cooperative Human Tissue Network (Columbus, OH). Diagnosis of DSRCT was confirmed on hematoxylin and eosin-stained frozen sections by pathologists T. K. and S. M. For detection of TALLA-1, frozen sections were incubated with 800-fold diluted B2D and then subjected to color reaction with the indirect immunoperoxidase method (Histofine Simple Stain, Nichirei, Japan).
Results
To explore comprehensively downstream genes with expression induced by EWS-WT1(-KTS), we established three clonal lines of U2OS cells that stably expressed EWS-WT1(-KTS). Microarray analysis of 9600 human genes identified a group of genes whose expression was reproducibly induced or repressed in these clones (Table 1) ▶ . These genes included those encoding IL2/15Rβ, adrenomedullin, and BAIAP3, which were reported previously. 10,11 We also found that expression of desmin, an established molecular marker for DSRCT, was induced by EWS-WT1(-KTS). We confirmed the change in expression of desmin by immunoblotting (data not shown).
Table 1.
Microarray Identification of Genes Activated or Repressed Downstream of EWS-WT1(-KTS)
| Gene symbol | Accession number | B12 | C9 | D9 | Average |
|---|---|---|---|---|---|
| RGS5 | NM_003617 | 4.8069 | 4.5771 | 4.9195 | 4.7678 |
| DKFZp434B227 | NM_032263 | 4.3451 | 4.6415 | 4.9425 | 4.6430 |
| DES (desmin) | NM_001927 | 4.8052 | 3.3748 | 5.1433 | 4.4411 |
| APOA1 | NM_000039 | 3.4595 | 5.0025 | 3.7444 | 4.0688 |
| CHI3L1 | NM_001276 | 3.4901 | 4.4559 | 3.8189 | 3.9216 |
| WT1 | NM_024424 | 3.9531 | 3.8653 | 3.8197 | 3.8794 |
| NK4 | NM_004221 | 2.7691 | 4.3032 | 4.1529 | 3.7417 |
| APOL2 | NM_030882 | 3.5625 | 4.3115 | 2.9113 | 3.5951 |
| FLJ20245 | NM_017723 | 3.4295 | 3.6399 | 2.9387 | 3.3360 |
| ADM (adrenomedullin) | NM_001124 | 3.2513 | 4.1579 | 2.4834 | 3.2975 |
| LOC135562 | XM_069429 | 3.0812 | 4.5899 | 2.0095 | 3.2269 |
| BAIAP3 | NM_003933 | 2.6486 | 4.0179 | 2.6717 | 3.1127 |
| BIK | NM_001197 | 2.3391 | 3.4097 | 3.3323 | 3.0270 |
| LOC90189 | XM_029748 | 2.9116 | 2.7470 | 2.8376 | 2.8321 |
| FLJ22671 | NM_024861 | 2.4854 | 2.7474 | 3.1525 | 2.7951 |
| KLK6 | NM_002774 | 2.8500 | 2.5292 | 2.7768 | 2.7187 |
| LCK | NM_005356 | 1.7054 | 2.2152 | 4.1992 | 2.7066 |
| IL8 | NM_000584 | 1.9838 | 3.7999 | 2.2517 | 2.6785 |
| DRP2 | NM_001939 | 1.9726 | 2.7053 | 3.2519 | 2.6433 |
| KREMEN2 | NM_024507 | 2.3643 | 2.9575 | 2.3405 | 2.5541 |
| TNFRSF21 | NM_014452 | 2.3449 | 2.9221 | 2.2719 | 2.5130 |
| SSTR3 | NM_001051 | 2.7267 | 2.5529 | 2.1279 | 2.4692 |
| FLJ20154 | NM_017787 | 1.8621 | 2.3683 | 2.9680 | 2.3995 |
| NFATC1 | NM_006162 | 2.3952 | 2.8337 | 1.9534 | 2.3941 |
| GAGE7 | NM_021123 | 3.0143 | 2.0046 | 2.1603 | 2.3931 |
| SNX9 | NM_016224 | 2.3430 | 2.3643 | 2.2830 | 2.3301 |
| TM4SF2 (Talla-1) | NM_004615 | 1.8004 | 2.4611 | 2.6285 | 2.2967 |
| LOC139728 | XM_060051 | 2.2622 | 2.3246 | 2.1933 | 2.2600 |
| TRIM29 | NM_012101 | 2.0576 | 2.8573 | 1.8001 | 2.2383 |
| SERPINB2 | NM_002575 | 2.1313 | 2.2954 | 2.2759 | 2.2342 |
| RELB | NM_006509 | 1.9808 | 2.8510 | 1.8384 | 2.2234 |
| FLJ23058 | NM_024696 | 2.1799 | 1.9497 | 2.5029 | 2.2108 |
| IL2RB (IL2 receptor β) | NM_000878 | 2.4299 | 2.5900 | 1.5083 | 2.1761 |
| HSPC022 | NM_014029 | 2.2705 | 2.4120 | 1.8273 | 2.1699 |
| MSLN | NM_005823 | 1.9397 | 2.2261 | 2.2971 | 2.1543 |
| OPRL1 | NM_000913 | 1.9394 | 2.7325 | 1.5796 | 2.0838 |
| MAGP2 | NM_003480 | 2.3341 | 2.0496 | 1.6075 | 1.9971 |
| NEDD4L | NM_015277 | 1.7847 | 2.4666 | 1.7302 | 1.9938 |
| SLC2A3 | NM_006931 | 1.6088 | 2.2489 | 2.1073 | 1.9883 |
| BGN | NM_001711 | 1.5921 | 1.9808 | 2.3423 | 1.9717 |
| COL6A3 | NM_004369 | 1.8971 | 2.3618 | 1.5310 | 1.9300 |
| S100A4 | NM_002961 | 1.8033 | 2.2082 | 1.6691 | 1.8935 |
| KRT17 | NM_000422 | 1.6139 | 2.2038 | 1.6792 | 1.8323 |
| MS4A6A | NM_022349 | 1.7096 | 2.0112 | 1.5267 | 1.7492 |
| C18orf1 | NM_004338 | −1.5796 | −1.5385 | −1.6386 | −1.5856 |
| H1F2 | NM_005319 | −1.9450 | −1.7604 | −1.8163 | −1.8406 |
| OSBPL10 | NM_017784 | −1.6272 | −2.2958 | −1.6578 | −1.8603 |
| FXYD6 | NM_022003 | −1.5180 | −2.1517 | −1.9351 | −1.8683 |
| HNOEL-iso | NM_020190 | −1.8806 | −2.7950 | −1.5057 | −2.0604 |
| P311 | NM_004772 | −2.0767 | −2.2441 | −1.9064 | −2.0757 |
| NECL1 | NM_021189 | −1.5862 | −3.3417 | −1.9093 | −2.2791 |
| FLJ20716 | NM_017938 | −2.0441 | −2.7866 | −2.2385 | −2.3564 |
| H2AFL | NM_003512 | −2.4452 | −2.2226 | −2.4156 | −2.3611 |
| BRAK | NM_004887 | −2.2034 | −2.7810 | −2.4533 | −2.4792 |
| CRTL1 | NM_001884 | −1.9105 | −3.5013 | −2.0543 | −2.4887 |
| SERPINF1 | NM_002615 | −1.8785 | −3.0730 | −2.6775 | −2.5430 |
| MMP2 | NM_004530 | −2.4785 | −2.8093 | −2.4158 | −2.5679 |
| LOC51316 | NM_016619 | −2.7566 | −3.4258 | −2.2395 | −2.8073 |
| SRCRB4D | NM_080744 | −3.4141 | −3.1876 | −3.4979 | −3.3665 |
| MVD | NM_002461 | −3.1784 | −3.7718 | −3.9392 | −3.6298 |
Expression levels (log2 values) of 9,600 human genes in the three EWS-WT1(-KTS)-expressing U2OS clones (B12, C9, and D9) relative to a mixture of control mock-transfected clones are calculated as expression ratios processed from the raw data of two individual experiments labeled reciprocally with two different fluors. Genes listed in the table show more or less than 1.5 of expression level (log2 values) in all three clones. A more detailed description of each gene is available on the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/).
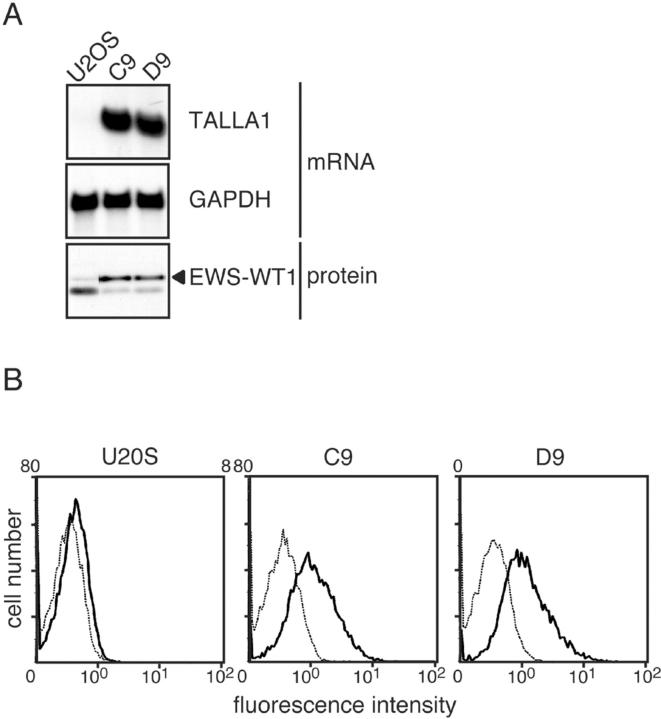
We also found that expression of Talla-1, a member of the tetraspanin family and a marker of T-cell acute lymphoblastic leukemia (T-ALL), 17 was up-regulated in clones expressing EWS-WT1(-KTS). Northern blotting confirmed the induction of Talla-1 mRNA in the clones (C9 and D9; Figure 1A ▶ ). Flow cytometric analysis showed that TALLA-1 protein was expressed on the plasma membranes of EWS-WT1(-KTS)-expressing cells (Figure 1B) ▶ . To exclude the possibility that these changes reflected clonal variation, we analyzed Talla-1 expression in cells that transiently expressed EWS-WT1(-KTS). For this purpose, we used a Nucleofector device, which enabled us to achieve approximately 80% transfection efficiency. EWS-WT1(-KTS) protein was detected at 8 hours after transfection, was sustained at 24 hours, and then gradually decreased (Figure 2A) ▶ . With transient transfection, Talla-1 mRNA was initially detected at 8 hours, and was expressed at high levels at 24 hours after transfection of vector encoding EWS-WT1(-KTS) (Figure 2A) ▶ . To examine if the induction of Talla-1 expression is specific to EWS-WT1(-KTS), we transfected vectors encoding EWS-WT1(+KTS), EWS-FLI1, WT1(-KTS), and WT1(+KTS) into U2OS cells. EWS-FLI1 is generated by chromosomal translocation t(11;22)(q24;q12), and is found in 85% of cases of Ewing’s sarcoma. The fusion protein consists of the N-terminal transcriptional activation domain of EWS and the C-terminal ETS DNA binding domain of FLI1. Thus, the primary target genes of Ewing’s sarcoma are thought to be different from those of DSRCT because of the difference in DNA recognition. Northern blot analysis revealed that only EWS-WT1(-KTS) induced Talla-1 expression (Figure 2B) ▶ .
Figure 1.
Expression of Talla-1 in cell clones stably expressing EWS-WT1(-KTS). A: Northern blotting analysis of Talla-1 mRNA. Total RNA (5 μg) was loaded in each lane. EWS-WT1(-KTS) protein (filled triangle) was detected by Western blotting with anti-WT1 antibody. B: Flow cytometric analysis of TALLA-1 protein. Living cells were incubated with anti-TALLA-1 (B2D) monoclonal antibody and Alexa-Fluor 488-labeled anti-mouse IgG and then subjected to flow cytometric analysis (solid line). Background profiles (dotted line) were measured by staining with Alexa-Fluor 488-labeled anti-mouse IgG only.
Figure 2.
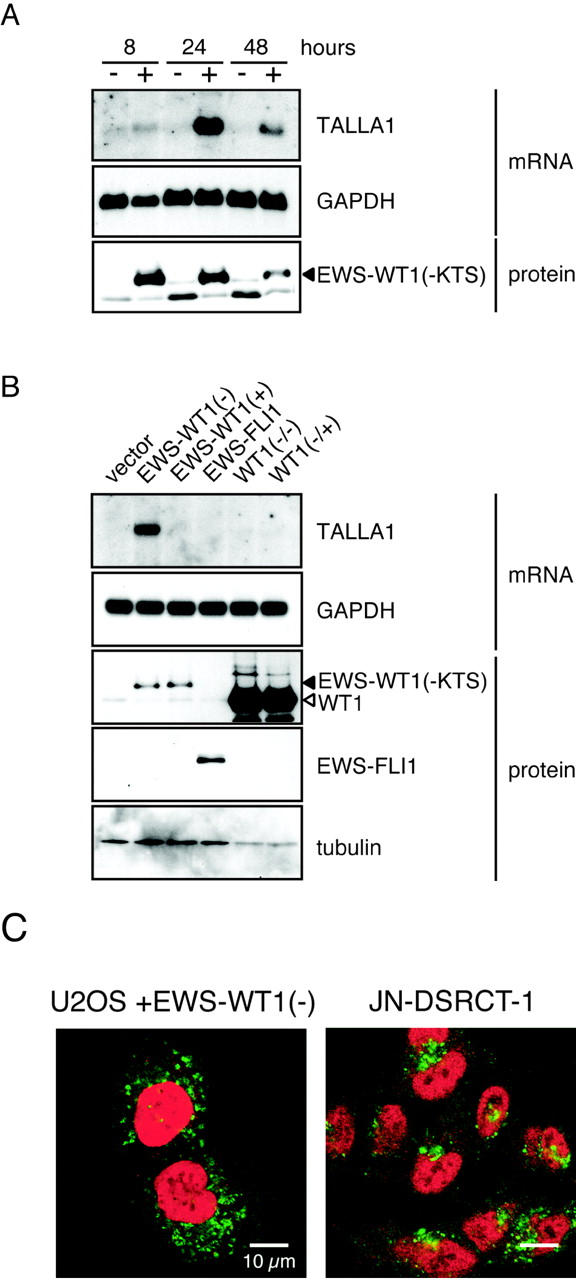
Specific expression of Talla-1 mRNA and subcellular localization of TALLA-1 protein in cells transiently expressing EWS-WT1(-KTS). A: Time course of Talla-1 mRNA induction after transfection of vector encoding EWS-WT1(-KTS). RNA and protein were prepared at the indicated times after transfection. B: Specific induction of Talla-1 expression by EWS-WT1(-KTS). Each expression vector was introduced into U2OS cells and then RNA and protein were prepared 24 hours after transfection. C: Immunofluorescent staining of EWS-WT1(-KTS)-transfected cells and JN-DSRCT-1. TALLA-1 and EWS-WT1 were stained with Alexa Fluor 488 (green) and Alexa Fluor 568 (red), respectively.
Recently a DSRCT cell line, JN-DSRCT-1, was established from the pleural effusion of a patient with pulmonary metastasis from a typical intraabdominal DSRCT. 16 We confirmed that Talla-1 mRNA and protein were expressed in this cell line (data not shown). Because some tetraspanins are localized not only on the plasma membrane but also at the intracellular compartments such as lysosome and exosome, we analyzed subcellular localization of TALLA-1 by immunohistochemistry. Figure 2C ▶ shows that TALLA-1 protein in U2OS cells was also localized in the cytoplasm when cells were permeabilized. This dot-like localization pattern is quite similar to that of endogenous TALLA-1 in JN-DSRCT-1 cells. Induction of Talla-1 mRNA was also observed when the EWS-WT1(-KTS) expression vector was transfected into human 293T cells, indicating that induction of Talla-1 expression by EWS-WT1(-KTS) was not specific to U2OS cells (data not shown). Taken together, these data suggest that EWS-WT1(-KTS) induces expression of Talla-1 mRNA and protein in human cells without cell type-specific cofactors.
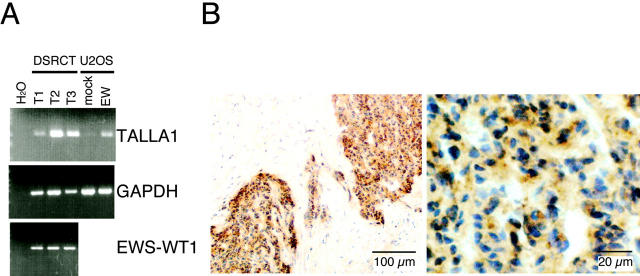
We then examined expression of TALLA-1 in DSRCT specimens. Because the supply and amount of tumor tissues were limited, we used RT-PCR analysis to detect Talla-1 mRNA. Under the condition we used, Talla-1 mRNA was detected in three of three independent DSRCT tissues (Figure 3A) ▶ . Immunohistochemical analysis showed localization of TALLA-1 protein in the nest of tumor cells in all three specimens (representative data shown in Figure 3B ▶ ). Less intense reaction was noted on the stromal endothelial cells. Other cellular components including fibroblasts, lymphocytes, fat cells, myothelial cells, or adventitial cells were basically unstained.
Figure 3.
Expression of TALLA-1 in DSRCT specimens. A: RT-PCR analysis of the Talla-1 mRNA in DSRCT. The Talla-1 mRNA was amplified from various sources by RT-PCR and then separated by electrophoresis on agarose gels. Sources are DSRCT tissues (T1, T2, T3), cells transiently expressing EWS-WT1(-KTS) (EW) or its negative control (mock), and various human tissues. Expression of EWS-WT1 fusion transcript was also detected in the three DSRCT tumors. PCR products were visualized by staining with ethidium bromide and UV transillumination. B: Immunohistochemical staining of a DSRCT specimen (T3). TALLA-1-positive cells are stained brown. Sections were counter-stained with hematoxylin.
Discussion
In the present study, we screened for downstream genes with expression induced by EWS-WT1(-KTS), which is thought to be involved in the tumorigenesis of DSRCT. The overall profile of gene expression by EWS-WT1(-KTS)-expressing cells was quite different from that of WT1(-KTS)-expressing cells (Ito E, unpublished data). In addition to the replacement of transcriptional regulatory domain of WT1 with that of EWS, the N-terminal first zinc finger that contributes to the binding specificity of WT1 is deleted during the generation of the EWS-WT1 fusion protein. 18,19 Moreover, EWS-WT1 has 10-times higher binding affinity for WT1-binding sequence than WT1. 20 Such differences may account for the unique expression profile of EWS-WT1.
We found that Talla-1, a member of the tetraspanin family, was expressed in a DSRCT cell line and tumor specimens, and was induced specifically by EWS-WT1(-KTS). Transcriptional regulatory sequence of the Talla-1 gene has not been identified so far; thus, mechanism of Talla-1 induction by EWS-WT1(-KTS) remains to be analyzed. Talla-1 was initially identified due to its preferential expression in T-ALL and neuroblastoma cell lines 12,17 In addition, Talla-1 is expressed at high levels in neurons of the brain, 21 and intriguingly its inactivation by chromosomal translocation (X;2) or by point mutations (Gly218 to stop codon, Pro172 to His) is associated with X-linked forms of nonsyndromic mental retardation (MRX). 22 Aberrant expression of Talla-1 in T-ALL is mediated cooperatively by two transcription factors, TAL1 and RBTN1, both of which are ectopically activated by chromosomal translocation in T-ALL, 23 thus Talla-1 may be a common target that is involved in oncogenesis. The biochemical and biological function of TALLA-1 is largely unknown except that TALLA-1 is associated with phosphoinositide 4-kinase. 24 Accumulating evidence indicate that tetraspanin proteins regulate integrin-extracellular matrix (ECM) interaction, 25 which includes migration, 26 invasion into collagen gels, 27 and morphology on Matrigel. 28 Furthermore, ligation of tetraspanin-α3β1 integrin complex by anti-tetraspanin or anti-integrin antibodies induces phosphoinositide 3-kinase-dependent production of matrix metalloproteinase 2 (MMP2) and long invasive protrusions within Matrigel, which is thought to increase invasive potential of tumor cells in the three dimensional extracellular matrix environment. 29 Recent biomechanical analysis directly showed that CD151, a member of the tetraspanin family, selectively strengthens α6β1 integrin-mediated adhesion to laminin-1. 30 Exogenous expression of tetraspanin proteins, Co-029 and CD151, increases the metastatic potentials of tumor cell lines 31,32 and anti-CD151 antibody inhibits metastasis and migration of a CD151-expressing tumor cell line. 31 Taken together, TALLA-1 may be involved in cellular processes, including cell adhesion and regulation of cytoskeleton through a phosphoinositide-dependent signaling pathway, which may reflect infiltration and desmoplastic character of DSRCT. This hypothesis is also attractive for pathogenesis of MRX because genes encoding Oligophrenin-1, 33 PAK3, 34 and ARHGEF6, 35 all of which are thought to regulate actin cytoskeleton, and are mutated in the cases of MRX. It should be noted that not all tetraspanin play a positive role in tumorigenesis or cancer metastasis. Some tetraspanins such as CD9, CD63, and CD82 are thought to function as negative regulators of metastasis. 15 In the case of CD82, a tetraspanin-associated protein, EWI-2 synergizes CD82 in inhibiting prostate cancer cell migration. 36 This discrepancy is not surprising, however, given that the function of the tetraspanin family depends on the components of tetraspanin-containing protein complex in individual cells.
TALLA-1 protein shows dot-like localization pattern in cytoplasm 37 (Figure 2C) ▶ . Tetraspanin proteins are localized in various types of intracellular vesicles such as late endosomes and Weibel-Palade bodies of endothelial cells, 38,39 multivesicular major histocompatibility class II-enriched compartments of B-lymphocytes, 40 and exosomes of dendritic cells. 41,42 These data point to a possible role of tetraspanin proteins in turnover and/or sorting of tetraspanin-associated proteins. A metastatic suppressor, CD82, attenuates the epidermal growth factor (EGF) signaling by accelerating EGF receptor endocytosis via its association with the EGF receptor. 43 TALLA-1 has a tyrosine-based sorting motif at the C terminus that might recruit clathrin adapter proteins to direct the TALLA-1-containing protein complex along various trafficking routes. 44 Thus TALLA-1 may perturb normal turnover of cell surface proteins in DSRCT cells.
To elucidate the function of TALLA-1, it is necessary to characterize the TALLA-1-containing protein complex on the plasma membrane and intracellular compartments. Using recent proteomics technology, tetraspanin-associated proteins have been isolated. 36,45,46 Our preliminary analysis shows that a number of cell surface proteins are associated with TALLA-1 in Jurkat (T-ALL), IMR32 (neuroblastoma), and JN-DSRCT-1 cells. Characterization of these proteins may help in understanding the functions of TALLA-1 in tumor cells.
Acknowledgments
We thank S. Kobayashi for secretarial assistance and A. Aono, N. Hoshina, and T. Kimura for technical assistance.
Footnotes
Address reprint requests to Kentaro Semba, Ph.D., Division of Cellular and Molecular Biology, Department of Cancer Biology, The Institute of Medical Science, The University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan. E-mail: ksemba@ims.u-tokyo.ac.jp.
Supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by a grant from the New Energy and Industrial Technology Development Organization (NEDO).
References
- 1.Ordóñez NG: Desmoplastic small round cell tumor: I: a histopathologic study of 39 cases with emphasis on unusual histological patterns. Am J Surg Pathol 1998, 22:1303-1313 [DOI] [PubMed] [Google Scholar]
- 2.Ordóñez NG: Desmoplastic small round cell tumor: I: an ultrastructural and immunohistochemical study with emphasis on new immunohistochemical markers. Am J Surg Pathol 1998, 22:1314-1327 [DOI] [PubMed] [Google Scholar]
- 3.Gerald WL, Ladanyi M, de Alava E, Cuatrecasas M, Kushner BH, LaQuaglia MP, Rosai J: Clinical, pathologic, and molecular spectrum of tumors associated with t(11;22)(p13;q12): desmoplastic small round-cell tumor and its variants. J Clin Oncol 1998, 16:3028-3036 [DOI] [PubMed] [Google Scholar]
- 4.Kim J, Pelletier J: Molecular genetics of chromosome translocations involving EWS and related family members. Physiol Genomics 1999, 1:127-138 [DOI] [PubMed] [Google Scholar]
- 5.Gerald WL, Rosai J, Ladanyi M: Characterization of the genomic breakpoint and chimeric transcripts in the EWS-WT1 gene fusion of desmoplastic small round cell tumor. Proc Natl Acad Sci USA 1995, 92:1028-1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim J, Lee K, Pelletier J: The desmoplastic small round cell tumor t(11;22) translocation produces EWS/WT1 isoforms with differing oncogenic properties. Oncogene 1998, 16:1973-1979 [DOI] [PubMed] [Google Scholar]
- 7.Lee SB, Kolquist KA, Nichols K, Englert C, Maheswaran S, Ladanyi M, Gerald WL, Haber DA: The EWS-WT1 translocation product induces PDGFA in desmoplastic small round-cell tumour. Nat Genet 1997, 17:309-313 [DOI] [PubMed] [Google Scholar]
- 8.Karnieli E, Werner H, Rauscher FJ, 3rd, Benjamin LE, LeRoith D: The IGF-I receptor gene promoter is a molecular target for the Ewing’s sarcoma-Wilms’ tumor 1 fusion protein. J Biol Chem 1996, 271:19304-19309 [DOI] [PubMed] [Google Scholar]
- 9.Finkeltov I, Kuhn S, Glaser T, Idelman G, Wright JJ, Roberts CT, Jr, Werner H: Transcriptional regulation of IGF-I receptor gene expression by novel isoforms of the EWS-WT1 fusion protein. Oncogene 2002, 21:1890-1898 [DOI] [PubMed] [Google Scholar]
- 10.Wong JC, Lee SB, Bell MD, Reynolds PA, Fiore E, Stamenkovic I, Truong V, Oliner JD, Gerald WL, Haber DA: Induction of the interleukin-2/15 receptor β-chain by the EWS-WT1 translocation product. Oncogene 2002, 21:2009-2019 [DOI] [PubMed] [Google Scholar]
- 11.Palmer RE, Lee SB, Wong JC, Reynolds PA, Zhang H, Truong V, Oliner JD, Gerald WL, Haber DA: Induction of BAIAP3 by the EWS-WT1 chimeric fusion implicates regulated exocytosis in tumorigenesis. Cancer Cell 2002, 2:497-505 [DOI] [PubMed] [Google Scholar]
- 12.Emi N, Kitaori K, Seto M, Ueda R, Saito H, Takahashi T: Isolation of a novel cDNA clone showing marked similarity to ME491/CD63 superfamily. Immunogenetics 1993, 37:193-198 [DOI] [PubMed] [Google Scholar]
- 13.Li SH, McInnis MG, Margolis RL, Antonarakis SE, Ross CA: Novel triplet repeat containing genes in human brain: cloning, expression, and length polymorphisms. Genomics 1993, 16:572-579 [DOI] [PubMed] [Google Scholar]
- 14.Stipp CS, Kolesnikova TV, Hemler ME: Functional domains in tetraspanin proteins. Trends Biochem Sci 2003, 28:106-112 [DOI] [PubMed] [Google Scholar]
- 15.Boucheix C, Rubinstein E: Tetraspanins. Cell Mol Life Sci 2001, 58:1189-1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishio J, Iwasaki H, Ishiguro M, Ohjimi Y, Fujita C, Yanai F, Nibu K, Mitsudome A, Kaneko Y, Kikuchi M: Establishment and characterization of a novel human desmoplastic small round cell tumor cell line, JN-DSRCT-1. Lab Invest 2002, 82:1175-1182 [DOI] [PubMed] [Google Scholar]
- 17.Takagi S, Fujikawa K, Imai T, Fukuhara N, Fukudome K, Minegishi M, Tsuchiya S, Konno T, Hinuma Y, Yoshie O: Identification of a highly specific surface marker of T-cell acute lymphoblastic leukemia and neuroblastoma as a new member of the transmembrane 4 superfamily. Int J Cancer 1995, 61:706-715 [DOI] [PubMed] [Google Scholar]
- 18.Nakagama H, Heinrich G, Pelletier J, Housman DE: Sequence and structural requirements for high-affinity DNA binding by the WT1 gene product. Mol Cell Biol 1995, 15:1489-1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drummond IA, Rupprecht HD, Rohwer-Nutter P, Lopez-Guisa JM, Madden SL, Rauscher FJ, 3rd, Sukhatme VP: DNA recognition by splicing variants of the Wilms’ tumor suppressor, WT1. Mol Cell Biol 1994, 14:3800-3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J, Lee K, Pelletier J: The DNA binding domains of the WT1 tumor suppressor gene product and chimeric EWS/WT1 oncoprotein are functionally distinct. Oncogene 1998, 16:1021-1030 [DOI] [PubMed] [Google Scholar]
- 21.Hosokawa Y, Ueyama E, Morikawa Y, Maeda Y, Seto M, Senba E: Molecular cloning of a cDNA encoding mouse A15, a member of the transmembrane 4 superfamily, and its preferential expression in brain neurons. Neurosci Res 1999, 35:281-290 [DOI] [PubMed] [Google Scholar]
- 22.Zemni R, Bienvenu T, Vinet MC, Sefiani A, Carrie A, Billuart P, McDonell N, Couvert P, Francis F, Chafey P, Fauchereau F, Friocourt G, Portes V, Cardona A, Frints S, Meindl A, Brandau O, Ronce N, Moraine C, Bokhoven H, Ropers HH, Sudbrak R, Kahn A, Fryns JP, Beldjord C, Chelly J: A new gene involved in X-linked mental retardation identified by analysis of an X;2 balanced translocation. Nat Genet 2000, 24:167-170 [DOI] [PubMed] [Google Scholar]
- 23.Ono Y, Fukuhara N, Yoshie O: Transcriptional activity of TAL1 in T cell acute lymphoblastic leukemia (T-ALL) requires RBTN1 or -2 and induces TALLA1, a highly specific tumor marker of T-ALL. J Biol Chem 1997, 272:4576-4581 [DOI] [PubMed] [Google Scholar]
- 24.Yauch RL, Hemler ME: Specific interactions among transmembrane 4 superfamily (TM4SF) proteins and phosphoinositide 4-kinase. Biochem J 2000, 351:629-637 [PMC free article] [PubMed] [Google Scholar]
- 25.Berditchevski F: Complexes of tetraspanins with integrins: more than meets the eye. J Cell Sci 2001, 114:4143-4151 [DOI] [PubMed] [Google Scholar]
- 26.Yauch RL, Berditchevski F, Harler MB, Reichner J, Hemler ME: Highly stoichiometric, stable, and specific association of integrin α3β1 with CD151 provides a major link to phosphatidylinositol 4-kinase, and may regulate cell migration. Mol Biol Cell 1998, 9:2751-2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yáñez-Mó M, Alfranca A, Cabañas C, Marazuela M, Tejedor R, Ursa MA, Ashman LK, de Landázuri MO, Sánchez-Madrid F: Regulation of endothelial cell motility by complexes of tetraspan molecules CD81/TAPA-1 and CD151/PETA-3 with α3β1 integrin localized at endothelial lateral junctions. J Cell Biol 1998, 141:791-804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang XA, Kazarov AR, Yang X, Bontrager AL, Stipp CS, Hemler ME: Function of the tetraspanin CD151-α6β1 integrin complex during cellular morphogenesis. Mol Biol Cell 2002, 13:1-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sugiura T, Berditchevski F: Function of α3β1-tetraspanin protein complexes in tumor cell invasion. Evidence for the role of the complexes in production of matrix metalloproteinase 2 (MMP-2). J Cell Biol 1999, 146:1375-1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lammerding J, Kazarov AR, Huang H, Lee RT, Hemler ME: Tetraspanin CD151 regulates α6β1 integrin adhesion strengthening. Proc Natl Acad Sci USA 2003, 100:7616-7621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Testa JE, Brooks PC, Lin JM, Quigley JP: Eukaryotic expression cloning with an antimetastatic monoclonal antibody identifies a tetraspanin (PETA-3/CD151) as an effector of human tumor cell migration and metastasis. Cancer Res 1999, 59:3812-3820 [PubMed] [Google Scholar]
- 32.Claas C, Seiter S, Claas A, Savelyeva L, Schwab M, Zoller M: Association between the rat homologue of CO-029, a metastasis-associated tetraspanin molecule and consumption coagulopathy. J Cell Biol 1998, 141:267-280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Billuart P, Bienvenu T, Ronce N, des Portes V, Vinet MC, Zemni R, Roest Crollius H, Carrie A, Fauchereau F, Cherry M, Briault S, Hamel B, Fryns JP, Beldjord C, Kahn A, Moraine C, Chelly J: Oligophrenin-1 encodes a rhoGAP protein involved in X-linked mental retardation. Nature 1998, 392:923-926 [DOI] [PubMed] [Google Scholar]
- 34.Allen KM, Gleeson JG, Bagrodia S, Partington MW, MacMillan JC, Cerione RA, Mulley JC, Walsh CA: PAK3 mutation in nonsyndromic X-linked mental retardation. Nat Genet 1998, 20:25-30 [DOI] [PubMed] [Google Scholar]
- 35.Kutsche K, Yntema H, Brandt A, Jantke I, Nothwang HG, Orth U, Boavida MG, David D, Chelly J, Fryns JP, Moraine C, Ropers HH, Hamel BC, van Bokhoven H, Gal A: Mutations in ARHGEF6, encoding a guanine nucleotide exchange factor for Rho GTPases, in patients with X-linked mental retardation. Nat Genet 2000, 26:247-250 [DOI] [PubMed] [Google Scholar]
- 36.Zhang XA, Lane WS, Charrin S, Rubinstein E, Liu L: EWI2/PGRL associates with the metastasis suppressor KAI1/CD82 and inhibits the migration of prostate cancer cells. Cancer Res 2003, 63:2665-2674 [PubMed] [Google Scholar]
- 37.Azorsa DO, Moog S, Cazenave JP, Lanza F: A general approach to the generation of monoclonal antibodies against members of the tetraspanin superfamily using recombinant GST fusion proteins. J Immunol Methods 1999, 229:35-48 [DOI] [PubMed] [Google Scholar]
- 38.Kobayashi T, Vischer UM, Rosnoblet C, Lebrand C, Lindsay M, Parton RG, Kruithof EK, Gruenberg J: The tetraspanin CD63/lamp3 cycles between endocytic and secretory compartments in human endothelial cells. Mol Biol Cell 2000, 11:1829-1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sincock PM, Fitter S, Parton RG, Berndt MC, Gamble JR, Ashman LK: PETA-3/CD151, a member of the transmembrane 4 superfamily, is localised to the plasma membrane and endocytic system of endothelial cells, associates with multiple integrins and modulates cell function. J Cell Sci 1999, 112:833-844 [DOI] [PubMed] [Google Scholar]
- 40.Escola JM, Kleijmeer MJ, Stoorvogel W, Griffith JM, Yoshie O, Geuze HJ: Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J Biol Chem 1998, 273:20121-20127 [DOI] [PubMed] [Google Scholar]
- 41.Lamparski HG, Metha-Damani A, Yao JY, Patel S, Hsu DH, Ruegg C, Le Pecq JB: Production and characterization of clinical grade exosomes derived from dendritic cells. J Immunol Methods 2002, 270:211-226 [DOI] [PubMed] [Google Scholar]
- 42.Zitvogel L, Regnault A, Lozier A, Wolfers J, Flament C, Tenza D, Ricciardi-Castagnoli P, Raposo G, Amigorena S: Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat Med 1998, 4:594-600 [DOI] [PubMed] [Google Scholar]
- 43.Odintsova E, Sugiura T, Berditchevski F: Attenuation of EGF receptor signaling by a metastasis suppressor, the tetraspanin CD82/KAI-1. Curr Biol 2000, 10:1009-1012 [DOI] [PubMed] [Google Scholar]
- 44.Bonifacino JS, Traub LM: Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem 2003, 72:395-447 [DOI] [PubMed] [Google Scholar]
- 45.Stipp CS, Kolesnikova TV, Hemler ME: EWI-2 is a major CD9 and CD81 partner and member of a novel Ig protein subfamily. J Biol Chem 2001, 276:40545-40554 [DOI] [PubMed] [Google Scholar]
- 46.Stipp CS, Orlicky D, Hemler ME: FPRP, a major, highly stoichiometric, highly specific CD81- and CD9-associated protein. J Biol Chem 2001, 276:4853-4862 [DOI] [PubMed] [Google Scholar]