Abstract
To elucidate the molecular events responsible for tumorigenesis and progression of ependymomas, we analyzed molecular alterations on the gene expression level in a series of newly diagnosed ependymal neoplasms (n = 39). To this aim, tumor RNA was hybridized to microarrays comprising 2600 different genes with relevance to mitosis, cell-cycle control, oncogenesis, or apoptosis. For CLU, IGF-2, and RAF-1, which are apparent candidate genes because they had been previously described to be involved in tumorigenesis of other human malignancies, we found a high expression on the mRNA as well as the protein level. We identified gene expression signatures for the differentiation of tumors with respect to location, grade, and patient age. Spinal ependymomas were characterized by high-expression levels of HOXB5, PLA2G, and CDKN2A and tumors in young patients (≤16 years of age) by high-expression levels of LDHB and STAM. Notably, we were able to classify supratentorial grade II and III tumors with 100% accuracy, whereas this did not apply for infratentorial ependymomas. The similar gene expression patterns of grade II and III infratentorial malignancies suggest that grade III tumors may develop through a secondary multistep transformation process involving genes that are related to cell proliferation (LDHA, cyclin B, MAT2A) or tumor suppression (PTEN). In summary, our results provide new insight in the biochemical pathways particularly intriguing in the pathomechanism of ependymomas and suggest that this entity comprises molecularly distinct diseases.
Ependymal tumors arise from the ependymal lining of the cerebral ventricles and from the remnants of the central canal of the spinal cord. This neoplasm constitutes ∼3 to 5% of all intracranial malignancies and is the third most common brain tumor in children and young adults. 1,2 In ependymomas, the morphological features and biological behavior vary considerably. Patients with spinal tumor location have usually a favorable prognosis after gross total resection, whereas local tumor progression is the predominant reason for death in patients with intracranial ependymomas, resulting in a 5-year overall survival of ∼60%. 3-5
Because ependymomas are characterized by tremendous variability in clinical behavior, the understanding of the complex changes taking place at the genomic level might lead to more precise understanding of the tumor biology. Cytogenetic studies revealed numerous chromosomal aberrations in ependymomas. In particular, a 30 to 50% incidence of aberrations involving chromosome 22, including monosomy 22 as well as deletions of 22q, prevailed the most frequent finding. 6-8 Recently, Hirose and co-workers 7 reported on different patterns of chromosomal abnormalities with respect to tumor location detected by comparative genomic hybridization. In intracranial tumors, gain of 1q and losses on 6q, 9, and 13 were frequent, whereas gains on chromosome 7 were recognized almost exclusively in spinal cord tumors and were associated with various other chromosomal aberrations including frequent loss of 22q, suggesting that intracranial and spinal cord ependymomas progress along substantially different pathways. As a hereditary form, neurofibromatosis type 2 is associated with spinal ependymomas, indicating a functional role of the NF2 tumor suppressor gene in these tumors. 9,10
In contrast to adults in which spinal tumors predominate, ∼90% of all pediatric ependymomas are of intracranial origin with most tumors arising infratentorially. 2 In addition, pediatric ependymoma patients tend to have a poorer outcome than adults. 1 Although this might be explained by age-dependent differences in tumor location or resectability, findings on the genomic level detected by comparative genomic hybridization suggest biological discrepancies as well. For example, 30 to 50% of pediatric ependymomas display balanced comparative genomic hybridization profiles in comparison to only 10% in adults. 7,8,11 This finding suggests that the development of ependymomas at younger age is often independent of chromosomal instability. The recent histological ependymoma classification has thus far proven to be an unreliable predictor of clinical outcome and relationship between ependymoma grade and specific chromosomal aberrations is also controversial. 2,7,8,11,12
By novel microarray technologies, the expression of thousands of genes can be studied simultaneously. In brain tumors, gene expression studies including astrocytic, 13,14 oligodendroglial, 15,16 as well as embryonal neuroepithelial tumors 17,18 have been performed recently. Statistical analyses identified subgroups of brain malignancies that differ according to tumor type, histological subclasses, metastatic stage, and survival. 14,17,18
Because the role of relevant genes on mRNA level is hardly understood in ependymomas, we analyzed the expression profile of 39 newly diagnosed ependymal neoplasms using cDNA-based microarrays containing some 2600 genes with relevance to mitosis, cell-cycle control, oncogenesis, and apoptosis. For a subset of genes the data were confirmed on the protein level by immunolabeling. The microarray data were used to identify signatures specific for tumor location, grade, and patient age.
Materials and Methods
Patient Characteristics
We collected all 39 tumor specimens used in this study at the Department of Neuropathology, Burdenko NN Neurosurgical Institute (Moscow, Russia). All patients were operated on between November 2000 and March 2002. As summarized in Table 1 ▶ , we analyzed 39 patients with a median age of 27 years (range, 1 to 55 years). The ependymomas were either located at the spine (n = 10) or brain (n = 29). Histological diagnoses were made according to the recent classification of tumors of the nervous system. 2 Additionally, 35 nonmyxopapillary tumors were graded as classic (grade II) and anaplastic (grade III) according to the criteria presented by Merchant and co-workers. 19
Table 1.
Clinical Characteristics of 39 Ependymoma Patients
| Histological grade | Location | Child/adult* | Median age | Male/female |
|---|---|---|---|---|
| Myxopapillary grade I (n = 4) | Spinal (n = 4) | 1/3 | 31.3 | 0/4 |
| Classic grade II (n = 19) | Spinal (n = 6) | 0/6 | 37.2 | 3/3 |
| Infratentorial (n = 9) | 4/5 | 24.7 | 4/5 | |
| Supratentorial (n = 4) | 1/3 | 30.1 | 1/3 | |
| Anaplastic grade III (n = 16) | Infratentorial (n = 9) | 4/5 | 23.4 | 7/2 |
| Supratentorial (n = 7) | 6/1 | 8.3 | 3/4 |
*Age of ≤16 years was used as a cut-off point for pediatric patients.
Sample Processing
Samples from each primary tumor were taken at the time of initial operation, immediately frozen in liquid nitrogen, and then continuously stored at −80°C until used for microarray experiments. To confirm the presence of viable cellular tumor (≥80% neoplastic cells), the cryosections of each ependymoma sample (∼5-μm thick) were stained with hematoxylin and eosin (Sigma Aldrich, Taufkirchen, Germany) and reviewed before RNA extraction. Additionally, tumor samples were stained with antibodies to synaptophysin (SY38, mouse, monoclonal, 1: 200; DAKO, Carpenteria, CA) to avoid contamination with nontumoral nervous tissue.
Total RNA was isolated according to a protocol that applies both Trizol reagent (Invitrogen, Karlsruhe, Germany) and RNeasy Midi spin columns (Qiagen, Hilden, Germany), followed by Oligotex resin (Qiagen) purification of poly-A RNA. The integrity and purity of the total RNA was analyzed on a 1% agarose gel. Total RNA quality and concentration were determined by absorption spectroscopy between 220 and 320 nm (1 OD260 = 40 μg/ml ssRNA).
cDNA Microarray
Microarrays containing replicate spots of 4211 different gene-specific fragments, representing 2600 different genes were processed as described previously. 20,21 The tumor RNA was co-hybridized with commercially available universal human reference RNA (Stratagene, La Jolla, CA), composed of total RNA from 10 human cancer cell lines. In detail, ∼1 μg of ependymoma and 1 μg of reference mRNA were labeled with Cy3 and Cy5, respectively, using the Omniscript Reverse Transcriptase kit (Qiagen) and hybridized with 10 μg of CotI DNA, 30 μg of bovine liver tRNA, and 10 μg of oligo-dT nucleotides in an automated hybridization chamber (GeneTac; Genomic Solutions, Ann Arbor, MI). For all samples we performed color switch experiments in which the tumor and reference DNA were labeled via Cy3- and Cy5-dUTP, respectively, and vice versa. Data sets for spots not recognized by the GenePix analysis software were excluded from further considerations. Additionally, all remaining data sets were ranked according to spot homogeneity (as assayed by the ratio of median and mean fluorescence intensities), spot intensity, and the SD of log ratios for replicate spots. Those data points ranked among the lower 20%, based on the criteria just described, were removed from the data set. For each hybridization, fluorescence ratios (Cy5:Cy3) were normalized by variance stabilization. 22 To combine experiments with switched dye labeling, the ratios of one experiment were inverted and averaged with the corresponding spots on the second array. The raw data of the microarray experiments are available under the following url address: http://www.dkfz.de/kompl_genome/Other/ependymoma.html.
Real-Time Quantitative Reverse Transcriptase-Polymerase Chain Reaction (RQ-PCR)
Each cDNA sample was analyzed in triplicate (aliquot of 1 μl each) using the ABI PRISM 7700 Sequence Detector (Applied Biosystems, Weiterstadt, Germany) as described previously. 23 To standardize the amount of sample cDNA, three endogenous control amplicons were used as housekeeping genes, coding for phosphoglycerate kinase 1 (PGK1), lamin B1 (LMNB1), and cyclophilin A (PPIA). Oligonucleotides, used for RQ-PCR, are listed in Table 2 ▶ . The relative quantification of each target gene in comparison to the reference genes was done by using the mathematical model developed by Pfaffl. 24
Table 2.
Primer Sequences Used for Real-Time Quantitative Reverse-Transcription PCR (RQ-PCR)
| Primer designation | Sequence |
|---|---|
| CLU forward | 5′-CTTGCTGGAGCAGCTGAAC-3′ |
| CLU reverse | 5′-AGAAGTGTGGGAAGCCACC-3′ |
| HOXB5 forward | 5′-GCACTCTGCCTGTCCGAG-3′ |
| HOXB5 reverse | 5′-CTGCCAGCTGTAGCCAGG-3′ |
| LMNB1 forward | 5′-CTGGAAATGTTTGCATCGAAGA-3′ |
| LMNB1 reverse | 5′-GCCTCCCATTGGTTGATCC-3′ |
| PGK1 forward | 5′-AAGTGAAGCTCGGAAAGCTTCTAT-3′ |
| PGK1 reverse | 5′-TGGGAAAAGATGCTTCTGGG-3′ |
| PPIA forward | 5′-GCTCGTGCCGTTTTGCA-3′ |
| PPIA reverse | 5′-GCAAACAGCTCAAAGGAGACG-3′ |
| ZFM1 forward | 5′-ATGGTGGACATCCCATGC-3′ |
| ZFM1 reverse | 5′-CGTACTTCCCAGGTACTGATCC-3′ |
Immunohistochemistry
Immunohistochemical analysis was performed on 5-μm paraffin sections mounted on poly-l-lysine-coated slides. The sections were microwaved in antigen unmasked solution (BD Biosciences, San Jose, CA). Antibodies (all produced by Santa Cruz Biotechnology, Santa Cruz, CA) against the following antigens were used: clusterin-a/b (H-330, rabbit, polyclonal, sc-8354, 1:200), RAF-1 protein (mouse, clone E-10, sc-7267, 1:100), and IGF-2 protein (H-103, rabbit, polyclonal, sc-5622, 1:200). The sections were incubated overnight at 4°C with all of the antibodies mentioned above. Immunostain visualization was achieved with the standard streptavidin-biotin peroxidase technique (LSAB kit, N K0675; DAKO Corp). The slides were stained with 3,3′-diaminobenzidine, counterstained with hematoxylin, and mounted. For the negative control procedure, primary antibodies were substituted according to commercially produced control reagents.
Statistical Analysis
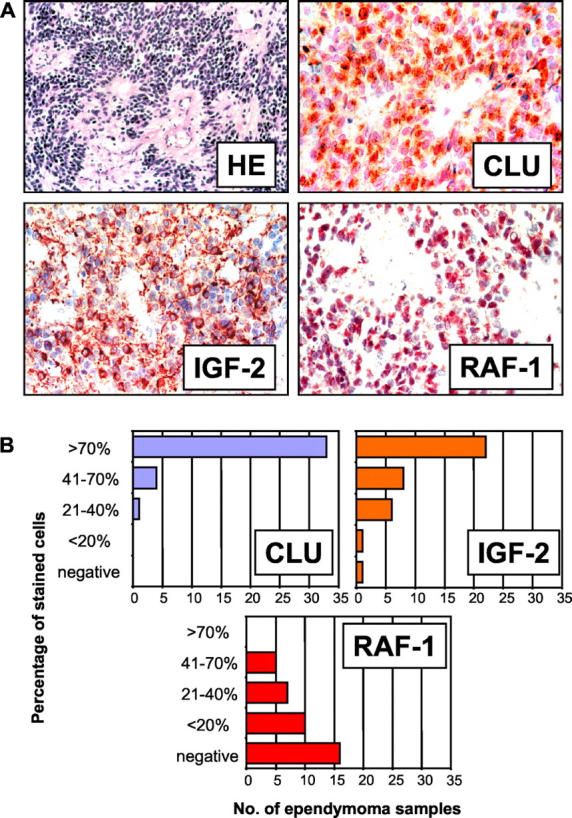
To identify subsets of genes that best characterize ependymoma samples based on location, patient age, and tumor grade, we used the nearest shrunken centroids classification as described by Tibshirani and colleagues. 25 This approach shrinks each class centroid toward the overall centroid. Classification of samples is then made to the nearest shrunken centroid. Let Ygj be the expression value for gene g, g = 1, …, N, and sample j, j = 1, …, n, and let Ck indicate the nk samples in class k, k = 1, …, K, and ∑k=1K nk=n. Corresponding to gene g the g-th component of the centroid for class k is Ȳgk=∑j∈Ck Ygj/nk, the mean relative expression in class k for gene g. The g-th component of the overall centroid is given by Ȳg=∑j=1n Ygj/n. In a first step, a t-statistic dgk is computed to compare the class k centroid with the overall centroid dgk=(Ȳgk−Ȳg)/(mksg), describing a standardized difference of class k and overall centroid, where sg is the pooled within-class SD for gene g and mk=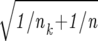 . Then, shrinkage is done by soft thresholding: For each gene dgk is shrunken by an amount of Δ toward zero, giving d′gk=sign(dgk)(|dgk|−Δ)+ where + means the positive part (t+ = t if t > 0 and zero otherwise). Shrunken centroids Ȳ′gk now result as Ȳ′gk=Ȳg+mksgd′gk, which are then used for classification. The amount of shrinkage is determined by 10-fold cross-validation. 26
. Then, shrinkage is done by soft thresholding: For each gene dgk is shrunken by an amount of Δ toward zero, giving d′gk=sign(dgk)(|dgk|−Δ)+ where + means the positive part (t+ = t if t > 0 and zero otherwise). Shrunken centroids Ȳ′gk now result as Ȳ′gk=Ȳg+mksgd′gk, which are then used for classification. The amount of shrinkage is determined by 10-fold cross-validation. 26
Pair-wise comparisons of quantitative data obtained by RQ-PCR experiments were done by the Mann-Whitney test. For categorical data the chi-square test was used. All tests were two-sided. An effect was considered statistically significant if the P value was 0.05 or less. The statistical analyses were performed using the software package R, version 1.5. 27
Results
Genes Abundantly Expressed in Ependymomas
To identify genes that are abundantly expressed in ependymomas, cDNAs derived from 39 tumor samples were co-hybridized with a standard reference RNA. After the filtering and normalization procedure (see Materials and Methods), 1980 of the initial set of 2600 genes were further evaluated because they were found transcribed in either the tumor or the reference cell population. In comparison to the reference RNA pool, 68 genes were highly expressed (more than twofold) in ependymomas, whereas 285 genes were expressed at low levels (<0.5-fold). The 16 most abundantly expressed genes (more than threefold) were CLU (88.5-fold), AK1 (9.2-fold), RAF1 (6.8-fold), MMP12 (6.5-fold), SERPING1 (6.4-fold), FOXJ1 (5.8-fold), MSX1 (4.6-fold), CRYAB (4.1-fold), PSAP (3.8-fold), PCDGCH3 (3.6-fold), ITIH2 (3.6-fold), CETN2 (3.4-fold), IGF2 (3.3-fold), HLA-DRA (3.1-fold), DNAJB2 (3.1-fold), and GRM5 (3.1-fold). The NF2 gene was 2.8-fold (range, 0.4 to 14.0) higher expressed in the ependymoma samples than in the reference RNA.
Gene Expression Signature Describing Tumor Location
The nearest shrunken centroid classification was applied to determine whether ependymomas of spinal and intracranial locations are characterized by specific gene expression signatures. 25 We identified a set of 21 genes that allowed an accurate classification with respect to ependymoma location (spinal versus intracranial) in 34 of 39 cases (Figure 1A) ▶ . All grade I ependymomas of spinal location (n = 4) displayed a common signature with high-expression levels of HOXB5, PLA2G5, and ITIH2, clearly separating these cases from all intracranial tumors. Whereas three of six grade II spinal ependymomas clustered with the four myxopapillary cases, the remaining three patients were classified to the group of intracranial tumors. Although the NF2 gene is not included in the gene expression signature, it was found to be threefold higher expressed in intracranial than spinal tumors (Mann-Whitney U-test, P = 0.002).
Figure 1.
Supervised hierarchical clustering analysis of differentially expressed genes according to tumor location (A, spinal versus intracranial) and patient age (B, ≤16 years versus >16 years). The genes were selected by the nearest shrunken centroid classification that identifies minimal combinations of genes that provide best discrimination between two known patient groups. For illustration, each gene was normalized to the mean expression between both patient groups. Misclassified patients are indicated by arrows. In addition, the NF2 gene expression of each individual patient is shown by bar plots (A). T, supratentorial; I, infratentorial; S, spinal.
Gene Expression Signature Describing Patient Age
To assess whether ependymomas in children and adults are characterized by different gene expression patterns, we used a cut-off point of 16 years for statistical analysis. A set of eight genes was identified that allowed a classification according to both patient age groups in 30 of 39 cases (Figure 1B) ▶ . Notably, the pediatric group can be subdivided additionally, because six supratentorial grade III ependymomas were characterized by high-expression levels of GPX3, STAM, COL6A1, PYCR1, HSPB1, and ARHGDIA.
Gene Expression Signature Describing Intracranial Tumor Grade
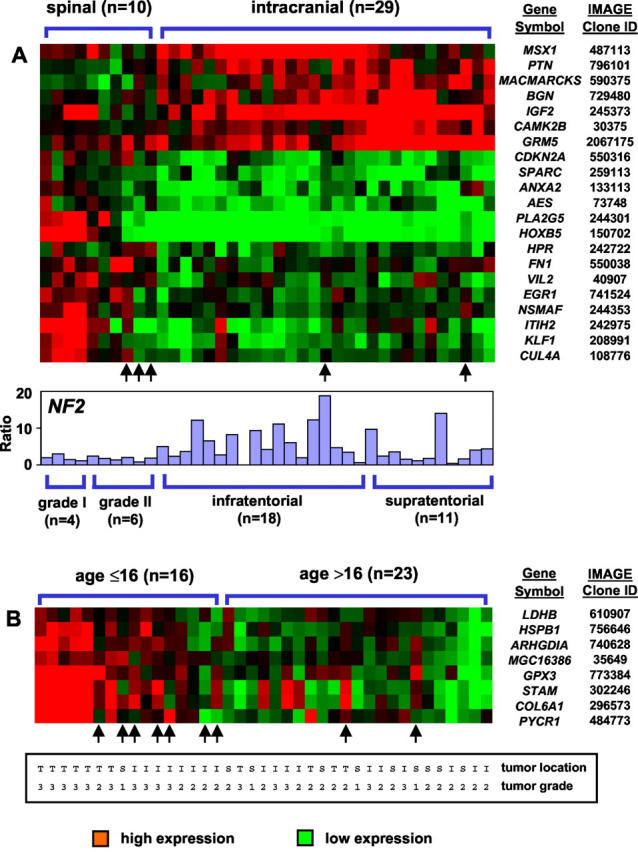
In intracranial ependymomas, differentially expressed genes between grade II and III tumors were analyzed separately for ependymomas with supra- and infratentorial location (Figure 2) ▶ . Whereas all supratentorial cases were classified correctly by a set of 26 genes (Figure 2A) ▶ , 5 of 18 infratentorial tumor samples were misclassified (Figure 2B) ▶ . Notably, these five patients were either among the youngest (age, 2 years) or oldest (age, 32, 45, 50, and 55 years).
Figure 2.
Supervised hierarchical clustering analysis of differentially expressed genes according to histological grade (grade II versus grade III) for tumors of supratentorial (A) and infratentorial (B) location. Whereas all supratentorial tumors were classified with 100% accuracy, 5 of 18 infratentorial ependymomas were misclassified as indicated by arrows. Notably, these five patients were either among the youngest (age, 2 years) or the oldest (age, 32, 45, 50, and 55 years) patients.
Correlation of cDNA Microarray Data with RQ-PCR
To assess the microarray data, three genes (CLU, ZMF1, HOXB5) were analyzed in further detail using RQ-PCR. As shown by microarray experiments, CLU was abundantly expressed in all ependymoma samples (median, 88.5-fold) tested, HOXB5 was differentially expressed between spinal and intracranial tumors (Figure 1A) ▶ and the transcription factor ZFM1 was lower expressed in all ependymoma samples as compared to the reference RNA (eightfold). In line with the results obtained by microarray experiments, all 12 ependymoma samples analyzed by RQ-PCR revealed high CLU expression levels (33-fold), whereas ZMF1 was lower expressed in 11 of 12 ependymoma samples (eightfold) as compared to the reference RNA. Consistent with the microarray experiments, the eight spinal ependymomas analyzed by RQ-PCR displayed statistically significant higher expression levels of HOXB5 as compared to eight intracranial neoplasms (Mann-Whitney U-test, P = 0.02). Within the group of spinal ependymomas, there was a trend (data not shown) toward higher HOXB5 expression measured by RQ-PCR for tumors with grade I histology as compared to grade II subtypes (Mann-Whitney U-test, P = 0.15).
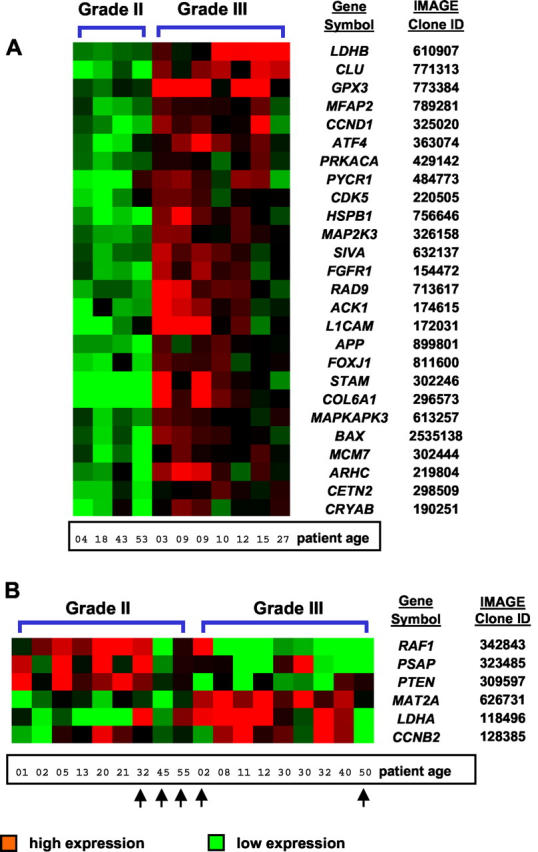
Correlation of Gene Expression Profiles with Immunohistochemical Data
To analyze whether the abundantly expressed genes CLU, IGF2, and RAF1 are also highly expressed on the protein level, we performed immunohistochemical studies in 38 of 39 ependymoma samples available (Figure 3) ▶ . Immunohistochemical analysis revealed strong cytoplasmic CLU reactivity in all 38 tumors examined (Figure 3A) ▶ and most of the samples (86.8%) showed a strong expression (stained cells, >70%) independent of tumor grade, location, and patient age. Antibodies to IGF-2 revealed nuclear and membranous-cytoplasmic immunoreactivity in all except one sample (Figure 3B) ▶ . In particular, ∼80% of ependymomas exhibited >40% of immunostained cells. Cytoplasmic immunoreactivity of RAF-1 was found only in 22 of 38 tumors examined and no ependymoma exhibited >70% of immunoreactive cells. The number of RAF-1-positive tumors was higher in intracranial tumors (65%) in comparison to spinal tumors (40%) (chi-square test, P < 0.0001).
Figure 3.
A: Photomicrographs of ependymomas showing H&E staining (HE) and patterns of immunohistochemical staining of clusterin (CLU), insulin-like growth factor protein-2 (IGF-2) and v-raf-1 murine leukemia viral oncogene homolog 1 (RAF-1). CLU, IGF-2, and RAF-1 were identified as abundantly expressed genes by microarray experiments. B: As shown by the bar plots their proteins are also expressed in most of the ependymoma samples as analyzed by immunohistochemistry.
Discussion
In the current study, cDNA-based microarrays were used to analyze genetic alterations of 39 newly diagnosed ependymomas on the gene expression level. Because nonmalignant ependymal tissue was not obtainable, the tumor RNA was co-hybridized to a reference RNA pool composed of 10 human tumor cell lines. The list of highly expressed genes in ependymomas includes CLU, IGF2, RAF1, MMP12, PSAP, and MSX1, which all had been described previously to be involved in tumorigenesis of other human malignancies. 28-33 The results obtained by cDNA microarray experiments were confirmed by RQ-PCR for three representative genes (CLU, ZFM1, HOXB5). Interestingly, gene expression levels could be correlated with frequently found chromosomal aberrations described in the literature. For example, increased expression (more than twofold) was found concerning PRELP, EPHX1, FY, and HSPA6, all located on chromosomal arm 1q, which frequently showed copy number gains in ependymoma. 7 Correspondingly, genes from frequently deleted regions 7,8,11 were found weakly expressed (<0.5-fold), eg, COX7A2, COL10A1, and TCP1 from chromosome 6q; CNTFR, RAGA, TXN, AMBP, and HSPA5 from chromosome 9; TUBA2, PRDX2, and LCP1 from chromosome 13; and RANBP1, MCM5, EP300, G22P1, BZRP, and MAPK12 from chromosomal arm 22q.
All 39 ependymomas analyzed in this study revealed a more than 10-fold expression of CLU, encoding for the glycoprotein clusterin (apolipoprotein J). In line with our results, Sasaki and co-workers 34 reported on intense clusterin protein and/or mRNA expression in nonmalignant human ependymal cells. Although the biological role of clusterin in tumorigenesis is still debated, the function of clusterin is related to maintenance of cell-cell contacts, lipid transportation, tissue remodeling, and apoptosis regulation. 28
In parallel to the cDNA microarray data, most of the patients showed also an intensive staining of the insulin-like growth factor-2 (IGF-2) and v-raf-1 murine leukemia viral oncogene homolog 1 (RAF-1) by immunohistochemistry. In brain tumors, IGF2 expression was found in human gliomas 35 and IGF-2 antibody reactivity was reported as a specific immunohistochemical pattern in ependymomas. 36 The proto-oncogene RAF-1 is a serine/threonine protein kinase that functions downstream of the Ras family of membrane associated GTPases and plays a major role in controlling cell proliferation, differentiation, and cell death. We found a statistically significant higher expression of RAF-1 in intracranial ependymomas by immunohistochemistry as compared to tumors with spinal location.
To analyze whether subgroups of ependymoma patients are characterized by specific gene expression patterns, which might explain pathophysiological processes on the gene expression level, we used the nearest shrunken centroid classification that identifies minimal combinations of genes that provide best discrimination between two known groups. 25 The identified gene expression patterns allowed an accurate classification of ependymoma patients according to tumor location (spinal versus intracranial) and patient age (≤16 years versus >16 years) in 87% and 77% of cases. Notably, our series did not include any spinal ependymoma of grade III histology, suggesting that the differences seen for tumor location are also related to tumor grade. Specifically, all grade I ependymomas of spinal location (n = 4) displayed a common signature with high-expression levels of HOXB5, PLA2G5, and ITIH2, clearly separating these cases from all intracranial tumors, whereas grade II spinal ependymomas clustered either to the myxopapillary cases or to the group of intracranial tumors, suggesting a molecular heterogeneity of this histologically homogeneous subset.
In comparison to intracranial tumor samples, we found CDKN2A among the highest expressed genes in spinal ependymomas. Recently, inactivation of both copies of the CDKN2A gene (p16INK4A and p14ARF) because of deletions, point mutations, or methylation has been demonstrated in ependymoma. 37,38 In a recent comparative genomic hybridization study by Hirose and colleagues, 7 intracranial ependymomas showed frequent losses of 9p (9 of 23 patients), whereas spinal ependymomas displayed gains of this chromosomal region (11 of 20 patients). This finding suggests that differential expression levels of CDKN2A in spinal and intracranial ependymomas found in our study might be because of chromosomal aberrations, which lead to deletions or additional copy numbers of CDKN2A. In addition, we revealed higher expression levels of NF2 in intracranial tumors as compared to spinal ependymomas. This might be because of NF2 gene deletions or mutations that are predominantly found in spinal cases 9,10,39 Moreover, the high NF2 gene expression levels found in 18 samples (more than threefold) might be explained by polysomies or gains of 22q that have been reported previously in ∼50% of ependymomas. 39
Although some age-related immunohistochemical patterns 5 and genetic alterations 7,8,11 have been found associated with the outcome of ependymoma patients, the underlying biological mechanisms are still unclear. By using an age of 16 years as a cut-off point, the identified gene expression signature includes LDHB and STAM as highly expressed genes characteristic for younger patients. While the gene product of LDHB functions as cell-cycle activator, STAM codes for an adaptor molecule that acts in signal transduction pathways of cytokine receptors. Thus, the high expression of both genes in ependymoma samples of younger patients might suggest an increased cell proliferation that could contribute to the unfavourable outcome observed in pediatric patients.
Although the relationships between ependymoma grade, specific chromosomal aberrations, 7,8,12,40 and patient outcome 3-5 are controversial, we were able to classify between supratentorial grade II and grade III tumors with 100% accuracy. The signature includes genes with relevance to oncogenic processes, ie, cell-cycle control (cyclin D1, CDK5, MCM7), signal transduction (MAP2K3, MAPKAPK3, STAM), apoptosis (SIVA, BAX), tumor invasiveness (COL6A1, COL9A3, L1CAM), and angiogenesis (FGFR1). Interestingly, grade II and III ependymomas of infratentorial location were more difficult to classify, 5 of 18 tumor samples were misclassified, including either the youngest (age, <3 years) or oldest (age, >30 years) patients. Whereas supratentorial ependymomas show a clear signature separating grade II and III tumors, the similar gene expression patterns of grade II and III infratentorial malignancies suggest that grade III tumors may develop through a secondary multistep transformation process involving genes that are related to cell proliferation (LDHA, cyclin B, MAT2A) or tumor suppression (PTEN).
In summary our results, based on gene expression experiments, provide new insight in the molecular pathways involved in the tumorigenesis and progression of ependymal neoplasms. Our results suggest that ependymoma patients have molecularly distinct diseases and that the heterogeneity seen on the chromosome level also applies at the gene expression level. The prognostic significance of these gene expression signatures has to be determined.
Acknowledgments
We thank Heidi Kramer and Daniel Goettel for their excellent technical assistance and Felix Kokocinski for the management of the microarray data base.
Footnotes
Address reprint requests to Prof. Dr. Peter Lichter, Deutsches Krebsforschungszentrum, Division of Molecular Genetics (B060), Im Neuenheimer Feld 280, D-69120 Heidelberg, Germany. E-mail: m.macleod@dkfz.de.
Supported by the Bundesministerium für Bildung und Forschung (FKZ 01 KW 9937 and NGFN 01 GR 0101).
K. N. is a scholar of the Deutsche José Carreras Leukämie-Stiftung e.V (DJCLS 2001/NAT-3).
A. K. and K. N. contributed equally to this work.
References
- 1.McLendon RE, Enterline DS, Tien RD, Thorstad WL, Bruner JM: Tumors of central epithelial origin. Bigner DD McLendon RE Bruner JM eds. Russel and Rubunstein’s Pathology of Tumors of the Nervous System. 1998:pp 387-418 Arnold London
- 2.Wiestler OD, Schiffer D, Coons SW, Prayson RA, Rosenblum MK: Ependymal tumors. Kleihues P Cavenee WK eds. Pathology and Genetics of Tumors of the Nervous System. 2000:pp 71-81 IARC Press Lyon
- 3.Pollack IF, Gerszten PC, Martinez AJ, Lo KH, Shultz B, Albright AL, Janosky J, Deutsch M: Intracranial ependymomas of childhood: long-term outcome and prognostic factors. Neurosurgery 1995, 37:655-666 [DOI] [PubMed] [Google Scholar]
- 4.Robertson PL, Zeltzer PM, Boyett JM, Rorke LB, Allen JC, Geyer JR, Stanley P, Li H, Albright AL, McGuire-Cullen P, Finlay JL, Stevens KR, Jr, Milstein JM, Packer RJ, Wisoff J: Survival and prognostic factors following radiation therapy and chemotherapy for ependymomas in children: a report of the Children’s Cancer Group. J Neurosurg 1998, 88:695-703 [DOI] [PubMed] [Google Scholar]
- 5.Korshunov A, Golanov A, Timirgaz V: Immunohistochemical markers for ependymal neoplasms. J Neuro-Oncol 2002, 58:255-270 [DOI] [PubMed] [Google Scholar]
- 6.Mazewsky C, Soukup S, Ballard E, Gotwals B, Lampkin B: Karyotype studies in 18 ependymomas with literature review of 107 cases. Cancer Genet Cytogenet 1999, 113:1-8 [DOI] [PubMed] [Google Scholar]
- 7.Hirose Y, Aldape K, Bollen A, James CD, Brat D, Lamborn K, Berger M, Feuerstein BG: Chromosomal abnormalities subdivide ependymal tumors into clinically relevant groups. Am J Pathol 2001, 158:1137-1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter M, Nicholson J, Ross F, Crolla J, Allibone R, Balaji V, Perry R, Walker D, Gilbertson R, Ellison D: Genetic abnormalities detected in ependymomas by comparative genomic hybridization. Br J Cancer 2002, 86:929-939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ebert C, von Haken M, Meyer-Puttlitz B, Wiestler OD, Reifenberger G, Pietsch T, von Deimling A: Molecular genetic analysis of ependymal tumors. NF2 mutations and chromosome 22q loss occur preferentially in intramedullary spinal ependymomas. Am J Pathol 1999, 155:627-632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamszus K, Lachenmayer L, Heinemann U, Kluwe L, Finckl U, Hoppner W, Stavrou D, Fillbrandt R, Westphal M: Molecular genetic alterations on chromosomes 11 and 22 in ependymomas. Int J Cancer 2001, 91:803-808 [DOI] [PubMed] [Google Scholar]
- 11.Dyer S, Prebble E, Davison V, Davies P, Ramani P, Ellison D, Grundy R: Genomic imbalances in pediatric intracranial ependymomas define clinically relevant groups. Am J Pathol 2002, 161:2133-2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeuken JWM, Sprenger SHE, Gilhuis J, Teepen HLJM, Grotenhuis AJ, Wesseling P: Correlation between localization, age, and chromosomal imbalances in ependymal tumors as detected by CGH. J Pathol 2002, 197:238-244 [DOI] [PubMed] [Google Scholar]
- 13.Ljubimova JY, Lakhter AJ, Loksh A, Yong WH, Riedinger MS, Miner JH, Sorokin LM, Ljubimov AV, Black KL: Overexpression of a 4 chain-containing laminin in human glial tumors identified by gene microarray analysis. Cancer Res 2001, 61:5601-5610 [PubMed] [Google Scholar]
- 14.Rickmann DS, Bobek MP, Misek DE, Kuik R, Blavias M, Kurnit DM, Taylor J, Hanash SM: Distinctive molecular profiles of high-grade and low-grade gliomas based on oligonucleotide microarray analysis. Cancer Res 2001, 61:6885-6891 [PubMed] [Google Scholar]
- 15.Watson MA, Perry A, Budhjara V, Hicks C, Shannon WD, Rich KM: Gene expression profiling with oligonucleotide microarrays distinguishes World Health Organization grade of oligodendrogliomas. Cancer Res 2001, 61:6885-6891 [PubMed] [Google Scholar]
- 16.Fuller GN, Hess KR, Rhee CH, Yung WK, Sawaya RA, Bruner JM: Molecular classification of human diffuse gliomas by multidimensional scaling analysis of gene expression profiles parallels morphology-based classification, correlates with survival, and reveals clinically relevant novel glioma subsets. Brain Pathol 2002, 12:108-116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacDonald TJ, Brown KM, LaFleur B, Peterson K, Lawlor C, Chen Y, Packer RJ, Cogen P, Stephan DA: Expression profiling of medulloblastoma: pDGFRA and the RAS/MAPK pathway as therapeutic targets for metastatic disease. Nat Genet 2001, 29:143-152 [DOI] [PubMed] [Google Scholar]
- 18.Pomeroy SL, Tamayo P, Gaasenbeek M, Sturla LM, Angelo M, McLaaughlin ME, Kim JY, Goumnerova LC, Black PM, Lau C, Allen JC, Zagzag D, Olson JM, Curran T, Weltmore C, Biegel JA, Poggio T, Mukherjee S, Rifkin R, Caligano A, Stolovitzky G, Louis DN, Mesirov JP, Lander ES, Golub TR: Prediction of nervous system embryonal tumor outcome based on gene expression. Nature 2002, 415:436-442 [DOI] [PubMed] [Google Scholar]
- 19.Merchant TE, Jenkins JJ, Burger PC, Sanford RA, Sherwood SH, Jones-Wallace D, Heideman RL, Thomson SJ, Helton KJ, Kun LE: Influence of tumor grade on time to progression after irradiation of localized ependymoma in children. Int J Oncol Biol Phys 2002, 53:52-57 [DOI] [PubMed] [Google Scholar]
- 20.Fritz B, Schubert F, Wrobel G, Schwaenen K, Wessendorf S, Nessling M, Korz C, Rieker RJ, Montgomery K, Kucherlapati R, Mechtersheimer G, Eils R, Joos S, Lichter P: Microarray based copy number and expression profiling in dedifferentiated and pleomorphic liposarcoma. Cancer Res 2002, 62:2993-2998 [PubMed] [Google Scholar]
- 21.Wrobel G, Schlingemann J, Hummerich L, Kramer H, Lichter P, Hahn M: Optimization of high-density cDNA-microarray protocols by “Design of Experiments.” Nucleic Acids Res 2003, 31:67e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huber W, von Heydebreck A, Sueltmann H, Poustka A, Vingron M: Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 2002, 18:96-104 [DOI] [PubMed] [Google Scholar]
- 23.Korz C, Pscherer A, Benner A, Mertens D, Schaffner C, Leupolt E, Dohner H, Stilgenbauer S, Lichter P: Evidence for distinct pathomechanisms in B-cell chronic lymphocytic leukemia and mantle cell lymphoma by quantitative expression analysis of cell cycle and apoptosis-associated genes. Blood 2002, 99:4554-4561 [DOI] [PubMed] [Google Scholar]
- 24.Pfaffl MW: A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001, 29:2002-2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tibshirani R, Hastie T, Narasimhan B, Chu G: Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci USA 2002, 99:6567-6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Efron B: Estimating the error rate of a prediction rule: improvement on cross-validation. J Am Stat Assoc 1983, 78:316-331 [Google Scholar]
- 27.Ihaka R, Gentleman R: A language for data analysis and graphics. J Comput Graph Stat 1996, 5:299-314 [Google Scholar]
- 28.Jones SE, Jomary C: Clusterin. Int J Biochem Cell Biol 2002, 34:427-431 [DOI] [PubMed] [Google Scholar]
- 29.Moschos SJ, Mantzoros CS: The role of the IGF system in cancer: from basic to clinical studies and clinical applications. Oncology 2002, 63:317-332 [DOI] [PubMed] [Google Scholar]
- 30.Kato-Stankiewicz J, Hakimi I, Zhi G, Zhang J, Serebriiskii I, Guo L, Edamatsu H, Koide H, Menon S, Eckl R, Sakamuri S, Lu Y, Chen QZ, Agarwal S, Baumbach WR, Golemis EA, Tamanoi F, Khazak V: Inhibitors of Ras/Raf-1 interaction identified by two-hybrid screening revert Ras-dependent transformation phenotypes in human cancer cells. Proc Natl Acad Sci USA 2002, 99:14398-14403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kerkela E, Ala-Aho R, Klemi P, Grenman S, Shapiro SD, Kahari VM, Saarialho-Kere U: Metalloelastase (MMP-12) expression by tumour cells in squamous cell carcinoma of the vulva correlates with invasiveness, while that by macrophages predicts better outcome. J Pathol 2002, 198:258-269 [DOI] [PubMed] [Google Scholar]
- 32.Misasi R, Sorice M, Di Marzio L, Campana WM, Molinari S, Cifone MG, Pavan A, Pontieri GM, O’Brien JS: Prosaposin treatment induces PC12 entry in the S phase of the cell cycle and prevents apoptosis: activation of ERKs and sphingosine kinase. FASEB J 2001, 15:467-474 [DOI] [PubMed] [Google Scholar]
- 33.Willert J, Epping M, Pollack JR, Brown PO, Nusse R: A transcriptional response to Wnt protein in human embryonic carcinoma cells. BMC Dev Biol 2002, 2:1-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sasaki K, Dohura K, Ironside JW, Iwaki T: Increased clusterin (apoliporotein J) expression in human and mouse brains infected with transmissible spongiform encephalopathies. Acta Neuropathol 2002, 103:199-208 [DOI] [PubMed] [Google Scholar]
- 35.Collins VP: Gene amplification in human gliomas. Glia 1995, 15:289-296 [DOI] [PubMed] [Google Scholar]
- 36.Ogino H, Kubo S, Abdul-Karim FW, Cohen ML: Comparative immunohistochemical study of insulin-like growth factor II and insulin-like growth factor receptor type 1 in pediatric brain tumors. Pediatr Dev Pathol 2001, 4:23-31 [DOI] [PubMed] [Google Scholar]
- 37.Bortolotto S, Chiado-Piat L, Cavalla P, Bosone I, Mauro A, Schiffer D: CDKN2A/p16 in ependymomas. J Neuro-Oncol 2001, 54:9-13 [DOI] [PubMed] [Google Scholar]
- 38.Rousseau E, Ruchoux MM, Scaravilli F, Chapon F, Godfraind CY, Vikkula MY: CDKN2A, CDKN2B and p14ARF are frequently and differentially methylated in ependymal tumors. Neuropathol Appl Neurobiol 2003, 29:196(Abstract) [DOI] [PubMed] [Google Scholar]
- 39.Singh PK, Gutmann DH, Fuller CE, Newsham IF, Perry A: Differential involvement of protein 4.1 family members DAL-1 and NF2 in intracranial and intraspinal ependymomas. Mod Pathol 2002, 15:526-531 [DOI] [PubMed] [Google Scholar]
- 40.Scheil S, Bruderlein S, Eicker M, Herms J, Herold-Mende C, Steiner HH, Barth TFE, Moller P: Low-frequence of chromosomal imbalancies in anaplastic ependymomas as detected by comparative genomic hybridization. Brain Pathol 2001, 11:133-143 [DOI] [PMC free article] [PubMed] [Google Scholar]


