Abstract
Id4 belongs to a family of helix-loop-helix (HLH) proteins that impact cellular growth and differentiation via regulation of basic HLH transcription factors. Herein the rat Id4 gene was cloned (GenBank Accession No. AF468681). The expression of rat Id4 was examined in rat mammary gland tumors inducedby 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP), a carcinogen found in the human diet. By real-time polymerase chain reaction analysis, relative expression of Id4 mRNA in carcinomas, adenomas, and normal tissue was 27, 6, and 1, respectively. Immunohistochemical analysis indicated statistically elevated nuclear expression for Id4 protein in carcinomas in comparison to adenomas and normal mammary gland. In carcinomas, Id4 nuclear expression was positively correlated with proliferation, invasiveness, and tumor weight (Fisher Exact Test or Spearman Correlation, P < 0.05). The consequence of enforced expression of Id4 on mammary epithelial cell proliferation, differentiation, and growth in soft agar was examined in HC11 cells, a well-characterized model for studying various aspects of mammary epithelial cell biology. After transient and stable transfection of HC11 cells, Id4 overexpression increased cell proliferation and inhibited lactogenic hormone-mediated differentiation as revealed by inhibition of β-casein promoter activity and β-casein expression. In addition, enforced expression of Id4 in HC11 cells induced a statistically significant increase in colony growth in soft agar. The results implicate Id4 in rat mammary gland carcinogenesis and suggest that Id4 may contribute to carcinogenesis by inhibiting mammary epithelial cell differentiation and stimulating mammary epithelial cell growth.
Basic helix-loop-helix (bHLH) transcription factors play an important role in cellular development and differentiation in virtually all eukaryotic organisms. 1 The Id proteins (“inhibitors of differentiation or DNA binding”) were identified as helix-loop-helix proteins lacking a basic domain that antagonize the action of bHLH transcription factors by forming DNA binding incompetent heterodimers. 1,2 To date, four Id family members (Id1–4) have been identified in mammals 1,2 . Id proteins coordinate many cellular functions including cell growth, differentiation, cell cycle progression, and invasiveness. 2-6 The mechanism of action of Id proteins is largely regarded to be via dominant-negative regulation of bHLH transcription factors. 2
Biochemical and genetic data suggest that Id proteins are in general positive regulators of cell proliferation and negative regulators of differentiation. 1-6 Id expression is regulated in a cell cycle-dependent manner and is necessary for G1-S transition. 1,7 In addition to binding to bHLH transcription factors, certain Id genes bind to other classes of transcription factors and cell cycle regulators. 1 One member of the Id family, Id2 has been shown to directly interact with the retinoblastoma tumor suppressor protein reversing the growth arrest and cell cycle block elicited by this protein. 8
Of the four identified Id proteins, the least is known about Id4. Mouse and human Id4 were cloned in the mid 1990s. 9-11 In adult mice, the expression of Id4 was shown to be distinct from Id1–3. 9,12,13 Id4 has been implicated in the regulation of differentiation of several tissue types including adipocytes, oligodendrocytes, astrocytes, and spermatogonia. 13-16 In addition, one previous study has shown elevated nuclear expression of Id4 during carcinogenesis specifically in seminomas, a malignant counterpart of early spermatogonia. 13
Id genes are regarded as prime candidates for oncogenesis. 1,2,4 Indeed, deregulated expression of Id mRNA and protein has been observed in various human malignancies, including breast cancer. 13,17-23 Both Id1 and Id2 have been shown to regulate mammary epithelial cell proliferation, differentiation, and apoptosis. 24-27 To date, Id1 has been implicated in the aggressive phenotype of human breast cancer cells and is more frequently expressed in infiltrating carcinomas than ductal carcinomas in situ. 21 Whether Id4 plays a role in human breast cancer is not yet known. However, Id4 has been recently shown to regulate the expression of Brca1, a gene linked to human breast cancer susceptibility. 28 Brca1 also appears to regulate the expression of Id4, and it has been speculated that a Brca1-Id4 regulatory loop may be disrupted in human breast cancer. 29 The co-regulation of Id4 and Brca1 raises the intriguing possibility that Id4 influences mammary gland carcinogenesis.
In previous studies using cDNA microarray analysis, we examined the expression profiles in rat mammary gland cancers induced by the experimental carcinogen 7,12-dimethylbenz[a]anthracene (DMBA) and the cooked meat-derived carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP). 30 An expressed sequence tag (EST) clone (RGIDV72) was highly overexpressed in both PhIP and DMBA-induced rat mammary gland carcinomas with nine of nine tumors showing at least fivefold higher mRNA expression than normal mammary gland. As described herein, this EST was cloned and identified as the rat Id4 gene (GenBank Accession No. AF468681). Rat Id4 was highly overexpressed in rat mammary gland carcinomas and its expression was correlated with tumor malignancy. Id4 was further shown to regulate the growth and differentiation of mammary gland epithelial cells in culture. The findings implicate Id4 in rat mammary gland carcinogenesis and suggest that Id4 overexpression may impact carcinogenesis by inhibiting mammary epithelial cell differentiation and augmenting cell growth.
Materials and Methods
Rat Mammary Gland Carcinomas and Normal Tissue
Archival Sprague-Dawley rat mammary gland tumors (tubulopapillary carcinomas and adenomas) induced previously with PhIP 30-32 were used in this study. Normal mammary gland tissue was collected from mature virgin (150 days old), mid-pregnant (10 to 15 days), and lactating (with 10- to 12-day old pups) Sprague-Dawley rats; in addition, the glands from Sprague-Dawley rats were collected on day 6 of involution. Tumor histopathology and microscopic invasion were assessed as described previously. 32
Cloning of the Rat Id4 Gene
Cloning of rat Id4 was carried out using a PCR-cloning strategy. Total RNA and high molecular weight DNA were isolated from normal mammary glands using TRIzol extraction reagent (Invitrogen, Carlsbad, CA) and phenol/chloroform method, respectively. First-strand cDNA was synthesized using SuperScript First-Strand Synthesis Kit (Invitrogen). To obtain the exon sequence, six pairs of overlapping primers were designed from rat ESTs which showed high homology (≥ 90%) to mouse Id4 by BLAST search as follows: F1 5′-acagcgatccaccttagtc-3′ and R1 5′-acaagcggtagagcgagct-3′; F2 5′-tcgatttctggagcttggaac-3′ and R2 5′-gagtgtagcagtcgttca-3′; F3 5′-tgtgcctgcagtgcgatatg-3′ and R3 5′-agctcagcggcagagaatg-3′; F4 5′-ttacggcgctcaacactgac-3′ and R4 5′-acggtgaatgcttgtgaactg-3′; F5 5′-acgttgtctcggcttgtcattc-3′ and R5 5′-tctgagcctcagatactagatac-3′; F6 5′-agtgagtgacatttcataccctg-3′ and R6 5′-aagacagcagttctgtagactc-3′. To amplify the intron sequences, the above F4 and R3 were used for intron 1 and the following one pair of primers was designed for intron II based on the exon sequence: iF 5′-acagcattctctgccgctga-3′ and iR 5′-gaatgacaagccgagacaac-3′. PCR fragments were cloned into the pCR 2.1 vector using the TA Cloning Kit (Invitrogen). Plasmid DNA was isolated using QIAprep Spin Miniprep Kit or QIAGEN Plasmid Maxi Kit (QIAGEN, Valencia, CA). Sequencing was performed using ABI PRISM Big Dye Terminator Sequencing Kit in an ABI PRISM 377 DNA sequencer (Applied Biosystems, Foster City, CA).
Immunohistochemistry
Paraffin-embedded sections from the tumors and normal mammary glands were immunohistochemically stained using LSAB2 system (DAKO, Carpinteria, CA). The optimal dilutions for antibodies against Id4 (Santa Cruz Biotechnology, Santa Cruz, CA) and proliferating cell nuclear antigen (PCNA) (DAKO) were 1:50 and 1:100, respectively. Omission of the primary antibody was used as a negative control. All slides were examined and scored in a blinded fashion without knowledge of the sample at the time of assessment. Nuclear immunoreactivity was expressed as the percentage of nuclear-stained positive cells by evaluating 5 to 10 randomly selected high-power fields (×400) (2000 to 5000 total cells counted).
Real-Time Quantitative PCR Analysis
Real-time PCR analysis was carried out using the TaqMan7 Gold RT-PCR Kit on the ABI Prism 7900 Sequence Detection System (PE Applied Biosystems, Foster City, CA) as described in detail previously. 33 Briefly, the primers and probes were designed using Perkin Elmer Primer Express7 software as forward primer, 5′-aggctcgtgcccaccat-3′; reverse primer, 5′-cgataacgtgctgcaggatct-3′; and probe, CCGCCCAACAAGAAAGTCAGCAAAGTG. The threshold cycles (CT) were recorded for Id4 and GAPDH. GAPDH was used for normalization. Relative gene expression (calculated as 2−Δ ΔCT) was derived by the method outlined in ABI Prism Sequence Detection System User Bulletin #2. For statistical analysis, comparison was done between Δ CT of the tumors and control tissue. The range for the relative expression was derived from the SE of the ΔCT values and calculated as the exponential function of −Δ Δ CT ± SE.
Cell Culture, Luciferase Assay of β-Casein Gene Promoter Activity, and Semi-Quantitative RT-PCR
HC11 mammary epithelial cell line (a subclone of COMMA-1D cells derived from mid-pregnant mice) and HC11-Lux cells (HC11 cells stably transfected with a β-casein promoter luciferase construct (p-344/-1βc-Lux)) were kindly provided by David S. Salomon (National Cancer Institute, Bethesda, MD) and Nancy E. Hynes (Friedrich Miescher Institute, Basel, Switzerland). 34,35 Both cell lines were routinely maintained in growth medium which consisted of RPMI 1640 (Invitrogen), 10% heat-inactivated fetal bovine serum (FBS, Invitrogen), 5 μg/ml bovine insulin (Invitrogen), 10 ng/ml epidermal growth factor (EGF, Invitrogen) and 50 μg/ml gentamicin (Invitrogen). To induce cell differentiation, cells were grown to confluence and maintained in growth medium for 1 or 2 days and then cultured for 48 hours in differentiation medium containing RPMI 1640, 10% heat-inactivated FBS, 5 μg/ml bovine insulin, 1 μmol/L dexamethasone (Sigma, St. Louis, MO), 5 μg/ml bovine prolactin (Sigma) and 50 μg/ml gentamicin. 34-36 β-casein gene expression, a marker of mammary epithelial cell differentiation, was analyzed using semi-quantitative RT-PCR and β-casein gene promoter activity. Semi-quantitative RT-PCR was performed for 20 cycles and the PCR fragments were separated on 2% agarose gel. The RT-PCR primers used for the β-casein gene were 5′-actgtatcctctgagactg-3 and 5′-tctaggtactgcagaaggtc-3. The GAPDH primers was purchased from Clontech (Palo Alto, CA) and used for internal control. β-casein gene promoter activity was assayed in triplicate using the luciferase assay system (Promega, Madison, WI) with activity being expressed as light units/μg protein. 34-36
Construction of Id4 Expression Vectors, Transient Transfection, and Generation of Stably Transfected Cell Lines
The coding region of the rat Id4 was amplified using primers 5′-aggaagcgcgcgatgaag-3′ and 5′-acacctggccaacgcagct-3′, and cloned into the Srf enzyme restriction site of the pCMV-Script mammalian expression vector (Stratagene, Cedar Creek, TX) in the sense-orientation. The inserted sequence and orientation were verified using SacII enzyme restriction and sequencing. Transfection was carried out for 24 hours in growth medium using Lipofectamine and Plus reagents with 1.6 μg of pCMV-Script-Id4 construct plasmid DNA according to Invitrogen’s protocol. Cells were cultured for an additional 24 hours in growth medium only and cell growth and colony-forming efficiency were subsequently assayed. After culturing in differentiation medium for 48 hours, the effect of transient Id4 overexpression on cell differentiation was evaluated by measuring β-casein promoter activity in HC11-Lux cells with the luciferase assay. Transfection was confirmed by Western blot analysis of Id4 protein and semi-quantitative RT-PCR analysis of the rat Id4.
To establish stably transfected cell lines, cells were selected using geneticin (G418, 600 μg/ml, Invitrogen). Individual cell clones were trypsinized using a cloning cylinder and re-cultured under continuous selection with G418. The expression levels of Id4 protein in these clones were determined by Western blot analyses. The effect of Id4 overexpression on cell growth and colony-forming efficiency was assayed as described below and the effect on cell differentiation was evaluated by Northern blot analysis of β-casein 36 expression.
Northern and Western Blot Analysis
Both Northern and Western blot analyses were carried out as described previously. 36 The β-casein gene probe was generated by RT-PCR using the primers described above, and the probe was 32P-labeled using ready-to-go DNA Labeling Beads (Amersham Biosciences, Piscataway, NJ) as described previously. 36 The optimal dilutions of Id4 primary and rabbit secondary antibodies in the Western blot analysis were 1:200 and 1:2000, respectively. As loading control, the membrane was stripped and re-probed using antibody against β-actin (Santa Cruz).
Cell Proliferation and Soft Agar Assay
Cell proliferation was measured using CellTiter 96 Aqueous One Solution Cell Proliferation Assay (Promega). Cells were seeded into the 96-well plates in growth medium at a starting density per well of 1 × 104 cells and cell proliferation measured at 24-hour intervals. Colony-forming efficiency of both transiently and stably transfected cells was analyzed using soft agar assay with a 0.66% agarose base and a 0.33% top soft agar layer (containing 4 × 105 cells) in 60-mm-diameter dishes. The dishes were incubated for 2 weeks and stained overnight with nitrobluetetrazolium before counting (Artec Colony Counter, Farmingdale, NY).
Statistical Analysis
Statistical analysis was carried out with SigmaStat Statistical Software Version 2 (Jandel, San Rafael, CA).
Results
Molecular Cloning of Rat Id4
In previous studies using cDNA microarray analysis, a rat EST (clone RGIDV72, GenBank Accession No. AW916745) was observed to be overexpressed in chemically induced rat mammary gland carcinomas. 30 BLAST searching with the forward and reverse sequences of the EST (634 bp) showed no known rat genes with high homology. However, the reverse sequence of this clone was highly homologous with a region on Id4 exon 3 from mouse (90%) and human (89%) suggesting that the unknown rat EST was a fragment of rat Id4. To clone the rat Id4 gene, a BLAST homology search was then performed for all published rat EST sequences in GenBank, EMBL, and DDBJ databases using the mouse Id4 cDNA sequence as a template (www.ncbi.nlm.nih.gov). A set of rat EST sequences was subsequently obtained which showed a high homology (>90%) to different areas of mouse Id4 cDNA, although several gaps in the sequence were evident among the aligned ESTs. Six pairs of overlapping primers were designed based on the aligned EST sequences and after sequencing these fragments, the cDNA sequence of rat Id4 was obtained. To determine the intron locations, the sequences for human, mouse and rat Id4 were compared, and PCR primers flanking the rat introns were designed. After PCR and cloning, 2881 nucleotides of the genomic sequence of rat Id4 were sequenced (Figure 1A) ▶ . The DNA sequence of rat Id4 was reported to GenBank (Accession No. AF468681).
Figure 1.

Rat Id4 gene sequence and structure (GenBank Accession No. AF468681). A: Nucleotide sequence of rat Id4. Rat Id4 contained three exons and two introns with stop codon in exon 2. A portion of the upstream promoter region (−1 to −311) was also cloned and sequenced. In bold are the start (ATG) and stop (TGA) codons, and the three potential polyadenylational signals (AATAAA). B: Comparison of the Id4 protein structure among human, mouse, and rat. The helix-loop-helix domain is depicted. As is characteristic of the Id family, there was no basic region adjacent to the HLH motif. The rat Id4 protein shows a high homology to mouse (97.52%) and human (95.65%) and the HLH domain is highly conserved. Only one amino acid in helix 1 of rat Id4 protein was different from mouse and human. The HLH domain is highly conserved within the rat Id family. Distinct from the other Id proteins, Id4 displayed an alanine-rich region adjacent to the HLH motif.
Genomic Sequence and Protein Structure of Id4
The Id4 gene encodes a 161 amino acid protein with the coding region encompassing exon 1 and a portion of exon 2 (Figure 1A) ▶ . The 3′ untranslated region includes part of exons 2 and 3. The first intron resides within the protein-coding region, and the second intron lay within the 3′ untranslated region. The 5′-flanking region is rich in GC bases and no TATAA or TATTA box was seen in the putative promoter region upstream of the gene. The helix-loop-helix (HLH) domain is situated in the first exon. As is characteristic of the Id family, there was no basic region adjacent to the HLH motif. In the 3′ untranslated region, three potential polyadenylational signals (AATAAA) were observed. The rat Id4 protein is highly homologous to mouse and human Id4 with a percent similarity of 97.5% and 95.7%, respectively (Figure 1B) ▶ . Among all rat Id family members (Id1–4), the HLH domain was highly conserved, particularly at helix 2. The conserved loop structure of different Id proteins indicated that the loop might play an important role in specifying the correct dimerization partner. Distinct from the other Id proteins, Id4 displayed an alanine-rich region adjacent to the HLH motif.
Id4 Expression in Normal Rat Mammary Gland and Carcinogen-Induced Tumors
Expression of Id4 protein in normal rat mammary gland and chemically induced rat mammary gland carcinomas and adenomas were examined by immunohistochemistry. Id4 protein, which could be observed in cytoplasm and nuclei, varied with the stage of mammary gland differentiation among virgin, pregnant, lactating, and involuting rats (Figure 2) ▶ . In mature virgin rats, Id4 protein was largely located in the cytoplasm of epithelial cells (Figure 2A) ▶ . Less than 5% of nuclei showed immunoreactivity (3.6 ± 0.9, mean ± SE, N = 10). However, nuclear immunoreactivity in epithelial cells of mammary glands from mid-pregnant rats was nearly 50% (47 ± 1.1, N = 3) (Figure 2B) ▶ . In lactating rats, less than 1% of epithelial cell nuclei were stained (0.7 ± 0.1, N = 3) and cytoplasmic staining was weak (Figure 2C) ▶ . In the involuting rat mammary glands, only a weak cytoplasmic staining (N = 5) was observed (Figure 2D) ▶ . In PhIP-induced rat mammary gland tumors, Id4 nuclear-positive cells were diffusely distributed throughout the carcinomas (Figure 2E) ▶ . In the majority of carcinomas examined (72%, 43 of 60 carcinomas), Id4 protein expression was detected in at least 10% of tumor epithelial cell nuclei. In contrast, only three adenomas (two tubular adenomas and one fibroadenoma) showed nuclear staining. The majority of adenomas (77%, 10 of 13) showed less than 10% of nuclei stained positive, and adenomas showed either no clear staining or cytoplasmic staining in comparison to the negative control (Figure 2, F and G ▶ , respectively). Consistent with immunohistochemistry, carcinomas had higher expression of Id4 message than did control mammary gland tissue. Microarray analysis of five PhIP-induced carcinomas showed increased relative expression of Id4 in comparison to normal mammary gland (Figure 2H) ▶ .
Figure 2.
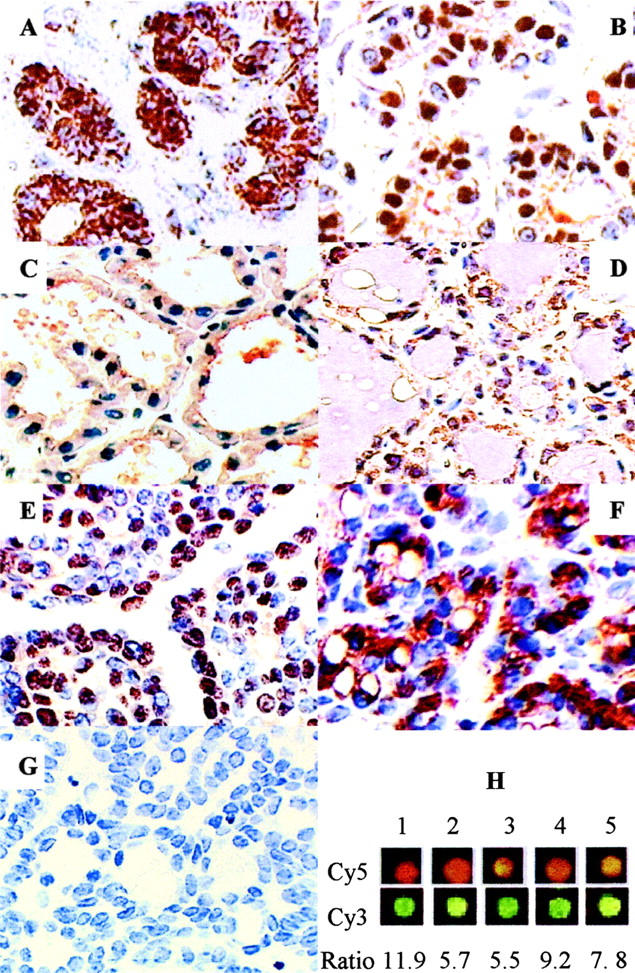
Immunohistochemical analysis of Id4 expression in normal rat mammary gland and tumors. A: Mammary gland from mature virgin rat shows largely cytoplasmic staining of Id4. B: Mammary gland from mid-pregnant rat shows ∼50% nuclear expression. C: Mammary gland from lactating rat (with 12-day-old pups) shows little nuclear staining and weak cytoplasmic staining. D: Mammary gland from involuted rat shows weak cytoplasmic staining. E: Representative carcinoma from PhIP-treated rats with high nuclear expression of Id4. F: Representative tubular adenoma from a PhIP-treated rat showing cytoplasmic Id4 staining. G: Negative control showing immunostaining without primary antibody of a representative carcinoma. The nuclei are counterstained using Mayer’s hematoxylin. Magnification, ×400. H: Microarray analysis of five PhIP-induced carcinomas showed increased relative expression of Id4 in comparison to normal mammary gland. Analysis included both Cy5 (top panel) or Cy3 (bottom panel) labeling of tumor RNA as described previously 30 and average increase in Id4 expression ranged from 5.5 to 11.9.
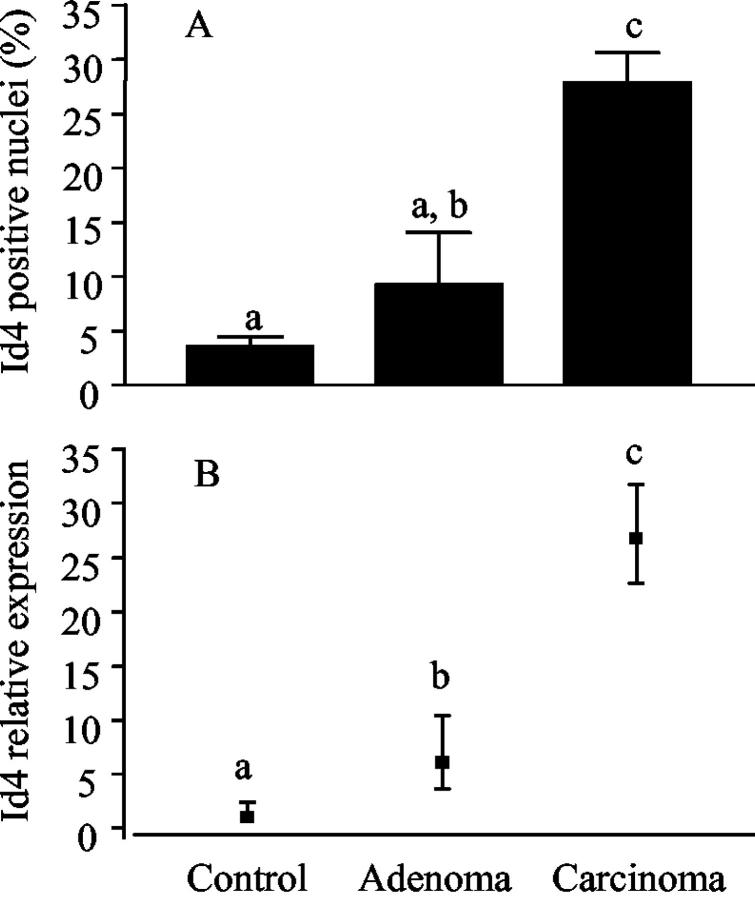
The percentage of Id4-positive nuclei and the mRNA expression detected by real-time PCR were further quantified in normal mammary gland, adenomas, and carcinomas (Figure 3) ▶ . The mean percentage of Id4-positive nuclei in carcinomas was over 3- and 7.5-fold higher than in adenomas and control mammary gland, respectively (Figure 3A) ▶ . By real-time PCR analysis, Id4 mRNA expression was highest in carcinomas, followed by adenomas, and lowest in control mammary gland with an average relative expression of 27, 6, and 1, respectively (Figure 3B) ▶ . In carcinomas, Id4 nuclear expression was positively correlated with the tumor cell proliferation as measured by PCNA labeling index (Table 1) ▶ . In addition, Id4 nuclear expression was statistically associated with a higher frequency of microscopic stromal invasion and a greater tumor weight.
Figure 3.
Expression of Id4 in normal rat mammary gland and PhIP-induced rat mammary gland tumors. A: Percentage of Id4-positive nuclei in control (normal rat mammary gland), adenomas, and carcinomas as detected immunohistochemically. Values are the means ± SE of 10, 13, and 60 samples, respectively. B: Real-time PCR analysis of Id4 expression in normal mammary gland, adenomas, and carcinomas. Values are the exponential function of −Δ ΔCT ± SE of N = 3, 5, and 23 samples for control, adenomas, and carcinomas, respectively. Distinct letters above the data indicate significantly different (one-way analysis of variance on ranks or one-way analysis of variance, P < 0.05).
Table 1.
Correlation Between Id4 Nuclear Expression and Tumor Cell Proliferation, Invasion, and Weight in PhIP-Induced Mammary Gland Carcinoma
| Tumor parameter | Id4 nuclear expression | Fisher test (p) | Spearman correlation | |
|---|---|---|---|---|
| >10% | ≤10% | |||
| PCNA labeling index (%) | ||||
| ≥30% (N = 26) | 92% | 8% | ||
| <30% (N = 34) | 56% | 44% | 0.003 | 0.424, p < 0.05 |
| Tumor invasion | ||||
| Yes (N = 14) | 93% | 7% | ||
| No (N = 26) | 58% | 42% | 0.03 | 0.317, p < 0.05 |
| Tumor weight (gram) | ||||
| ≥ 2.0 (N = 34) | 79% | 21% | ||
| < 2.0 (N = 20) | 50% | 50% | 0.035 | 0.293, p < 0.05 |
Fisher Exact Test was applied to tumors showing a cut-off of > or ≤ 10% Id4 nuclear expression and categorized for labeling index, invasion and weight as indicated. Spearman Rank Order Correlation was applied to all Id4 nuclear expression values without categorization. N, number of carcinomas examined.
Effect of Id4 Overexpression on Mammary Epithelial Cell Growth and Differentiation
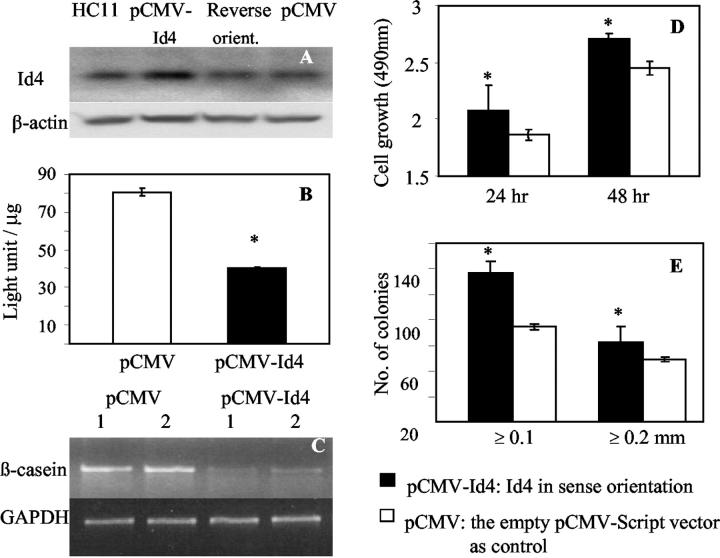
To examine in more detail the consequence of Id4 overexpression on mammary epithelial cells, studies were carried out in HC11 cells transiently and stably transfected with Id4. This mammary epithelial cell line was chosen because it is a non-tumorigenic, well-characterized cell line capable of in vitro differentiation. In the presence of lactogenic hormones (ie, differentiation medium), HC11 cells undergo differentiation with the production of milk proteins such as β-casein. The increased expression of β-casein is a marker of HC11 cell differentiation, and the HC11-Lux cells (HC11 cells harboring the β-casein promoter-luciferase construct) provide a facile means to evaluate β-casein expression. 34,35 By Western blot analysis, endogenous Id4 expression was detected in HC11 cells cultured in growth medium, however, after treatment with differentiation medium, Id4 protein declined to barely detectable levels (data not shown). HC11-Lux cells transiently transfected with the pCMV-Id4 construct showed elevated expression of the Id4 protein, whereas cells transfected with Id4 in the reverse orientation or empty vector control (pCMV) did not show elevated Id4 expression (Figure 4A) ▶ . After culturing in differentiation medium, the β-casein promoter activity (Figure 4B) ▶ and mRNA level (Figure 4C) ▶ were significantly lower in cells transfected with pCMV-Id4 than in cells transfected with the empty vector indicating that enforced expression of Id4 inhibited mammary epithelial cell differentiation. Cell proliferation was significantly higher in cells transiently transfected with pCMV-Id4 than with the empty vector control (Figure 4D) ▶ . Furthermore, colony-forming efficiency of pCMV-Id4 transfected cells in the soft agar assay was 1.6- and 1.3-fold higher than empty vector control for ≥0.1- and ≥0.2-mm diameter colonies, respectively (Figure 4E) ▶ .
Figure 4.
Effect of Id4 overexpression on HC11 cell growth and differentiation after transient transfection. A: Western blot analysis of Id4 showing higher expression of Id4 after transient transfection of HC11-Lux cells with Id4 (pCMV-Id4) than with pCMV empty vector or pCMV with Id4 in the reverse orientation (orient.). B: Luciferase assay of β-casein promoter activity in HC11-Lux cells cultured in differentiation medium showing inhibition by enforced Id4 expression (values are means ± SD, N = 5). C: Semi-quantitative RT-PCR showing decreased expression of β-casein mRNA. Duplicate determinations were carried out on separate pools of transfected cells. D: Cell growth assay showing higher proliferation of HC11 cells transfected with Id4. Cells were cultured in growth medium and proliferation was assayed at 24 and 48 hours, respectively (values are the means ± SD, N = 5). E: Soft agar assay indicating increased colony-forming efficiency in HC11 overexpressing Id4 in comparison to HC11 cells carrying empty vector (values are the means ± SD, N = 3). Colonies sizes were counted at ≥0.1 mm and at ≥0.2 mm. Asterisk indicates statistically significant differences (Student’s t-test, P < 0.05).
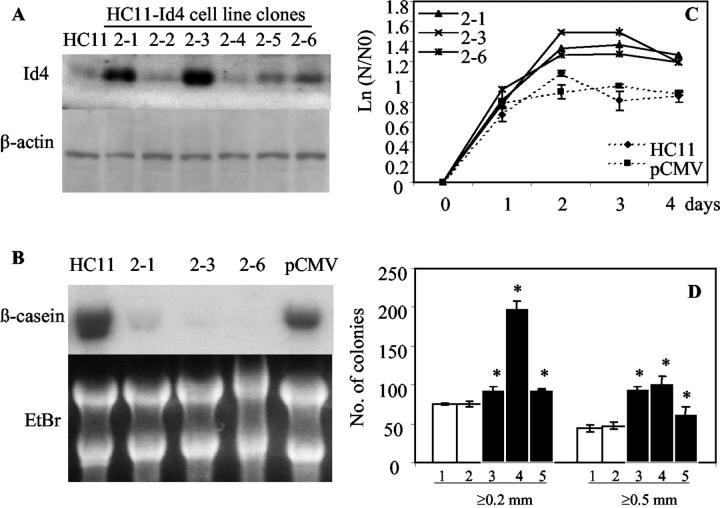
To further confirm the effect of enforced expression of Id4 on cell growth and differentiation, clonal lines of HC11 cells stably transfected with pCMV-Id4 were established. By Western blot analysis, these clones showed higher Id4 protein expression than the parental HC11 cells (Figure 5A) ▶ . Three lines (2–1, 2–3, and 2–6) that showed the highest expression of Id4 were selected for further analysis. After lactogenic hormone treatment, the induction of β-casein gene expression was markedly lower in clones expressing Id4 than in parental HC11 cells or HC11 cells stably transfected with the empty vector indicating that overexpression of Id4 inhibited differentiation (Figure 5B) ▶ . As was observed after transient transfection, all Id4-overexpressing clones showed a statistically significantly higher rate of proliferation in comparison to parental HC11 cells or HC11-pCMV cells carrying the empty vector (Figure 5C) ▶ . No difference in proliferation was observed between HC11 and HC11-pCMV cells. In the soft agar assay, colony formation was 2- to 3-fold higher in Id4-overexpressing clones than in parental or HC11-pCMV cells for both ≥0.2 and ≥0.5 mm diameter colonies; differences that were statistically significant (Figure 5D) ▶ . The total number of colonies formed by each clone appeared to be proportional to the level of Id4 protein expressed.
Figure 5.
Effect of Id4 overexpression on HC11 cell growth and differentiation after stable transfection. A: Western blot analysis of Id4 expression in established stably transfected clonal cell lines and in parental HC11 cells. The highest Id4 expression was seen in clones 2–1, 2–3, and 2–6. B: Northern blot analysis of β-casein gene expression in HC11 cells, Id4-overexpressing clones, and in a representative clone harboring the pCMV empty vector. Cells were cultured in differentiation medium for 48 hours before analysis. Equal loading was confirmed by ethidium bromide (EtBr) staining. C: Growth of cell lines was determined in growth medium over a 4-day period. Values are the means ± SD, N = 5. At 2, 3, and 4 days, proliferation was statistically higher in all Id4-overexpressing clones than in parental HC11 cells or HC11-pCMV cells with empty vector (analysis of variance, P < 0.05). N, absorbance at each time point; N0, absorbance at 2 hours after cell plating. D: Soft agar assay indicating higher colony-forming efficiency in clones overexpressing Id4 than in parental HC11 cells or HC11 cells with empty vector (values are the means ± SD, N = 3). Bars labeled 1 through 5 correspond to parental HC11 cells, HC11 cells with empty vector, clone 2–1, clone 2–3, and clone 2–6, respectively. Colonies sizes were counted at ≥0.2 mm and at ≥0.5 mm. Asterisk indicates statistically significant differences from parental or empty vector cells (analysis of variance, P < 0.05).
Discussion
Id4 has recently been proposed to be a gene involved in breast cancer because of a reciprocal regulation of Id4 and Brca1. 28,29 In the current study we demonstrate that Id4 is overexpressed both at the mRNA and protein level in PhIP-induced rat mammary gland carcinomas relative to age-matched normal mammary gland and adenomas. Furthermore, we demonstrate that the overexpression of Id4 correlates with malignancy and with proliferation, growth, and invasiveness of rat mammary gland carcinomas. These findings support a link between Id4 and breast cancer and raise the possibility that overexpression of Id4 may play a role in the development of this malignancy. Preliminary immunohistochemical studies carried out by us using human tissue microarrays (National Cancer Institute, Bethesda, MD) suggested elevated nuclear expression of Id4 protein in human breast cancers (unpublished observations). Further studies are needed to ascertain whether Id4 is similarly involved in human breast cancer.
To better understand the impact of Id4 overexpression on the mammary gland, the effect of enforced expression of Id4 was examined in HC11 mammary gland epithelial cells. Although several studies have described the ability of various Id genes to inhibit differentiation and induce cell growth in several different cell lines, 15-17,24,25,37 to our knowledge no previous study has examined the effect of enforced Id4 expression on growth and differentiation of mammary epithelial cells. The HC11 cell line proliferates in culture in the growth medium and at confluency can be induced to differentiate in the presence of medium containing lactogenic hormones. 34,35 Enforced expression of Id4 in HC11 cells was shown to enhance cell proliferation and growth in soft agar. This enhanced growth in vitro is consistent with the observed in vivo correlation of Id4 expression with proliferation of rat mammary gland carcinomas. Similar to our findings, overexpression of Id4 has been shown to enhance the growth of an ovarian cancer cell line in soft agar. 28 In addition to the effects of Id4 on cell growth, Id4 inhibited hormone-mediated differentiation of mammary epithelial cells as shown by the inhibition of β-casein promoter activity and gene expression in HC11 cells transiently and stably transfected with Id4. It is further notable that reduction in the expression of Id4 coincided with differentiation of HC11 cells and with lactation in the rat mammary gland, a differentiated state of the gland. Therefore our data support the notion that Id4 may be a regulatory factor for mammary gland differentiation.
It is well recognized that the stage of mammary gland differentiation is an important determinant in rat mammary gland carcinogenesis. In the rat model, susceptibility to chemical carcinogenesis is highest during pubescence when the gland is less differentiated and undergoing rapid growth. 38 Therefore, the ability of Id4 to inhibit differentiation concomitant with increasing proliferation may facilitate mammary gland carcinogenesis. Rat mammary gland adenomas showed significantly lower expression of Id4 than did carcinomas. Adenomas arise from more differentiated cell types such as those in alveolar buds and lobules whereas carcinomas are derived largely from less differentiated ductal epithelium. 38,39 It is tempting to speculate that tumor malignancy in the rat model may in part be influenced by Id4-mediated regulation of mammary epithelial cell differentiation.
The molecular mechanism of action of Id proteins are, in a general sense, known to involve regulation of bHLH transcription factors and effects on cell cycle regulatory proteins. Id proteins including Id4 are likely to inhibit the commitment or differentiation that the bHLH proteins promote. 1,2 Nuclear localization appears to influence the action of Id4 in the mammary gland. Nuclear expression of Id4 was higher in carcinomas than in normal tissue, and the nuclear expression was altered during normal differentiation of the rat mammary gland. Movement of Id4 between the nucleus and cytosol has been observed with differentiation of oligodendrocytes and associated with cell cycle withdrawal. 16 Id4 has no nuclear localization sequences, and therefore its presence in the nucleus is indicative of possible heterodimerization with bHLH proteins. 40 Preliminary microarray analysis of Id4-overexpressing HC11 cells (versus parental HC11) show differences in expression of several genes regulating cell growth (unpublished observations).
An inverse association between Brca1 and Id4 expression has been previously suggested. 28 PhIP-induced carcinomas have been reported to express reduced levels of Brca1 protein. 41 However, Brca1 is not mutated in chemically induced rat mammary gland carcinomas. 41,42 Although it has been suggested that a Brca1-Id4 regulatory loop may be disrupted in some human breast cancers resulting in the down-regulation of both genes 29 , the elevated Id4 and low Brca1 expression in PhIP-induced carcinomas suggests that this regulatory loop is not disrupted in the rat model. Further studies are required to ascertain the possible impact of Id4-Brca1 regulation on the development of rat mammary gland cancer.
Studies now suggest that Id genes are strongly expressed in undifferentiated, growing, and tumor cells. 1-6,21,22 Id4 fits the paradigm of the Id gene family. We have shown that Id4 is highly overexpressed in rat mammary gland cancer, its expression varies with stage of mammary gland differentiation in vivo, and it is involved in the regulation of mammary epithelial cell differentiation, proliferation, and growth. The mechanisms contributing to Id4 overexpression in rat mammary gland cancers are not yet known. We analyzed Id4 for mutations, and although a single nucleotide polymorphism (T/C) was detected in the third nucleotide from the stop codon in rat DNA, no mutations were specific to carcinomas (data not shown). Further studies are warranted to better define the molecular mechanisms of Id4 in rat mammary gland carcinogenesis and to ascertain its potential role in human breast cancer.
Acknowledgments
We thank Drs. David S. Salomon and Nancy E. Hynes for providing the HC11 cells, and Dr. Snorri S. Thorgeirsson for helpful discussions.
Footnotes
Address reprint requests to Elizabeth G. Snyderwine, Chemical Carcinogenesis Section, Laboratory of Experimental Carcinogenesis, Center for Cancer Research, National Cancer Institute, Building 37, Room 4146, 37 Convent Drive, Bethesda, MD 20892-4262. E-mail elizabeth_snyderwine@nih.gov.
References
- 1.Norton JD: ID helix-loop-helix proteins in cell growth, differentiation, and tumorigenesis. J Cell Sci 2000, 113:3897-3905 [DOI] [PubMed] [Google Scholar]
- 2.Norton JD, Deed RW, Craggs G, Sablitzky F: Id helix-loop-helix proteins in cell growth and differentiation. Trends Cell Biol 1988, 8:58-65 [PubMed] [Google Scholar]
- 3.Yokota Y: Id and development. Oncogene 2001, 20:8290-8298 [DOI] [PubMed] [Google Scholar]
- 4.Rivera R, Murre C: The regulation and function of the Id proteins in lymphocyte development. Oncogene 2001, 20:8308-8316 [DOI] [PubMed] [Google Scholar]
- 5.Lasorella A, Uo T, Iavarone A: Id proteins at the cross-road of development and cancer. Oncogene 2001, 20:8326-8333 [DOI] [PubMed] [Google Scholar]
- 6.Benezra R, Rafii S, Lyden D: The Id proteins and angiogenesis. Oncogene 2001, 20:8334-8341 [DOI] [PubMed] [Google Scholar]
- 7.Norton JD, Atherton GT: Coupling of cell growth and apoptosis functions of Id proteins. Mol Cell Biol 1998, 18:2371-2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toma JG, El-Bizri H, Barnabé-Heider F, Aloyz R, Miller FD: Evidence that helix-loop-helix proteins collaborate with retinoblastoma tumor suppressor protein to regulate cortical neurogenesis. J Neurosci 2000, 20:7648-7656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riechmann V, van Crüchten I, Stablitzky F: The expression pattern of Id4, a novel dominant helix-loop-helix protein is distinct from Id1, Id2 and Id3. Nucleic Acids Res 1994, 22:749-755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mantani A, Hernandez M-C, Kuo W-L, Israel MA: The mouse Id2 and Id4 genes: structural organization and chromosomal localization. Gene 1998, 222:229-235 [DOI] [PubMed] [Google Scholar]
- 11.Pagliuca A, Bartoli PC, Saccone S, Valle GD, Lania L: Molecular cloning of Id4, a novel dominant-negative helix-loop-helix human gene on chromosome 6p21.3-p22. Genomics 1995, 27:200-203 [DOI] [PubMed] [Google Scholar]
- 12.Chaudhary J, Johnson J, Kim G, Skinner MK: Hormonal regulation and differential actions of the helix-loop-helix transcriptional inhibitors of differentiation (Id1, Id2, Id3, and Id4) in Sertoli cells. Endocrinology 2001, 142:1727-1736 [DOI] [PubMed] [Google Scholar]
- 13.Sablitzky F, Moore A, Bromley M, Deed RW, Newton JS, Norton JD: Stage- and subcellular-specific expression of Id proteins in male germ and Sertoli cells implicates distinctive regulatory roles for Id proteins during meiosis, spermatogenesis, and Sertoli cell function. Cell Growth Differ 1998, 9:1015-1024 [PubMed] [Google Scholar]
- 14.Chen H, Weng Y-C, Schatteman GC, Sanders L, Christy RJ, Christy BA: Expression of the dominant-negative regulator Id4 is induced during adipocyte differentiation. Biochem Biophys Res Commun 1999, 256:614-619 [DOI] [PubMed] [Google Scholar]
- 15.Andres-Barquin PJ, Hernandez M-C, Israel MA: Id4 expression induces apoptosis in astrocytic cultures and is down-regulated by activation of the cAMP-dependent signal transduction pathway. Exp Cell Res 1999, 247:347-355 [DOI] [PubMed] [Google Scholar]
- 16.Kondo T, Raff M: The Id4 HLH protein and the timing of oligodendrocyte differentiation. EMBO J 2000, 19:1998-2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desprez P-Y, Lin CQ, Thomasset N, Sympson CJ, Bissell MJ, Campisi J: A novel pathway for mammary epithelial cell invasion induced by the helix-loop-helix protein Id-1. Mol Cell Biol 1998, 18:4577-4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polsky D, Young AZ, Busam KJ, Alani RM: The transcriptional repressor of p16/Ink4a, Id1, is up-regulated in early melanomas. Cancer Res 2001, 61:6008-6011 [PubMed] [Google Scholar]
- 19.Langlands K, Down GA, Kealey T: Id proteins are dynamically expressed in normal epidermis and dysregulated in squamous cell carcinomas. Cancer Res 2000, 60:5929-5933 [PubMed] [Google Scholar]
- 20.Maruyama H, Kleeff J, Wildi S, Friess H, Büchler MW, Israel MA, Korc M: Id-1 and Id-2 are overexpressed in pancreatic cancer and in dysplastic lesions in chronic pancreatitis. Am J Pathol 1999, 155:815-822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin CQ, Singh J, Murata K, Itahana Y, Parrinello S, Liang SH, Gillett CE, Campisi J, Desprez PY: A role for Id-1 in the aggressive phenotype and steroid hormone response of human breast cancer cells. Cancer Res 2000, 60:1332-1340 [PubMed] [Google Scholar]
- 22.Wilson JW, Deed RW, Inoue T, Balzi M, Becciolini A, Faraoni P, Potten CS, Norton JD: Expression of Id helix-loop-helix proteins in colorectal adenocarcinomas correlates with p53 expression and mitotic index. Cancer Res 2001, 61:8803-8801 [PubMed] [Google Scholar]
- 23.Ouyang XS, Wang X, Lee DTW, Tsao SW, Wong YC: Overexpression of Id1 in prostate cancer. J Urol 2002, 167:2598-2602 [PubMed] [Google Scholar]
- 24.Parrinello S, Lin CQ, Murata K, Itahana Y, Singh J, Krtolica A, Campisi J, Desprez P-Y: Id-1, ITF-2, and Id-2 comprise a network of helix-loop-helix proteins that regulate mammary epithelial cell proliferation, differentiation, and apoptosis. J Biol Chem 2001, 276:39213-39219 [DOI] [PubMed] [Google Scholar]
- 25.Desprez P-Y, Hara E, Bissell MJ, Campisi J: Suppression of mammary epithelial cell differentiation by the helix-loop-helix protein Id-1. Mol Cell Biol 1995, 15:3398-3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh J, Itahana Y, Parrinello S, Murata K, Desprez P-Y: Molecular cloning and characterization of a zinc finger protein involved in Id-1-stimulated mammary epithelial cell growth. J Biol Chem 2001, 276:11852-11858 [DOI] [PubMed] [Google Scholar]
- 27.Mori S, Nishikawa S-I, Yokota Y: Lactation defect in mice lacking the helix-loop-helix inhibitor Id2. EMBO J 2000, 19:5772-5781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beger C, Pierce LN, Krüger M, Marcusson EG, Robbins JM, Welcsh P, Welch PJ, Welte K, King M-C, Barber JR, Wong-Staal F: Identification of Id4 as a regulator of BRCA1 expression by using a ribozyme-library-based inverse genomics approach. Proc Natl Acad Sci USA 2001, 98:130-135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welcsh PL, Lee MK, Gonzalez-Hernandez RM, Black DJ, Mahadevappa M, Swisher EM, Warrington JA, King M-C: BRCA1 transcriptionally regulates genes involved in breast tumorigenesis. Proc Natl Acad Sci USA 2002, 99:7560-7565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shan L, He M, Yu M, Qiu C, Lee NH, Liu ET, Snyderwine EG: cDNA microarray profiling of rat mammary gland carcinomas induced by 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine and 7,12-dimethylbenz[a]anthracene. Carcinogenesis 2002, 23:1561-1568 [DOI] [PubMed] [Google Scholar]
- 31.Ghoshal A, Preisegger KH, Takayama S, Thorgeirsson SS, Snyderwine EG: Induction of mammary tumors in female Sprague-Dawley rats by the food-derived carcinogen 2-amino-1-methyl-6-phenylimidazo[4, 5-b]pyridine and effect of dietary fat. Carcinogenesis 1994, 15:2429-2433 [DOI] [PubMed] [Google Scholar]
- 32.Snyderwine EG, Thorgeirsson UP, Venugopal M, Roberts-Thomson SJ: Mammary gland carcinogenicity of 2-amino-1-methyl-6-phenylimidazo[4, 5-b]pyridine in Sprague-Dawley rats on high- and low-fat diets. Nutr Cancer 1998, 31:160-167 [DOI] [PubMed] [Google Scholar]
- 33.Qiu C, Yu M, Shan L, Snyderwine EG: Allelic imbalance and altered expression of genes in chromosome 2q11–2q16 from rat mammary gland carcinomas induced by 2-amino-1-methyl-6-phenylimidazo[4, 5-b]pyridine. Oncogene 2003, 22:1253-1260 [DOI] [PubMed] [Google Scholar]
- 34.Ball RK, Friis RR, Schoenenberger CA, Doppler W, Groner B: Prolactin regulation of β-casein gene expression and of a cytosolic 120-kd protein in a cloned mouse mammary epithelial cell line. EMBO J 1988, 7:2098-2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merlo GR, Graus-Porta D, Cella N, Marte BM, Taverna D, Hynes NE: Growth, differentiation, and survival of HC11 mammary epithelial cells: diverse effects of receptor tyrosine kinase-activating peptide growth factors. Eur J Cell Biol 1996, 70:97-105 [PubMed] [Google Scholar]
- 36.Shan L, Rouhani SA, Schut HAJ, Snyderwine EG: 2-amino-1-methyl-6-phenylimidazo[4, 5-b]pyridine (PhIP) modulates lactogenic hormone-mediated differentiation and gene expression in HC 11 mouse mammary epithelial cells. Cell Growth Differ 2001, 12:649-656 [PubMed] [Google Scholar]
- 37.Lister J, Forrester WC, Baron MH: Inhibition of an erythroid differentiation switch by the helix-loop-helix protein Id1. J Biol Chem 1995, 270:17939-17946 [DOI] [PubMed] [Google Scholar]
- 38.Russo IH, Russo J: Developmental stage of the rat mammary gland as determinant of its susceptibility to 7, 12-dimethylbenz[a]anthracene. J Natl Cancer Inst 1978, 61:1439-1449 [PubMed] [Google Scholar]
- 39.Russo J, Gusterson BA, Rogers AE, Russo IH, Wellings SR, van Zwieten MJ: Biology of disease: comparative study of human and rat mammary tumorigenesis. Lab Invest 1990, 62:244-278 [PubMed] [Google Scholar]
- 40.Pagliuca A, Cannada-Bartoli P, Lania L: A role for Sp and helix-loop-helix transcription factors in the regulation of the human Id4 gene promoter activity. J Biol Chem 1998, 273:7668-7674 [DOI] [PubMed] [Google Scholar]
- 41.Okochi E, Miyamoto K, Wakazono K, Shima H, Sugimura T, Ushijima T: Reduced Brca1 protein expression in 2-amino-1-methyl-6-phenylimidazo[4, 5-b]pyridine-induced rat mammary carcinomas. Mol Carcinog 2002, 34:211-218 [DOI] [PubMed] [Google Scholar]
- 42.Chen K-S, Shepel LA, Haag JD, Heil GM, Gould MN: Cloning, genetic mapping, and expression studies of the rat Brca1 gene. Carcinogenesis 1996, 17:1561-1566 [DOI] [PubMed] [Google Scholar]