Abstract
The INT6 gene has been implicated in human breast cancer formation, but its function is unknown. We isolated an Int6 homolog from fission yeast, Yin6, by its binding to a conserved protein in the Ras pathway, Moe1. Yin6 and Moe1 converge on the same protein complex to promote microtubule instability/disassembly. Yin6 and Moe1 interact cooperatively: when either protein is absent, the other becomes mislocalized with decreased protein levels. Furthermore, whereas full-length human Int6 rescues the phenotypes of the yin6-null (yin6Δ) mutant cells and binds human Moe1, truncated Int6 proteins found in tumors do not. Importantly, yin6Δ alone impairs chromosome segregation weakly, but yin6Δ together with ras1Δ causes severe chromosome missegregation. These data support a model in which INT6 mutations in humans either alone or together with additional mutations, such as a RAS mutation, may contribute to tumorigenesis by altering genome stability.
Keywords: signal transduction, G proteins, eukaryotic initiation factor 3, cell cycle, mouse mammary tumor virus
The INT genes were identified as key loci in breast cancer formation because they are frequent sites of integration by the mouse mammary tumor virus (MMTV; reviewed in ref. 1). So far a total of nine INT genes have been identified, including three families of very important and conserved signaling molecules: Wnt (encoded by WNT1/INT1, WNT3/INT4, and WNT10b), Notch (encoded by INT3), and Fgf (encoded by FGF3/INT2, FGF4, and FGF8). The functions of INT6 are the least known among all of the INT genes.
Both human and mouse INT6 genes play critical roles in breast tumorigenesis. In mouse mammary tumors, MMTV insertions into INT6 lead to the expression of C-terminally truncated Int6 proteins (2). It is not known whether these truncated proteins are gain-of-function, dominant-negative, or loss-of-function versions of full-length Int6. In the only reported incidence in humans, Miyazaki et al. (3) have shown that human INT6 (h-INT6) is frequently lost in primary breast carcinomas (30% of all of the examined patients), which supports a hypothesis that a loss of function in h-INT6 contributes to tumorigenesis.
We use the fission yeast Schizosaccharomyces pombe as a model system to study signal transduction pathways mediated by the Ras1 G protein, a homolog of the mammalian Ha-Ras protooncoproteins (4, 5). The Ras1 pathways in yeast bear great similarities to those of animal cells. In particular, both yeast and mammalian Ras proteins are important for proper organization of the cytoskeleton, and both do so through Rho-like GTPases, such as Cdc42 (reviewed in ref. 6). In S. pombe, Ras1 regulates Cdc42 by interacting with its putative cognate guanine nucleotide exchange factor, Scd1 (ref. 7; also known as Ral1; ref. 8).
To better understand the mechanism by which Ras1 regulates organization of the cytoskeleton, we conducted a two-hybrid screen using Scd1 as bait, and we isolated a conserved protein, Moe1 (9). We have determined that Moe1 modulates microtubule stability/assembly in a negative fashion. Null mutants of moe1 (moe1Δ) contain abnormally stable microtubules that are longer and more abundant than normal. We show further that moe1Δ in combination with a loss-of-function mutation affecting the Ras1–Cdc42 pathway results in cell death (9). The resulting double mutants are defective in proper spindle formation and chromosome segregation.
In this study, we isolated an S. pombe Int6 homolog as a Moe1-binding protein, and named it Yin6 (for Yeast int6). We show here that the actions of Yin6 and Moe1 converge on the same protein complex, which functions to negatively modulate microtubule polymerization and/or stability. Yin6 and Moe1 are mutually dependent on each other for proper protein levels and subcellular localization. Importantly, yin6Δ weakly impairs chromosome segregation, but yin6Δ together with ras1Δ severely impairs chromosome segregation. Moreover, full-length h-Int6, but not truncated h-Int6, found in tumors, effectively rescues the phenotype of yin6Δ cells and binds h-Moe1, suggesting that human Int6 and Moe1 are also part of a protein complex that mediates microtubule functioning and chromosome segregation. Our results support a model in which a loss of function in h-INT6, either alone or in combination with mutations in RAS, may alter chromosome segregation in humans.
Methods and Materials
Strains and Microbial Manipulation.
All strains used were derived from SP870 (h90, ade6-M210, leu1-32, ura4-D18), unless otherwise indicated. The rich and synthetic media used are the yeast extract medium (YEAU) and MM medium, respectively (9). We carried out all of the experiments with cells pregrown to early logarithmic phase (2–5 × 106 cells per ml). The minichromosome (Ch16) stability was measured by the half-sectoring assay (10, 11).
Yeast Two-Hybrid System and Cloning of yin6.
Four cDNA clones encoding full-length Yin6 were isolated from the same screen as described in ref. 12, one of which was named pGADYIN6 and used for all of the two-hybrid assays. We did not detect any two-hybrid interactions between Yin6 and other components in the Ras1 morphogenic pathway: Ras1, Scd1, Scd2, or Cdc42. A 3.1-kb SalI–SpeI DNA fragment containing the genomic yin6, isolated from cosmid 646 (13), was cloned into pAL1 (see below), and the resulting plasmid (named pAL1YIN6g) could fully rescue the phenotypes of yin6Δ cells (see Results). yin6 has been identified independently by the S. pombe Genome Sequencing Project at the Sanger Centre (Cambridge, U.K.) and by Crane et al. (GenBank accession no. AF117648). The quantitative β-galactosidase assay was performed with o-nitrophenyl β-d-galactoside as substrate (7).
Plasmid Constructions.
pAL1 was created by cloning an EcoRI fragment of ars1 and a ClaI fragment of leu1 into pBluescript (Stratagene). pLBDHMOE1 was constructed by cloning a BamHI fragment encoding the human Moe1 (9) into pVJL11 (7). A SalI–SpeI fragment of genomic yin6 was cloned into pBluescript to create pBSYIN6g. A 1.5-kb BamHI–SphI yin6 fragment was removed from pBSYIN6g and replaced with the kanR (14) or ura4 gene to generate pYIN6K and pYIN6U. DNA fragments encoding GFP-KanR, HA-KanR, and MYC-KanR (GFP, green fluorescent protein; KanR, kanamycin-resistance protein; HA, hemagglutinin epitope) were amplified by PCR using pFA6a-GFP(S65T)-KanMX6, pFA6a-3HA-KanMX6, and pFA6a-MYC-KanMX6 (14) as templates to create novel SphI sites for cloning into pBSYIN6g. The resulting vectors were named pBSYIN6-GFPKanR, pBSYIN6-HAKanR, and pBSYIN6-MYCKanR, respectively. A SmaI–ApaI fragment containing the yin6 cDNA was isolated from pGADYIN6 and inserted into pSLF173 (15), giving pHAYIN6. A BglII--XhoI fragment of yin6 was released from pHAYIN6 and cloned into pREP41-GFP (unpublished work) and pRP259 (7) to create pREP41GFPYIN6 and pGSTYIN6. Both pSLF173 and pREP41 (16) contain nmt1 promoters, and the former allows for the expression of proteins that are N-terminally tagged with 3× HA epitope. cDNAs encoding full-length and truncated h-Int6, T22 and T1139 (residues 1–317 and 1–157; the h-Int6 sequence is shown in Fig. 9, which is published as supplemental data on the PNAS web site, www.pnas.org), were amplified by PCR from a cDNA library of human erythroleukemia K562 cell line (CLONTECH). The amplified fragments were sequenced and subcloned into pSLF173, named pHAHINT6, pHAT1139, and pHAT22, and into pGADGH (7), named pGADHINT6, pGADT1139, and pGADT22. The coding sequence for GFP was excised from pALG (11) and cloned into pTrcHisC (CLONTECH) at the PstI/KpnI sites to create pHISGFP.
Strain Constructions.
To create yin6Δ strains (YIN6K and YIN6U), EcoO109I fragments containing yin6∷KanR or yin6∷ura4 were released from pYIN6K and pYIN6U and used to transform strain SP870. Stable G418-resistant clones were selected (14), and the deletion of yin6 was confirmed by PCR and Northern blots. yin6 was deleted (yin6∷kanR) from strain MOE1L (9) to create a yin6Δ moe1Δ strain (ECP17). yin6 was deleted (yin6∷kanR) from diploid strain SPRUD (17), and the tetrads from the resulting diploid cells were dissected to select for a yin6Δ ras1Δ strain (ECP19). To tag chromosomal yin6 at the 3′ end with the coding sequences for GFP, 3× HA and c-Myc, EcoO109I DNA fragments released from pBSYIN6-GFPKanR, pBSYIN6-HAKanR, and pBSYIN6-MYCKanR, respectively, were used to transform strains SP870 and MOE1L. Fusion of these tags results in the removal of the last four amino acids from Yin6. The resulting strains were named ECP20 (yin6-gfp), ECP21 (yin6-HA), ECP32 (yin6-myc), ECP22 (moe1Δ yin6-gfp), and ECP23 (moe1Δ yin6-HA). Chromosomal moe1 was tagged with GFP (strain ECP24, moe1-gfp) by a PCR-based method (14); yin6 was deleted (yin6∷ura4) from ECP24 to create ECP25 (yin6Δ moe1-gfp). Proper tagging was confirmed by PCR and Western blotting.
Fluorescence Microscopy.
The general procedures for calcofluor and 4′,6-diamidino-2-phenylindole (DAPI) staining were as described (www.bio.uva.nl/pombe/handbook). The immunostaining for microtubule was as described (9), and cells were fixed in formaldehyde (3%) and glutaraldehyde (0.05%).
In Vitro Protein Binding.
Escherichia coli BL21(DE3)pLysS strain was transformed with pGSTYIN6, pHTMOE1 (9), pHISGFP, or pRP259 to express GST-Yin6, His-Moe1, His-GFP, or GST, respectively. Purified His-tagged proteins (2 μg) were separated by SDS/PAGE and then transferred to a nitrocellulose membrane. The proteins on the membrane were first denatured by 6 M guanidine⋅HCl (in 250 mM NaCl/50 mM MgCl2/250 mM Hepes–KOH, pH 7.7) for 20 min, and then refolded by gradually decreasing the concentration of guanidine⋅HCl. The membrane was blocked in 5% nonfat dry milk/250 mM NaCl/50 mM MgCl2/250 mM Hepes–KOH, pH 7.7, for 3 h, and then probed with 1 μg of purified glutathione S-transferase (GST)-Yin6 or GST at 4°C for 4 h.
Preparation of Yeast Lysates, Immunoprecipitation, and Western Blotting.
S. pombe cell lysates were prepared in the PEM buffer (www.bio.uva.nl/pombe/handbook). Total crude extracts containing equal amounts of proteins were analyzed by immunoblotting. For coprecipitation, crude lysates were centrifuged at 2,700 × g for 20 min. Supernatants were mixed with antibodies that were conjugated to staphylococcal protein A beads at 4°C for 3 h, and the beads were later washed with PEM buffer containing 0.5% Triton X-100 and 0.5% Nonidet P-40. Moe1 was detected by a polyclonal antibody (12). Antibodies against GFP (1:500) and Cdc8 (1:400) were from CLONTECH and M. Balasubramanian (University of Singapore, Singapore). The detection of GST, His, and HA tags was as described in ref. 9.
Results
Isolation of yin6.
The yin6 gene was isolated from a two-hybrid screen using full-length moe1 as bait (Methods and Materials). yin6 is predicted to encode 501 amino acids. The degree of identity in amino acid sequences among Int6 homologs is striking: 100% identity between human and mouse (3) and 43% identity between human and yeast proteins over the entire length of the proteins (see supplemental Fig. 9, www.pnas.org). This high degree of identity in primary sequence suggests that the functions of Yin6 are also conserved (see below).
Yin6 and Moe1 Bind Directly.
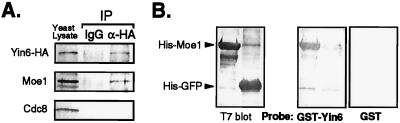
We investigated whether Yin6 and Moe1 form a complex in fission yeast. We tagged the endogenous Yin6 at its C terminus with either the HA or the c-MYC epitope. The tagged Yin6 proteins are as functional as wild-type Yin6, as the resulting strains are phenotypically indistinguishable from the wild-type strain. Our data show that Yin6-HA specifically precipitates Moe1 (Fig. 1A), but not Cdc8 (18), and Moe1 specifically precipitates Yin6-Myc (not shown). Next, using recombinant proteins expressed in bacteria, we examined whether Yin6 binds directly to Moe1. In a Far-Western (filter overlay) assay, purified and immobilized His-tagged Moe1, but not the His-GFP control, bound specifically to purified GST-tagged Yin6 (GST-Yin6), but not to the GST control (Fig. 1B). These biochemical data suggest that Yin6 and Moe1 interact directly.
Figure 1.
Physical interactions between Yin6 and Moe1. (A) Yeast lysates were prepared from strain ECP21 (yin6-HA) and treated with the antibody against the HA tag and the antibody control, IgG, as indicated on the top. Yeast lysates and immunoprecipitated proteins (IP) were analyzed by Western blotting using antibodies that recognize specific proteins, as indicated on the left. (B) Purified His-tagged proteins on the nitrocellulose membranes (two blots on the Right) were probed with purified GST and GST-Yin6. The bound GST-proteins were revealed by Western blotting. A duplicated immunoblot on the Left shows the amounts of His-tagged proteins used.
yin6 and moe1 Null Mutants Display Similar Phenotypes.
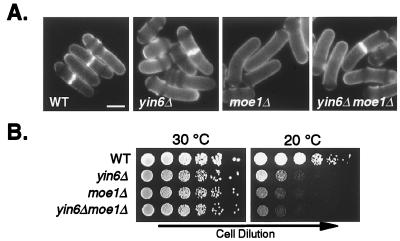
To determine the biological function of Yin6 and its relationship with Moe1, we created yin6Δ cells lacking almost the entire yin6 coding region (thus removing amino acid residues 3–497). Remarkably, we found that yin6Δ and moe1Δ cells are nearly indistinguishable phenotypically. Both strains display the same abnormal cell morphology (bent cells that are also wider and longer than normal; Fig. 2A) and cold-dependent growth defect (Fig. 2B). Additionally, yin6Δ/yin6Δ diploid cells, as well as moe1Δ/moe1Δ cells, frequently contain abnormal asci with fewer than the normal four spores (not shown). Because the phenotypes of yin6Δ and moe1Δ cells are very similar, we propose that Yin6 and Moe1 participate in the same biological process.
Figure 2.
Growth and morphology of various cells. (A) Various strains were pregrown in YEAU at 30°C and stained with calcofluor. The average length and width of the septated (constant volume) yin6Δ cells are 18.5 ± 1.7 and 5.1 ± 0.2 μm (mean ± SD, n = 20 cells), respectively, compared with 15.4 ± 0.5 and 4.0 ± 0.2 μm (mean ± SD, n = 10 cells) of wild-type cells. We note that the bent cell shape is more pronounced at 30°C than 20°C. (Bar = 5 μm.) (B) Cells after a series of 1:5 dilutions were spotted on YEAU plates, which were incubated at either 30°C or 20°C. At 20°C, the doubling times of yin6Δ and wild-type (WT) cells in rich medium are 8.5 and 5.5 h (and 22.8 and 7.9 h in minimal medium). Strains tested were SP870 (WT), YIN6K (yin6Δ), MOE1L (moe1Δ), and ECP17 (yin6Δ moe1Δ).
Yin6 Negatively Affects Microtubule Stability/Assembly.
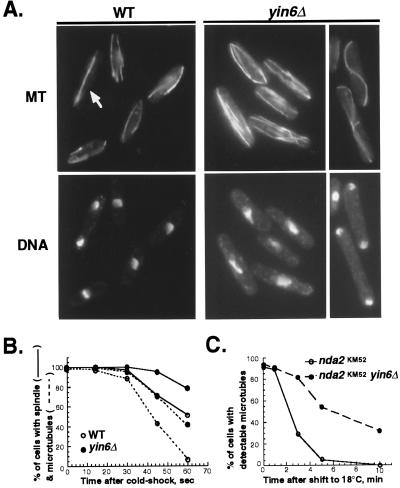
The bent cell shape and cold-dependent growth defect are two phenotypes common among S. pombe mutants defective in microtubule function, including moe1Δ cells (9). Consistent with the hypothesis that Yin6 and Moe1 participate in the same biological process, we found that yin6Δ cells display the same microtubule abnormalities as moe1Δ cells. As shown in Fig. 3A, interphase microtubule bundles in yin6Δ cells are abnormally abundant (4–6 vs. 2–3 bundles per cell), and the spindles in 45% of the anaphase cells appear too long and curve into an S shape. Both abnormalities are more severe at 20°C (i.e., only 15% anaphase cells at 30°C contain S-shaped spindles). S-shaped spindles are not detectable in wild-type cells regardless of the temperature (<1%). As compared with wild-type cells, microtubules in yin6Δ cells are also more stable against treatments that promote microtubule depolymerization—i.e., in the presence of microtubule poisons, thiabendazole (TBZ; Table 1) and benomyl (not shown), and cold shock (Fig. 3B). Additionally, yin6Δ leads to stabilization of microtubules in both α1 and β tubulin mutant strains, carrying the nda2-KM52 and nda3-KM311 mutations (Fig. 3C and data not shown; refs. 19 and 20), respectively, suggesting that yin6Δ is equally effective in stabilizing microtubules made of either defective α1- or β-tubulin. We excluded the possibility that the described microtubule abnormalities are caused by elevated tubulin levels by using Western blots (not shown). Instead, we propose that Yin6, like Moe1, plays a key role in promoting microtubule depolymerization and/or instability.
Figure 3.
Abnormal microtubules in yin6Δ cells. (A) Wild-type (WT) and yin6Δ (YIN6K) cells were pregrown in YEAU at 30°C to early logarithmic phase, resuspended in fresh medium, and after 3 h were shifted to 20°C to grow for another 17 h. Microtubules (MT) and DNA were revealed by immunostaining and 4′,6-diamidino-2-phenylindole (DAPI) staining. The numbers of microtubules in the cell were counted by adjusting focal planes, and a total of 40 cells were examined. An arrow marks a mitotic wild-type cell. (B) Cells were pregrown at 30°C in YEAU to early logarithmic phase and then were placed in an ice bath. Samples were immunostained for microtubules over time, as indicated. Cells containing at least one microtubule bundle that was ≥1/3 of the cell length or an intact spindle were scored. (C) The α1-tubulin single mutant (nda2-KM52) and the Yin6 α1-tubulin double mutants (yin6∷kanR nda2-KM52) were pregrown at 32°C and then shifted to 18°C. Their microtubules were visualized over time after immunostaining. Cells containing at least one visible microtubule were scored. Approximately 500 cells were examined for each time point.
Table 1.
Cell survival in TBZ
| Relevant genotype* | %
survival† in TBZ
|
|
|---|---|---|
| 15 μg/ml | 20 μg/ml | |
| Wild type | 53 ± 6 | 0 |
| yin6Δ | 77 ± 6 | 43 ± 6 |
| moe1Δ | 55 ± 3 | 11 ± 2 |
| yin6Δ moe1Δ | 74 ± 5 | 41 ± 1 |
Cells were pregrown at 30°C in YEAU. Equal numbers of cells were spread on YEAU plates containing 0, 15, or 20 μg/ml TBZ and incubated at 30°C for 10 days.
See Fig. 1 for strains tested.
The number of colonies formed by each strain on the control plate (containing no TBZ) was taken as 100%. The cells were seeded in triplicate; the numbers shown are mean ± SD (n = 3).
Yin6 and Moe1 Converge on the Same Target.
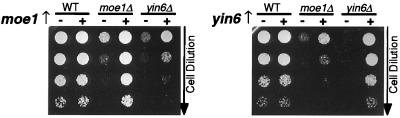
We performed genetic analyses to further study the relationship between Yin6 and Moe1. We found that cells lacking both yin6 and moe1 are phenotypically indistinguishable from either one of the single mutants: all have the same morphology, grow slowly at 20°C, are resistant to TBZ (Fig. 2 and Table 1), and display the same abnormal appearance in microtubules and spindles as those shown in Fig. 3A. Hence, Yin6 and Moe1 either act in a linear pathway or converge on the same protein target. To differentiate between these two possibilities, we overexpressed moe1 under the control of its endogenous promoter and found that it modestly rescued the growth defect of yin6Δ cells; reciprocally, overexpressed yin6 also modestly rescued the phenotypes of moe1Δ cells (Fig. 4). Overexpressing moe1 also partially restored the TBZ response of yin6Δ cells and vice versa (not shown). These data support the hypothesis that the actions of Yin6 and Moe1 converge on the same protein target.
Figure 4.
Genetic interactions between Yin6 and Moe1. Wild-type (WT; SP870), yin6Δ (YIN6K), and moe1Δ (MOE1U) cells were transformed with either a vector control (−, pAL1) or a plasmid carrying the genomic version (+) of either yin6 (pAL1YIN6g, Right) or moe1 (pALMOE1g, Left). Serially diluted cells were spotted on MM plates and incubated at 22°C.
Yin6 and Moe1 Are Mutually Dependent on Each Other for Proper Protein Levels and Subcellular Localization.
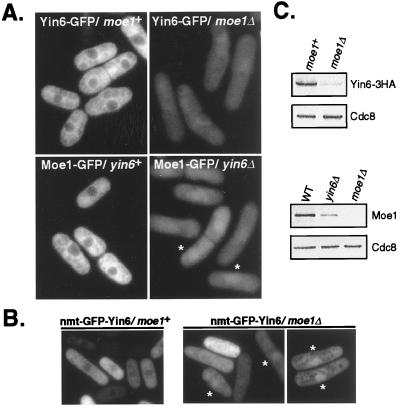
We investigated the subcellular localization of Yin6 and whether this localization is affected by Moe1. To visualize Yin6, we tagged the endogenous Yin6 at its C terminus with GFP (or the HA epitope, see Fig. 1). Our data show that Yin6-GFP is largely cytosolic and slightly concentrated near the nuclear periphery (Fig. 5A), as has been reported for h-Int6 (21, 22). We obtained the same result by immunostaining with antibodies specific for GFP or HA (not shown). Next, we examined Yin6 localization in moe1Δ cells. Surprisingly, we observed that the levels of Yin6-GFP decreased substantially in moe1Δ cells, which was further confirmed by Western blots (Fig. 5 A and C). Moreover, Yin6-GFP was no longer predominantly cytosolic but appeared evenly distributed between the nucleus and the cytosol. The nuclear accumulation of Yin6 in moe1Δ cells is more dramatic when it is overexpressed. As shown in Fig. 5B, whereas GFP-Yin6 remains cytosolic when overexpressed in wild-type cells, it is distinctly nuclear in 20% of moe1Δ cells (and evenly distributed between the cytoplasm and the nucleus in the other 80%). These data indicate that Moe1 plays a role in maintaining proper protein levels and subcellular localization of Yin6.
Figure 5.
Subcellular localizations and protein levels of Yin6 and Moe1. (A) The relevant genotypes of tested cells and the GFP-tagged proteins under investigation are indicated. Moe1-GFP concentrates more efficiently in the nucleus of yin6Δ cells; two such cells are marked by asterisks. The images were captured by the camera under identical conditions to reveal the drastically different intensities among these samples. As a control, we examined cells without a GFP tag and found autofluorescence signal that is primarily cytosolic and too weak to be photographed. The strains tested were ECP20 (yin6-gfp), ECP22 (yin6-gfp moe1Δ), ECP24 (moe1-gfp), and ECP25 (moe1-gfp yin6Δ). (B) yin6Δ or moe1Δ cells (YIN6K or MOE1U) transformed with pREP41GFPYIN6 were inoculated in thiamin-free MM medium and photographed. Asterisks mark those cells whose GFP-Yin6 is clearly concentrated in the nucleus. Cells were also transformed with a control vector expressing the GFP control, which diffuses throughout the cell (9). (C) The same amount of total proteins from each strain was analyzed by Western blotting to reveal the protein levels of Yin6-HA, Moe1, and Cdc8. The strains tested were, from the upper left to the lower right, ECP21 (yin6-HA moe1+), ECP22 (yin6-HA moe1Δ), SP870 (WT), YIN6K (yin6Δ), and MOE1L (moe1Δ).
We carried out the reciprocal experiments to examine chromosomal Moe1-GFP localization and found that, like Yin6-GFP, it is predominantly cytosolic and slightly more concentrated around the nucleus in wild-type cells (Fig. 5A). In yin6Δ cells, Moe1 protein levels decrease markedly (Fig. 5 A and C), and it is no longer predominantly cytosolic. In fact, in >80% of yin6Δ cells, the levels of Moe1-GFP are substantially higher in the nucleus than in the cytosol (Fig. 5A). We obtained similar results by examining endogenous Moe1 with immunostaining using an anti-Moe1 antibody (not shown). Note that the Cdc8 protein levels remain unchanged in yin6Δ and moe1Δ cells (Fig. 5C), suggesting that Yin6 and Moe1 affect each other's protein levels with substantial specificity. We did not detect any obvious changes in the protein levels and localization of Moe1 and Yin6 throughout the cell cycle. On the basis of these results, we conclude that Yin6 and Moe1 require each other to maintain proper protein levels and subcellular localization.
Yin6 Interacts with Ras1 to Affect Chromosome Segregation.
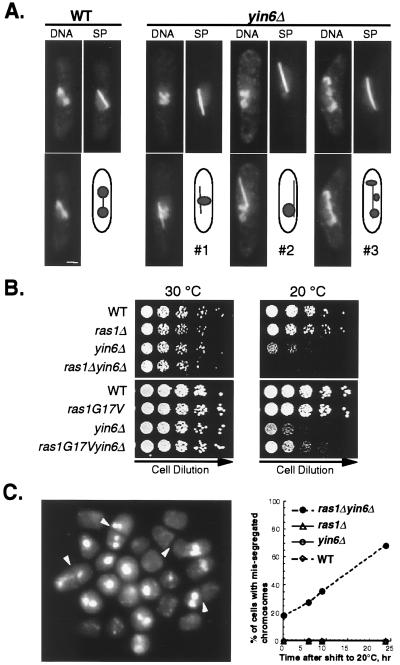
moe1Δ in combination with a defect in the Ras1 pathway leads to pronounced chromosome missegregation (9); hence, we investigated whether Yin6 and Ras1 genetically interact. First, we examined whether yin6Δ alone can lead to chromosome missegregation. Our data show that yin6Δ renders a linear minichromosome unstable: it is lost at a frequency of 7% per division (567 of 8,556 cells examined), nearly 90-fold over that of the control (0.08%, 11 of 14,322 cells). Furthermore, the regular chromosomes in yin6Δ cells segregated inefficiently at 20°C. As shown in Fig. 6A, yin6Δ cells frequently contain either a single mass of DNA as they enter anaphase (spindle length 3–5 μm; cells 1 and 2) or what appear to be chromosomes scattered along the spindle (cell 3). These abnormal anaphase cells occur more frequently in the cold (60% at 20°C but only 20% at 30°C). In wild-type cells, by contrast, these abnormal anaphase cells are not detectable at 30°C (<1%) and appear less frequently at 20°C (13%). Despite the presence of unseparated chromosomes in yin6Δ cells during early anaphase, the majority of cells in late anaphase (spindle length >7 μm) contain properly segregated chromosomes. Furthermore, inviable cells with fragmented chromosomes do not accumulate at 20°C for up to three generations (Fig. 6C and data not shown). These results suggest that the observed chromosome missegregation occurs only transiently.
Figure 6.
Ras1 interacts with Yin6 to affect chromosome segregation. (A) Cells were pregrown at 30°C and then shifted to either 30°C or 20°C for another 17 h (see legend for Fig. 3A). Spindle (SP) and DNA staining are merged and shown below each panel. Three representative patterns of chromosome missegregation occurring frequently at 20°C are shown. (Bar = 2 μm.) (B) Various strains (relevant genotypes are indicated on the left) were serially diluted and spotted on YEAU plates and incubated at 30°C or 20°C. Strains used in the upper plates were SP870 (WT), YIN6K (yin6Δ), SPRU (ras1Δ; ref. 15), and ECP19 (yin6Δ ras1Δ). The strains used in the lower plates were from the same tetrad after a fusion between strain SP562 (carrying ras1G17V, ref. 5) and YIN6U (yin6Δ). (C) ras1Δ yin6Δ cells were pregrown at 30°C and then shifted to 20°C. The percentages of cells with missegregated chromosomes were scored over time after shifting to 20°C (Right). A representative mixed population of cells after 24 h at 20°C with missegregated chromosomes is shown on the Left. Arrowheads mark the septa.
Although yin6Δ alone impairs chromosome segregation weakly, yin6Δ together with ras1Δ induces severe chromosome missegregation. As shown in Fig. 6B, ras1Δ yin6Δ cells are unable to form colonies at 20°C. These cells appear to lose viability because of chromosome missegregation, since the percentage of cells with missegregated chromosomes steadily increases over time at 20°C, reaching 61% after 24 h, a defect undetectable in either single mutants (<0.1%; Fig. 6C). Interestingly, in contrast to ras1Δ, replacing wild-type ras1 with ras1G17V, which encodes a mutant Ras1 protein that is constitutively active, rescues the growth defect of yin6Δ cells (Fig. 6B). These data support the hypothesis that proper chromosome segregation requires an interaction between the Yin6–Moe1 complex and the Ras1 pathway.
The Functions of Yin6 Are Conserved.
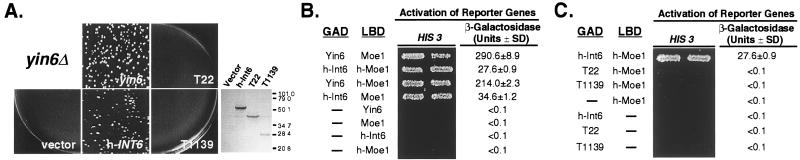
To determine whether the function of Yin6 has been conserved during evolution, we expressed h-Int6 and found that it rescues the growth defects of yin6Δ cells (Fig. 7A). h-Int6 also restores Moe1 protein levels and the response to TBZ in yin6Δ cells to nearly those of wild-type cells (not shown). Additionally, we determined that h-Int6 and h-Moe1 form a protein complex (Fig. 7B), which is consistent with the fact that h-Moe1 and h-Int6 coprecipitate from human cells (23). We conclude that the interaction between Yin6 and Moe1 has been conserved extensively in evolution.
Figure 7.
h-Int6 and Moe1 interaction is conserved during evolution. (A) yin6Δ cells (YIN6K), transformed with either a vector control (pSLF173) or various plasmids (pHAHINT6, pHAT1139, pHAYIN6, and pHAT22) carrying the indicated genes, were spread on MM medium containing 0, 0.2, 2, or 20 μM thiamin at 23°C. We obtained the same results regardless of the thiamin concentration tested, and show herein only those with 20 μM. On the right is a Western blot showing the expression of full-length and truncated h-Int6 in wild-type cells. (B) Protein–protein interactions as tested by the yeast two-hybrid system. The β-galactosidase activity (Miller units) was measured on three independent colonies per transformation. The plasmids tested were pGADGH, pGADHINT6, pGADT1139, pGADT22, pGADYIN6, pVJL11, pLBDMOE1, and pLBDHMOE1.
Because mouse mammary tumor virus insertions lead to truncations in the C terminus of Int6, we made analogous truncations in h-Int6 (T22 and T1139) to test whether the truncated Int6 proteins are biologically active. Our data show that the truncated h-Int6 can neither rescue the growth defects of yin6Δ cells (Fig. 7A) nor bind h-Moe1 (Fig. 7B). Western blots confirmed that T22 and T1139 were expressed in yeast (Fig. 7A). Furthermore, yin6Δ cells (as well as wild-type cells) containing a vector expressing either T22 or T1139 grow at the same rate as those containing the control vector (not shown), indicating that the truncated h-Int6 proteins do not impair cell growth. These results suggest that the truncated h-Int6 proteins found in tumors lack domains critical for the functions of Int6, one of which may be binding to Moe1.
Discussion
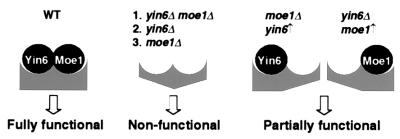
Our data support a hypothesis that Yin6 interacts directly with Moe1 in the cell, and the actions of Yin6 and Moe1 converge on the same target. Some of the functions regulated by this protein complex, such as promoting microtubule instability/disassembly and maintaining proper chromosome segregation, have been revealed. We present a model in Fig. 8 to illustrate how Yin6 and Moe1 modulate the function of this complex. This complex is postulated to be inactive if both Yin6 and Moe1 are disassociated from it. This may occur in yin6Δ moe1Δ cells as well as in yin6 and moe1 single mutant because Yin6 and Moe1 affect each other's protein level and localization. This complex is hypothesized to be partially active if either Yin6 or Moe1 remains associated with it; for example, when Yin6 is overexpressed in moe1Δ cells (and vice versa). Because h-Int6 can rescue the phenotype of yin6Δ cells and forms a complex with h-Moe1, we believe that the functions of the Yin6-Moe1 complex have been conserved in humans.
Figure 8.
The model.
h-Int6 has been linked to tumorigenesis, but its precise role is poorly understood. Loss of h-INT6 heterozygosity and mRNA expression have been shown to occur frequently in human breast tumors (3). Additionally, our data indicate that the two truncated forms of Int6 found in mouse mammary tumors appear to be loss-of-function (but not gain-of-function or dominant-negative) forms. Hence, collectively, these results suggest that a loss-of-function in h-INT6 can potentiate the risk for tumor formation. Furthermore, we found (unpublished work) that the growth defect and TBZ resistance in a yin6−/yin6+ diploid are both intermediate between those of yin6−/yin6− and yin6+/yin6+ cells, suggesting that Yin6 may interact with its partners synergistically and stoichiometrically (Fig. 8). Therefore, we surmise that the loss of only a single copy of h-INT6 in human cells can result in haploinsufficiency.
Chromosome missegregation in yeast is analogous to the genomic instability that is frequently associated with solid tumors. Genomic instability can allow for the creation of multiple mutations critical for tumorigenesis (24). Because yin6Δ leads to chromosome missegregation in yeast, we speculate that a loss-of-function h-INT6 mutation may similarly compromise mitotic fidelity, ultimately leading to genetic instability in humans. In keeping with this possibility, h-Int6 has been implicated in maintaining genomic stability by its binding to a viral oncoprotein, Tax, which is associated with the spindle checkpoint (25). We note that the effects of yin6Δ on chromosome missegregation are further modulated by the Ras1 pathway; particularly, ras1Δ severely worsens the phenotype of yin6Δ cells. Thus, it would be of interest to determine in human breast tumors whether loss-of-function mutations affecting both h-Int6 and Ras frequently coexist.
h-Int6, as well as h-Moe1, has been shown to form a complex with subunits of the eukaryotic initiation factor, eIF3 (23, 26), the indispensable element for translation initiation. However, whether h-Int6 and Moe1 are in fact involved in translation remains unknown. Because Yin6 and Moe1 are not essential for viability and are absent from budding yeast, it is quite unlikely that the Int6–Moe1 complex is essential for translation.
Our localization data suggest that Yin6 and Moe1 can shuttle in and out of the nucleus. Yin6 contains both a nuclear export signal (NES) and a bipartite nuclear localization signal (NLS), which are present in all known Int6-like proteins, including h-Int6 (Fig. 9; ref. 21). The NES in h-Int6 has been shown to mediate h-Int6 nuclear export because h-Int6 mutants defective in NES are nuclear, not cytosolic (4). We surmise that nuclear trafficking of the Yin6–Moe1 complex is similarly mediated by the balanced actions of the NES and NLS in Yin6. We have examined Moe1 localization previously by using high-copy plasmids, and we concluded that it can efficiently accumulate in the nucleus (9). Here we show that endogenous Moe1 is cytosolic in yin6+ cells but nuclear in yin6− cells. It seems likely that Moe1 can accumulate in the nucleus when its levels are in excess of those of Yin6.
The mechanisms by which Yin6 affects chromosome segregation are likely to be complex. yin6Δ renders microtubules abnormally stable. The S. pombe spindle is composed of several types of microtubules, and both the length and type of these microtubules change throughout mitosis (27). Excessive microtubule stability can conceivably block preexisting microtubular structures from depolymerizing, thus impeding the formation of new ones that are necessary for proper spindle functioning. So far we have been unable to detect any physical association between Yin6 and microtubules, both in vitro and in vivo (Fig. 5 and unpublished results). Hence, it is likely that Yin6 affects microtubule or spindle function indirectly by transiently associating with microtubule-binding proteins. Indeed, we have reported a conserved interaction between the Yin6–Moe1 complex and Mal3, a member of the EB1 family of microtubule-binding proteins, that is necessary for proper spindle formation (ref. 12 and data not shown). Alternatively, Yin6, as well as h-Int6, contains a PCI domain (Fig. 9), which is commonly found in components of the 26S proteasome complex (28). The 26S proteasome complex plays a central role in proper chromosome segregation by degrading “inhibitors,” such as Cut2, that block sister-chromatid separation (reviewed in ref. 29). It is enticing to speculate that Yin6 may interact with the proteasome complex to regulate chromosome segregation and Moe1 protein levels. In addition, Yin6 may affect proper attachment between the chromosomes and the spindle, since the chromosomes in yin6Δ cells are frequently seen nearly detached from the spindle (i.e., cell 2 in Fig. 6).
Supplementary Material
Acknowledgments
We express our appreciation for M. Balasubramanian, U. Fleig, K. Gull, P. Nurse, J. Pringle, and the Genome Sequencing Project at the Sanger Centre for providing materials critical for our study. We also thank A. Changou and J. Chen for technical assistance and C. Desplan, J. Hubbard, T. Holmes, P. Furmanski, and members of the Chang lab for discussion. Furthermore, we greatly appreciate U. Maitra, T. Matsumoto, and C. Norbury for sharing unpublished results on yeast Int6. This study was supported by the American Cancer Society (RPG-97–137-01-MGO) and by Whitehead and Goddard Fellowships from New York University.
Abbreviations
- GFP
green fluorescent protein
- KanR
kanamycin-resistance protein
- HA
hemagglutinin epitope
- GST
glutathione S-transferase
- TBZ
thiabendazole
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF260238).
References
- 1.Tekmal R R, Keshava N. Front Biosci. 1997;2:519–526. doi: 10.2741/a209. [DOI] [PubMed] [Google Scholar]
- 2.Marchetti A, Buttitta F, Miyazaki S, Gallahan D, Smith G, Callahan R. J Virol. 1995;69:1932–1938. doi: 10.1128/jvi.69.3.1932-1938.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miyazaki S, Imatani A, Ballard L, Marchetti A, Buttitta F, Albertsen H, Nevanlinna H A, Gallahan D, Callahan R. Genomics. 1997;46:155–158. doi: 10.1006/geno.1997.4996. [DOI] [PubMed] [Google Scholar]
- 4.Fukui Y, Kozasa T, Kaziro Y, Takeda T, Yamamoto M. Cell. 1986;44:329–336. doi: 10.1016/0092-8674(86)90767-1. [DOI] [PubMed] [Google Scholar]
- 5.Nadin-Davis S A, Nasim A, Beach D. EMBO J. 1986;5:2963–2971. doi: 10.1002/j.1460-2075.1986.tb04593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Aelst L, D'Souza-Schorey C. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- 7.Chang E C, Barr M, Wang Y, Jung V, Xu H, Wigler H M. Cell. 1994;79:131–141. doi: 10.1016/0092-8674(94)90406-5. [DOI] [PubMed] [Google Scholar]
- 8.Fukui Y, Yamamoto M. Mol Gen Genet. 1988;215:26–31. doi: 10.1007/BF00331298. [DOI] [PubMed] [Google Scholar]
- 9.Chen C-R, Li Y-C, Chen J, Hou M C, Papadaki P, Chang E C. Proc Natl Acad Sci USA. 1999;96:517–522. doi: 10.1073/pnas.96.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allshire R C, Nimmo E R, Ekwall K, Javerzat J-P, Granston G. Genes Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- 11.Li Y-c, Chen C-r, Chang E C. Genetics. 2000;156:995–1004. doi: 10.1093/genetics/156.3.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen C-r, Chen J, Chang E C. Mol Biol Cell. 2000;11:4067–4077. doi: 10.1091/mbc.11.12.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizukami T, Chang W I, Garkavtsev I, Kaplan N, Lombardi D, Matsumoto T, Niwa O, Kounosu A, Yanagida M, Marr T G, Beach D. Cell. 1993;73:121–132. doi: 10.1016/0092-8674(93)90165-m. [DOI] [PubMed] [Google Scholar]
- 14.Bähler J, Wu J Q, Longtine M S, Shah N G, McKenzie A, 3rd, Steever A B, Wach A, Philippsen P, Pringle J R. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 15.Forsburg S L, Sherman D A. Gene. 1997;191:191–195. doi: 10.1016/s0378-1119(97)00058-9. [DOI] [PubMed] [Google Scholar]
- 16.Basi G, Schmid E, Maundrell K. Gene. 1993;123:131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- 17.Xu H P, White M, Marcus S, Wigler M. Mol Cell Biol. 1994;14:50–58. doi: 10.1128/mcb.14.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balasubramanian M K, Helfman D M, Hemmingsen S M. Nature (London) 1992;360:84–87. doi: 10.1038/360084a0. [DOI] [PubMed] [Google Scholar]
- 19.Hiraoka Y, Toda T, Yanagida M. Cell. 1984;39:349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- 20.Toda T, Adachi Y, Hirakoa Y, Yanagida M. Cell. 1984;37:233–242. doi: 10.1016/0092-8674(84)90319-2. [DOI] [PubMed] [Google Scholar]
- 21.Guo J, Sen G. J Virol. 2000;74:1892–1899. doi: 10.1128/jvi.74.4.1892-1899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diella F, Levi G, Callahan R. DNA Cell Biol. 1997;16:839–847. doi: 10.1089/dna.1997.16.839. [DOI] [PubMed] [Google Scholar]
- 23.Asano K, Vornlocher H-P, Richter-Cook N J, Merrick W C, Hinnebusch A G, Hershey J W B. J Biol Chem. 1997;272:27042–27052. doi: 10.1074/jbc.272.43.27042. [DOI] [PubMed] [Google Scholar]
- 24.Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 25.Jin D Y, Spencer F, Jeang K T. Cell. 1998;93:81–91. doi: 10.1016/s0092-8674(00)81148-4. [DOI] [PubMed] [Google Scholar]
- 26.Asano K, Merrick W C, Hershey J W. J Biol Chem. 1997;272:23477–23480. doi: 10.1074/jbc.272.38.23477. [DOI] [PubMed] [Google Scholar]
- 27.Ding R, McDonald K L, McIntosh J R. J Cell Biol. 1993;120:141–151. doi: 10.1083/jcb.120.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hofmann K, Bucher P. Trends Biochem Sci. 1998;23:204–205. doi: 10.1016/s0968-0004(98)01217-1. [DOI] [PubMed] [Google Scholar]
- 29.Yanagida M. Genes Cells. 2000;5:1–8. doi: 10.1046/j.1365-2443.2000.00306.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.